Abstract
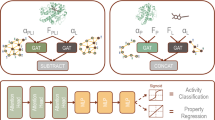
In drug design, compound potency prediction is a popular machine learning application. Graph neural networks (GNNs) predict ligand affinity from graph representations of protein–ligand interactions typically extracted from X-ray structures. Despite some promising findings leading to claims that GNNs can learn details of protein–ligand interactions, such predictions are also controversially viewed. For example, evidence has been presented that GNNs might not learn protein–ligand interactions but memorize ligand and protein training data instead. We have carried out affinity predictions with six GNN architectures on community-standard datasets and rationalized the predictions using explainable artificial intelligence. The results confirm a strong influence of ligand—but not protein—memorization during GNN learning and also show that some GNN architectures increasingly prioritize interaction information for predicting high affinities. Thus, while GNNs do not comprehensively account for protein–ligand interactions and physical reality, depending on the model, they balance ligand memorization with learning of interaction patterns.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data generated in this study are freely available on GitHub https://github.com/AndMastro/protein-ligand-GNN. The ligand interaction graph data were taken from Volkov et al.22. PDBbind data are available at: http://www.pdbbind.org.cn/. Source data are provided with this paper.
Code availability
The code generated in this study is freely available on GitHub https://github.com/AndMastro/protein-ligand-GNN, with an archived version also available through Zenodo54 at https://doi.org/10.5281/zenodo.8358539. A reproducible code capsule is available through CodeOcean at https://codeocean.com/capsule/9675097 (ref. 55), EdgeSHAPer code can be accessed on Zenodo56 https://doi.org/10.5281/zenodo.8358595 and GitHub https://github.com/AndMastro/EdgeSHAPer.
References
Akamatsu, M. Current state and perspectives of 3D-QSAR. Curr. Top. Med. Chem. 2, 1381–1394 (2002).
Lewis, R. A. & Wood, D. Modern 2D QSAR for drug discovery. WIREs Comp. Mol. Sci. 4, 505–522 (2014).
Drucker, H., Burges, C. J. C., Kaufman, L., Smola, A. & Vapnik, V. Support vector regression machines. Adv. Neur. Inform. Proc. Syst. 9 (1996).
Smola, A. J. & Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 14, 199–222 (2004).
Svetnik, V. et al. Random forest: a classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 43, 1947–1958 (2003).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Lavecchia, A. Deep learning in drug discovery: opportunities, challenges and future prospects. Drug Discov. Today 24, 2017–2032 (2019).
Kim, J., Park, S., Min, D. & Kim, W. Comprehensive survey of recent drug discovery using deep learning. Int. J. Mol. Sci. 22, 9983 (2021).
Bajorath, J. Deep machine learning for computer-aided drug design. Front. Drug Discov. 2, 829043 (2022).
Guedes, I. A., Pereira, F. S. S. & Dardenne, L. E. Empirical scoring functions for structure-based virtual screening: applications, critical aspects, and challenges. Front. Pharmacol. 9, 1089 (2018).
Liu, J. & Wang, R. Classification of current scoring functions. J. Chem. Inf. Model. 55, 475–482 (2015).
Li, H., Sze, K.-H., Lu, G. & Ballester, P. J. Machine-learning scoring functions for structure-based virtual screening. WIREs Comp. Mol. Sci. 11, e1478 (2021).
Gleeson, M. P. & Gleeson, D. QM/MM calculations in drug discovery: a useful method for studying binding phenomena? J. Chem. Inf. Model. 49, 670–677 (2009).
Williams-Noonan, B. J., Yuriev, E. & Chalmers, D. K. Free energy methods in drug design: prospects of ‘alchemical perturbation’ in medicinal chemistry. J. Med. Chem. 61, 638–649 (2018).
Gomes, J., Ramsundar, B., Feinberg, E. N. & Pande, V. S. Atomic convolutional networks for predicting protein-ligand binding affinity. Preprint at https://doi.org/10.48550/arXiv.1703.10603 (2017).
