Abstract
Biomedical image analysis algorithm validation depends on high-quality annotation of reference datasets, for which labelling instructions are key. Despite their importance, their optimization remains largely unexplored. Here we present a systematic study of labelling instructions and their impact on annotation quality in the field. Through comprehensive examination of professional practice and international competitions registered at the Medical Image Computing and Computer Assisted Intervention Society, the largest international society in the biomedical imaging field, we uncovered a discrepancy between annotators’ needs for labelling instructions and their current quality and availability. On the basis of an analysis of 14,040 images annotated by 156 annotators from four professional annotation companies and 708 Amazon Mechanical Turk crowdworkers using instructions with different information density levels, we further found that including exemplary images substantially boosts annotation performance compared with text-only descriptions, while solely extending text descriptions does not. Finally, professional annotators constantly outperform Amazon Mechanical Turk crowdworkers. Our study raises awareness for the need of quality standards in biomedical image analysis labelling instructions.
Similar content being viewed by others
Main
Machine learning (ML) is in the process of revolutionizing medicine, with deep learning (DL) as a key enabling technology1. High-quality annotated datasets are a critical bottleneck for supervised DL, and the quality of the annotated data is crucial for algorithm performance2,3,4,5. Recent work reflects an increasing awareness of widespread problems in commonly used image benchmarks, which are subject to errors6,7 and biases8,9, and calls for a fundamental change in dataset culture10. Annotation-related problems may be particularly relevant in the field of biomedical image analysis, where data are typically sparse11, inter-rater variability is naturally high12,13, labelling ambiguities occur14 and medical experts have their individual style of annotations13,15.
In addition to these limitations, domain expert resources are typically limited and costly13. As a result, an increasingly popular approach to generating image annotations involves outsourcing the labelling task to crowdsourcing platforms16,17 or professional annotation companies11. Historically, outsourcing was first performed on general labour markets such as Amazon Mechanical Turk (MTurk)18, which is still the predominant choice in health research 17. With rising demand for annotations, professional annotation companies catering to the specific needs of their target domain emerged. An overview of data annotation from a surgical data science perspective is provided in ref. 11.
While crowdsourcing has successfully been applied in a number of medical imaging applications, the high variation in labelling quality is a key issue16,19,20,21,22. Poor annotation quality can generally be attributed to two main factors:
-
(1)
Lack of motivation and time pressure: Driven by subpar compensation policies23 and power dynamics24, MTurk suffers from workers who perform sloppy annotations with the goal of completing tasks as fast as possible and thus maximizing the monetary reward. This has led to a notable decline in the annotation quality in recent years25.
-
(2)
Lack of knowledge/expertise: A worker depends on the provided information to create the desired outcome for a given task26,27. A lack of labelling instructions leads to workers filling knowledge gaps with their own interpretations28.
While the first problem has been addressed in literature20,29,30,31,32, the notion of training workers has been given almost no attention in the context of medical imaging16,17. Regardless of the annotator type (general crowdsourcing or professional annotation companies), knowledge transfer is typically achieved via so-called labelling instructions, which specify how to correctly label the (imaging) data. Such labelling instructions are not only needed for training non-experts but can also help reduce experts’ inter-rater variability by defining a shared standard.
Given the importance of annotation quality for algorithm performance and the fundamental role of labelling instructions in this process, it may be surprising that extremely limited effort has been invested into the question of how to best generate labelling instructions in a way that annotation quality is maximized. While previous work on instructions has dealt with instructions for annotation systems with a focus on natural language processing tasks33,34,35,36,37 and standardization of dataset reporting38,39, we are not aware of any work dedicated to labelling instructions in the field of biomedical image analysis and involving professional annotation companies. Closing this gap in the literature, our contribution is twofold:
-
(1)
Analysis of common practice (Fig. 1a): Through comprehensive examination of professional annotating practice and major international biomedical image analysis competitions, we systematically investigated the importance of labelling instructions, their quality and their availability.
