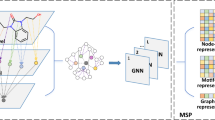
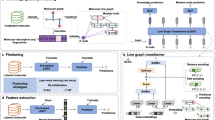
Abstract
Improving drug discovery efficiency is a core and long-standing challenge in drug discovery. For this purpose, many graph learning methods have been developed to search potential drug candidates with fast speed and low cost. In fact, the pursuit of high prediction performance on a limited number of datasets has crystallized their architectures and hyperparameters, making them lose advantage in repurposing to new data generated in drug discovery. Here we propose a flexible method that can adapt to any dataset and make accurate predictions. The proposed method employs an adaptive pipeline to learn from a dataset and output a predictor. Without any manual intervention, the method achieves far better prediction performance on all tested datasets than traditional methods, which are based on hand-designed neural architectures and other fixed items. In addition, we found that the proposed method is more robust than traditional methods and can provide meaningful interpretability. Given the above, the proposed method can serve as a reliable method to predict molecular interactions and properties with high adaptability, performance, robustness and interpretability. This work takes a solid step forward to the purpose of aiding researchers to design better drugs with high efficiency.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Code availability
All code of GLAM is freely available at https://github.com/yvquanli/GLAM with an MIT licence. The version used for this publication is available at https://doi.org/10.5281/zenodo.637116443.
References
Schneider, G. Automating drug discovery. Nat. Rev. Drug Discov. 17, 97–113 (2018).
Schneider, P. et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 19, 353–364 (2020).
Inglese, J. & Auld, D. S. in Wiley Encyclopedia of Chemical Biology (ed. Begley, T. P.) (Wiley, 2008); https://doi.org/10.1002/9780470048672.wecb223
Sliwoski, G., Kothiwale, S., Meiler, J. & Lowe, E. W. Computational methods in drug discovery. Pharmacol. Rev. 66, 334–395 (2014).
Fleming, N. How artificial intelligence is changing drug discovery. Nature 557, S55–S57 (2018).
Zheng, S., Li, Y., Chen, S., Xu, J. & Yang, Y. Predicting drug–protein interaction using quasi-visual question answering system. Nat. Mach. Intell. 2, 134–140 (2020).
Shen, W. X. et al. Out-of-the-box deep learning prediction of pharmaceutical properties by broadly learned knowledge-based molecular representations. Nat. Mach. Intell. 3, 334–343 (2021).
Kotsias, P.-C. et al. Direct steering of de novo molecular generation with descriptor conditional recurrent neural networks. Nat. Mach. Intell. 2, 254–265 (2020).
Méndez-Lucio, O., Baillif, B., Clevert, D. A., Rouquié, D. & Wichard, J. De novo generation of hit-like molecules from gene expression signatures using artificial intelligence. Nat. Commun. 11, 10 (2020).
Chen, H., Engkvist, O., Wang, Y., Olivecrona, M. & Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 23, 1241–1250 (2018).
Jiang, S. & Balaprakash, P. Graph neural network architecture search for molecular property prediction. In Proc. IEEE International Conference on Big Data 1346–1353 (IEEE, 2020).
Cai, S., Li, L., Deng, J., Zhang, B., Zha, Z. J., Su, L., & Huang, Q. Rethinking Graph Neural Architecture Search from Message-passing. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 6653–6662. https://doi.org/10.1109/CVPR46437.2021.00659 (2021).
Zhang, Z., Wang, X., & Zhu, W. Automated Machine Learning on Graphs: A Survey. IJCAI International Joint Conference on Artificial Intelligence, 4704–4712. https://doi.org/10.24963/ijcai.2021/637 (2021)
Ekins, S. et al. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 18, 435–441 (2019).
Sculley, D. et al. Hidden technical debt in machine learning systems. In Proc. Advances in Neural Information Processing SystemsVol. 2015-January, 2503–2511 (NIPS, 2015).