Jimenez, J., Skalic, M., Martinez-Rosell, G. & De Fabritiis, G. K(DEEP): protein-ligand absolute binding affinity prediction via 3D-convolutional neural networks. J. Chem. Inf. Model. 58, 287–296 (2018).
Stepniewska-Dziubinska, M. M., Zielenkiewicz, P. & Siedlecki, P. Development and evaluation of a deep learning model for protein-ligand binding affinity prediction. Bioinformatics 34, 3666–3674 (2018).
Scarselli, F., Gori, M., Tsoi, A. C., Hagenbuchner, M. & Monfardini, G. The graph neural network model. IEEE Trans. Neural Netw. 20, 61–80 (2008).
Jiang, D. et al. Could graph neural networks learn better molecular representation for drug discovery? A comparison study of descriptor-based and graph-based models. J. Chem. Inform. 13, 12 (2021).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. Proc. Mach. Learn. Res. 70, 1263–1272 (2017).
Volkov, M. et al. On the frustration to predict binding affinities from protein–ligand structures with deep neural networks. J. Med. Chem. 65, 7946–7958 (2022).
Shen, H., Zhang, Y., Zheng, C., Wang, B. & Chen, P. A Cascade graph convolutional network for predicting protein–ligand binding affinity. Int. J. Mol. Sci. 22, 4023 (2021).
Xiong, J., Xiong, Z., Chen, K., Jiang, H. & Zheng, M. Graph neural networks for automated de novo drug design. Drug Discov. Today 26, 1382–1393 (2021).
Son, J. & Kim, D. Development of a graph convolutional neural network model for efficient prediction of protein-ligand binding affinities. PLoS ONE 16, e0249404 (2021).
Nguyen, T. et al. GraphDTA: predicting drug-target binding affinity with graph neural networks. Bioinformatics 37, 1140–1147 (2021).
Wang, J. & Dokholyan, N. V. Yuel: improving the generalizability of structure-free compound–protein interaction prediction. J. Chem. Inf. Model. 62, 463–471 (2022).
Yang, J., Shen, C. & Huang, N. Predicting or pretending: artificial intelligence for protein-ligand interactions lack of sufficiently large and unbiased datasets. Front. Pharmacol. 11, 69 (2020).
Kipf, T. N. & Welling M. Semi-supervised classification with graph convolutional networks. Preprint at https://doi.org/10.48550/arXiv.1609.02907 (2016).
Velickovic, P. et al. Graph attention networks. Preprint at https://doi.org/10.48550/arXiv.1710.10903 (2017).
Xu, K., Hu, W. Leskovec J. & Jegalka S. How powerful are graph neural networks? Preprint at https://doi.org/10.48550/arXiv.1810.00826 (2018).
Hu, W. et al. Strategies for pre-training graph neural networks. Preprint at https://doi.org/10.48550/arXiv.1905.12265 (2019).
Hamilton, W., Ying, Z. & Leskovec, J. Inductive representation learning on large graphs. Adv. Neur. Inform. Proc. Syst. 31 (2017).
Morris, C. et al. Weisfeiler and Leman go neural: higher-order graph neural networks. In Proc. AAAI Conference on Artificial Intelligence Vol. 33, 4602–4609 (2019).
Wang, R., Fang, X., Lu, Y., Yang, C. Y. & Wang, S. The PDBbind database: methodologies and updates. J. Med. Chem. 48, 4111–4119 (2005).
Liu, Z. et al. PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics 31, 405–412 (2015).
Liu, Z. et al. Forging the basis for developing protein-ligand interaction scoring functions. Acc. Chem. Res. 50, 302–309 (2017).
Schmitt, S., Kuhn, D. & Klebe, G. A new method to detect related function among proteins independent of sequence and fold homology. J. Mol. Biol. 323, 387–406 (2002).
