Abstract
CRISPR technology, which is widely used for plant genome editing, will accelerate the breeding of food crops beyond what was imaginable before its development. Here we provide a brief overview of CRISPR technology, its most important applications for crop improvement and several technological breakthroughs. We also make predictions of the applications of CRISPR technology to food crops, which we believe would provide the potential for synthetic biology and domestication of crops. We also discuss the implications of regulatory policy for deployment of the technology in the developing world.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


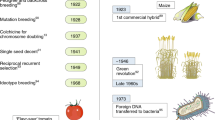
Blickwinkel / Alamy Stock Photo (a); Bilder ur Nordens Flora, Stockholm (b); David Cobb / Alamy Stock Photo (c); DE AGOSTINI PICTURE LIBRARY // gettyimages (d)
Similar content being viewed by others
References
Hickey, L. T. et al. Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754 (2019).
Ray, D. K., Mueller, N. D., West, P. C. & Foley, J. A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8, e66428 (2013).
Chen, K., Wang, Y., Zhang, R., Zhang, H. & Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697 (2019).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Symington, L. S. & Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 (2011).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Zhang, Y., Massel, K., Godwin, I. D. & Gao, C. Applications and potential of genome editing in crop improvement. Genome Biol. 19, 210 (2018).
Li, M. et al. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7, 377 (2016).
Waltz, E. CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 34, 582 (2016).
Wang, Y. et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951 (2014).
Nekrasov, V. et al. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482 (2017).
Khanday, I., Skinner, D., Yang, B., Mercier, R. & Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95 (2019).
Wang, C. et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 37, 283–286 (2019).
Henikoff, S. & Comai, L. Single-nucleotide mutations for plant functional genomics. Annu. Rev. Plant Biol. 54, 375–401 (2003).
Zong, Y. et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953 (2018).
Zhang, R. et al. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 5, 480–485 (2019).
Tian, S. et al. Engineering herbicide resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 37, 1353–1356 (2018).
Li, C. et al. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19, 59 (2018).
Long, S. P., Marshall-Colon, A. & Zhu, X. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66 (2015).
Rodriguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E. & Lippman, Z. B. Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480 (2017).
Zhang, H. et al. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898 (2018).
Lowe, K. et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015 (2016).
Kelliher, T. et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 37, 287–292 (2019).
Wang, B. et al. Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol. Plant 12, 597–602 (2019).
Maher, M. F. et al. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89 (2020).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Wurtzel, E. T. et al. Revolutionizing agriculture with synthetic biology. Nat. Plants 5, 1207–1210 (2019).
Salesse-Smith, C. E. et al. Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants 4, 802–810 (2018).
Wersch, S. & Li, X. Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci. 24, 688–699 (2019).
Engqvist, M. K. M. & Rabe, K. S. Applications of protein engineering and directed evolution in plant research. Plant Physiol. 179, 907–917 (2019).
Butt, H. et al. CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 20, 73 (2019).
Li, C. et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. https://doi.org/10.1038/s41587-019-0393-7 (2020).
Zsögön, A. et al. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216 (2018).
Li, T. et al. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163 (2018).
Lemmon, Z. H. et al. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770 (2018).
Cui, L. et al. Development of perennial wheat through hybridization between wheat and wheatgrasses: a review. Engineering 4, 507–513 (2018).
DeHaan, L., Christians, M., Crain, J. & Poland, J. Development and evolution of an intermediate wheatgrass domestication program. Sustainability 10, 1499 (2018).
Venske, E. et al. Bread wheat: a role model for plant domestication and breeding. Hereditas 156, 16 (2019).
Sánchez-Pérez, R. et al. Mutation of a bHLH transcription factor allowed almond domestication. Science 364, 1095–1098 (2019).
Nour-Eldin, H. H. et al. Reduction of antinutritional glucosinolates in Brassica oilseeds by mutation of genes encoding transporters. Nat. Biotechnol. 35, 377–382 (2017).
Podevin, N., Davies, H. V., Hartung, F., Nogue, F. & Casacuberta, J. M. Site-directed nucleases: a paradigm shift in predictable, knowledge-based plant breeding. Trends Biotechnol. 31, 375–383 (2013).
Importation, Interstate Movement, and Environmental Release of Certain Genetically Engineered Organisms (US Department of Agriculture Animal and Health Inspection Service, 2017); https://go.nature.com/2VBr64V
Whelan, A. I. & Lema, M. A. Regulatory framework for gene editing and other new breeding techniques in Argentina. GM Crops Food 6, 253–265 (2015).
Lema, M. A. Regulatory aspects of gene editing in Argentina. Transgenic Res. 28, 147–150 (2019).
Normile, D. Gene-edited foods are safe, Japanese panel concludes. Science (19 March 2019); https://doi.org/10.1126/science.aax3903
Thygesen, P. Clarifying the regulation of genome editing in Australia: situation for genetically modified organisms. Transgenic Res. 28, 151–159 (2019).
Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 560, 16 (2018).
Huang, S., Weigel, D., Beachy, R. N. & Li, J. A proposed regulatory framework for genome-edited crops. Nat. Genet. 48, 109–111 (2016).
Acknowledgements
We apologize to those colleagues whose work was not cited due to restrictions on the number of references. C.G. was supported by the National Natural Science Foundation of China (grant no. 31788103) and the Strategic Priority Research Program of the Chinese Academy of Sciences (Precision Seed Design and Breeding, grant no. XDA24000000), Y.Z. and M. Pribil by the Innovation Fund Denmark grant no. 8055-00038A, and M. Palmgren by the Novo Nordisk Foundation Challenge grant no. NNF19OC005658.
Author information
Authors and Affiliations
Contributions
Y.Z. drafted the first version of the manuscript. M. Pribil revised the manuscript, and C.G. and M. Palmgren designed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Y., Pribil, M., Palmgren, M. et al. A CRISPR way for accelerating improvement of food crops. Nat Food 1, 200–205 (2020). https://doi.org/10.1038/s43016-020-0051-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43016-020-0051-8
This article is cited by
-
CRISPR–Cas13d in plant biology: an insight
Plant Biotechnology Reports (2024)
-
Accelerating crop domestication through genome editing for sustainable agriculture
Journal of Plant Biochemistry and Biotechnology (2023)
-
Genetic manipulation of photosynthesis to enhance crop productivity under changing environmental conditions
Photosynthesis Research (2023)
-
Stimuli-responsive nanoformulations for CRISPR-Cas9 genome editing
Journal of Nanobiotechnology (2022)
-
Genetically modified organisms: adapting regulatory frameworks for evolving genome editing technologies
Biological Research (2022)