Jiang, M. et al. Drug–target affinity prediction using graph neural network and contact maps. RSC Adv. 10, 20701–20712 (2020).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. In Proc. 2017 International Conference on Learning Representations (ICLR, 2017).
Veličković, P. et al. Graph attention networks. In Proc. 2018 International Conference on Learning Representations 1–12 (ICLR, 2018).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. In Proc. International Conference on Machine Learning Vol. 3, 2053–2070 (ACM, 2017).
Xiong, Z. et al. Pushing the boundaries of molecular representation for drug discovery with graph attention mechanism. J. Med. Chem. https://doi.org/10.1021/acs.jmedchem.9b00959 (2019).
Xu, K., Jegelka, S., Hu, W. & Leskovec, J. How powerful are graph neural networks? In Proc. 7th International Conference on Learning Representations, ICLR 2019 (ICLR, 2019).
Källberg, M. et al. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012).
Li, H., Leung, K. S., Wong, M. H. & Ballester, P. J. Improving AutoDock Vina using Random Forest: the growing accuracy of binding affinity prediction by the effective exploitation of larger data sets. Mol. Informatics 34, 115–126 (2015).
Chen, L. et al. TransformerCPI: improving compound-protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics 36, 4406–4414 (2020).
Huang, K., Xiao, C., Hoang, T., Glass, L. & Sun, J. CASTER: predicting drug interactions with chemical substructure representation. Proc. AAAI Conf. Artif. Intell. 34, 702–709 (2020).
Yang, Y.-Y., Rashtchian, C., Zhang, H., Salakhutdinov, R. & Chaudhuri, K. A closer look at accuracy vs. robustness. In Proc. 34th International Conference on Neural Information Processing Systems Vol. 720, 8588–8601 (NIPS, 2020).
Tetko, I. V., Tanchuk, V. Y. & Villa, A. E. P. Prediction of n-octanol/water partition coefficients from PHYSPROP database using artificial neural networks and E-state indices. J. Chem. Inf. Comput. Sci. 41, 1407–1421 (2001).
Zeng, Y., Chen, X., Luo, Y., Li, X. & Peng, D. Deep drug–target binding affinity prediction with multiple attention blocks. Briefings Bioinform. 22, bbab117 (2021).
Withnall, M., Lindelöf, E., Engkvist, O. & Chen, H. Building attention and edge message passing neural networks for bioactivity and physical-chemical property prediction. J. Cheminform. 12, 1–18 (2020).
Feurer, M., Eggensperger, K., Falkner, S., Lindauer, M. & Hutter, F. Auto-Sklearn 2.0: the next generation (2020); https://www.researchgate.net/publication/342801746_Auto-Sklearn_20_The_Next_Generation
Erickson, N., Mueller, J., Shirkov, A., Zhang, H., Larroy, P., Li, M., & Smola, A. AutoGluon-Tabular: Robust and Accurate AutoML for Structured Data. ICML Workshop on Automated Machine Learning (2020).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Xiong, J., Xiong, Z., Chen, K., Jiang, H. & Zheng, M. Graph neural networks for automated de novo drug design. Drug Discov. Today 26, 1382–1393 (2021).
Dai, H. et al. Retrosynthesis prediction with conditional graph logic network. In Proc. 33rd International Conference on Neural Information Processing Systems Vol. 796, 8872–8882 (NIPS, 2020).
Wang, X. et al. RetroPrime: a diverse, plausible and transformer-based method for single-step retrosynthesis predictions. Chem. Eng. J. 420, 129845 (2021).
Kuznetsov, M. & Polykovskiy, D. MolGrow: a graph normalizing flow for hierarchical molecular generation. In Proc. AAAI Conference on Artificial Intelligence Vol. 35, 8226–8234 (AAAI, 2021).
Luo, Y., Yan, K. & Ji, S. GraphDF: a discrete flow model for molecular graph generation. In Proc. 38th International Conference on Machine Learning, PMLR Vol. 139, 7192–7203 (PMLR, 2021).