Desaphy, J., Raimbaud, E., Ducrot, P. & Rognan, D. Encoding protein-ligand interaction patterns in fingerprints and graphs. J. Chem. Inf. Model. 53, 623–637 (2013).
Mastropietro, A., Pasculli, G., Feldmann, C., Rodríguez-Pérez, R. & Bajorath, J. EdgeSHAPer: bond-centric Shapley value-based explanation method for graph neural networks. iScience 25, 105043 (2022).
Mastropietro, A., Pasculli, G. & Bajorath, J. Protocol to explain graph neural network predictions using an edge-centric Shapley value-based approach. STAR Protoc. 3, 101887 (2022).
Lundberg, S. M. & Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neur. Inform. Proc. Syst. 30 (2017).
Shapley, L. S. in Contributions to the Theory of Games (AM-28) Vol. II (eds Kuhn, H. W. & Tucker, A. W.) 307–317 (Princeton Univ. Press, 1953).
Ying, Z., Bourgeois, D., You, J., Zitnik, M. & Leskovec, J. GNNExplainer: generating explanations for graph neural networks. Adv. Neur. Inform. Proc. Syst. 32, 9240–9251 (2019).
Pfungst, O. Clever Hans (the horse of Mr. Von Osten): contribution to experimental animal and human psychology. J. Philos. Psychol. Sci. Method 8, 663–666 (1911).
Lapuschkin, S. et al. Unmasking Clever Hans predictors and assessing what machines really learn. Nat. Commun. 10, 1096 (2019).
Da Silva, F., Desaphy, J. & Rognan, D. IChem: a versatile toolkit for detecting, comparing, and predicting protein-ligand interactions. Chem. Med. Chem. 13, 507–510 (2018).
Hagberg, A. A., Schult, D. A. & Swart, P. J. Exploring network structure, dynamics, and function using NetworkX. In Proc. 7th Python in Science Conference (SciPy008) (eds. Varoquaux, G. et al.) 11–15 (2008).
Ahsan, M. M., Mahmud, M. P., Saha, P. K., Gupta, K. D. & Siddique, Z. Effect of data scaling methods on machine learning algorithms and model performance. Technologies 9, 52 (2021).
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. Adv. Neur. Inform. Proc. Syst. 32, 8024–8035 (2019).
Fey, M. & Lenssen J. E. Fast graph representation learning with PyTorch Geometric. Preprint at https://doi.org/10.48550/arXiv.1903.02428 (2019).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint at https://doi.org/10.48550/arXiv.1412.6980 (2014).
Mastropietro, A. & Pasculli, G. AndMastro/protein-ligand-GNN: v.1.0.0. Zenodo https://doi.org/10.5281/zenodo.8358539 (2023).
Mastropietro, A., Pasculli, G. & Bajorath, J., Predicting affinities from simplistic protein-ligand interaction representations–what do graph neural networks learn? CodeCapsule. Code Ocean codeocean.com/capsule/8085311 (2023).
Mastropietro, A., Feldmann, C. & Pasculli, G. EdgeSHAPer: v.1.1.0. Zenodo https://doi.org/10.5281/zenodo.8358595 (2023).
Acknowledgements
This work was partly supported (A.M.) by the EC H2020RIA project ‘SoBigData++’ (grant no. 871042), PNRR MUR project no. PE0000013-FAIR and PNRR MUR project no. IR0000013-SoBigData.it.
Author information
Authors and Affiliations
Contributions
Conceptualization was done by J.B. Methodology was done by A.M., G.P. and J.B. Data and code were written by A.M. and G.P. The investigation was carried out by A.M. and G.P. Analysis was done by A.M. and J.B. The original draft was written by A.M. and J.B. Review and editing of the draft were done by A.M. and J.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Sai Pooja Mahajan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Table 1
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mastropietro, A., Pasculli, G. & Bajorath, J. Learning characteristics of graph neural networks predicting protein–ligand affinities. Nat Mach Intell 5, 1427–1436 (2023). https://doi.org/10.1038/s42256-023-00756-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-023-00756-9