Liu, M., Yan, K., Oztekin, B. & Ji, S. GraphEBM: molecular graph generation with energy-based models. Proc. ILCR Workshop on Energy Based Models 1–16 (2021).
Tran-Nguyen, V. K., Jacquemard, C. & Rognan, D. LIT-PCBA: an unbiased data set for machine learning and virtual screening. J. Chem. Inf. Model. 60, 4263–4273 (2020).
Gilson, M. K. et al. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–D1053 (2016).
Wishart, D. S. et al. DrugBank: a knowledge base for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–D906 (2008).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Li, Y. Code for ‘An adaptive graph learning method for automated molecular interactions and properties predictions’ (Zenodo, 2022); https://doi.org/10.5281/zenodo.6371164
Halgren, T. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759 (2004).
Fey, M. & Lenssen, J. E. Fast graph representation learning with PyTorch Geometric. In Proc. ICLR 2019 Workshop on Representation Learning on Graphs and Manifolds (ICLR, 2019); https://arxiv.org/abs/1903.02428
Acknowledgements
This work was supported by the National Natural Science Foundation of China (22173038 and 21775060). We thank the Supercomputing Center of Lanzhou University for providing high-performance computing resources. We acknowledge help from J. Xu, the author of RaptorX22, as well as help from M. Jiang, the author of DGraphDTA16.
Author information
Authors and Affiliations
Contributions
Y.L., C.-Y.H. and X.Y. conceived the project. Y.L., C.-Y.H., R.L., X.G., X.W. and P.L. designed and conducted the experiments. C.-Y.H., S.L., Y.T., D.J., J.Y., Q.B. and H.L. evaluated the experiments and contributed ideas. S.Z., C.-Y.H. and X.Y. managed and supervised the project. All authors co-wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks William McCorkindale and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Design space for blocks of the architectures.
a, Feed-forward Block. It takes a tensor as input and outputs a tensor. Abbreviations and their full name correspond as follows: Norm(Normalization), ReLU(Rectified linear units), CeLU(Continuously differentiable exponential linear units). b, Message Passing Block. It takes a graph as input and outputs a graph. Abbreviations and their full name correspond as follows: GCN(Graph convolutional networks), GAT(Graph attention networks), MPN(Message-passing neural networks), Tri-MPN(Triplet message-passing neural networks), Light Tri-MPN(Light triplet message-passing neural networks). c, Fusion Block. It takes a graph as input and outputs a tensor. Dot means the dot multiplication operation. d, Global Pooling Block. It takes a graph as input and outputs a tensor.
Extended Data Fig. 2 Cases of node-level interpretation.
a, Case studies of solubility prediction. The atoms in the hydrophilic group tend to be bluer in our visualization, which means their weights are closer to 1. In contrast, the atoms in the lipophilic group tend to be redder in our visualization, which means their weights are closer to −1. b, Case studies of drug-drug interactions. The visualization results show the models in predictor pay more attention to the nitrates of isosorbide dinitrate and nicorandil, and pay more attention to the N-methyl of sildenafil and udenafil.
Supplementary information
Supplementary Information
Supplementary Tables 1–5.
Rights and permissions
About this article
Cite this article
Li, Y., Hsieh, CY., Lu, R. et al. An adaptive graph learning method for automated molecular interactions and properties predictions. Nat Mach Intell 4, 645–651 (2022). https://doi.org/10.1038/s42256-022-00501-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-022-00501-8
This article is cited by
-
Relative molecule self-attention transformer
Journal of Cheminformatics (2024)
-
GraphsformerCPI: Graph Transformer for Compound–Protein Interaction Prediction
Interdisciplinary Sciences: Computational Life Sciences (2024)
-
TransG-net: transformer and graph neural network based multi-modal data fusion network for molecular properties prediction
Applied Intelligence (2023)
-
Simple nearest-neighbour analysis meets the accuracy of compound potency predictions using complex machine learning models
Nature Machine Intelligence (2022)