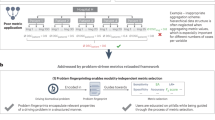
Fig. 1: Hypotheses of this work and overview of methodology. a, Annotators’ needs for labelling instructions (left) were captured via a survey. The availability of labelling instructions (right) was captured via international competitions conducted in the scope of the MICCAI conference. b, To assess the impact of labelling instructions on annotation quality, three types of labelling instructions were created for the same dataset: (1) instructions using minimal text, (2) extended text and (3) extended text including pictures were issued to a total of 864 annotators from five different annotation providers. The resulting dataset of 14,040 annotated images was analysed with a two-part beta mixed model. The dials qualitatively represent the observed annotation performance.
-
(2)
Experiments on the impact of biomedical image labelling instructions (Fig. 1b): On the basis of the 14,040 images annotated by a total of 864 annotators from five different annotation providers, we experimentally determined the effect of varying information density in labelling instructions. In this context, we also investigated annotation quality of professional annotators in comparison with the current status quo in scalable annotations, MTurk crowdworkers.
Of note, varying annotation quality impacts training and validation/testing of ML models to different extents. In safety-critical applications, it is particularly the test data that ultimately determine the real-world (for example, clinical) applicability of an algorithm, thus requiring a higher level of quality compared with training sets. Hence, the focus of our study has been placed on test data.
Results
Given the lack of (1) awareness of the importance of labelling instructions and (2) quantitative research investigating how to best perform the labelling, we initiated our study by systematically analysing the perspective and work characteristics of professional annotators, and common practice of labelling instructions in leading biomedical imaging competitions. Subsequently, we investigated the impact of labelling instructions with varying levels of information density on the annotation quality, and the effect of different annotator types on biomedical imaging data.
Professional annotators request better labelling instructions
To motivate our empirical study on annotation quality, we conducted an international survey among 363 (298 after filtering noisy answers) professional annotators employed by five different internationally operating annotation companies. Depicted in Fig. 2a,b, the results reveal that the majority of annotators request more time and resources to be spent in the generation of labelling instructions. In fact, poor labelling instructions were identified as the primary cause of problems related to annotation work followed by concentration issues (50%) and poor input data (45%).
a, Professional annotators agree that unclear instructions consistently cause delay and rework. Comprehensive text descriptions and images are perceived to improve annotation quality. b, Professional annotators attribute labelling instructions as the primary cause for problems related to their daily annotation work, followed by concentration issues and poor input data. Answers for a and b were processed from 298 annotators from five annotation companies. c, Seventy-six per cent of the recent MICCAI conference competition tasks, matching the inclusion criteria, do not report any labelling instructions. The analysis includes all 96 registered MICCAI competition tasks between 2020 and 2021.
The importance of labelling instructions may be underrated
Despite their apparent importance, earlier research revealed that labelling instructions are typically not provided and/or reported in the field of biomedical image analysis40. This even holds true for international image analysis competitions, although these can be expected to provide particularly high quality with respect to validation standards. To address this issue, the Medical Image Computing and Computer Assisted Intervention Society (MICCAI), the largest international society in the field, took action and developed a comprehensive reporting guideline39 for biomedical image analysis competitions. The guideline comprises an entire paragraph on reporting the annotation process, including the labelling instructions. Before conducting a competition in the scope of a MICCAI conference, researchers must put the report for their competition online41 to foster transparency and reproducibility, and to prevent cheating42. To capture the state of the art regarding labelling instructions in biomedical image analysis, we analysed all MICCAI competitions officially registered in the past 2 years (Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement43; Supplementary Note 1). Although the reporting guideline explicitly asks for (a link to) the labelling instructions, 76% of the recent MICCAI competitions do not report any labelling instructions (Fig. 2c). Given that MICCAI competitions make up around 50% of the biomedical image analysis competitions in a year40, this can be regarded as a widely spread phenomenon.
In current biomedical image analysis practice, labelling instructions are thus often neither of sufficient quality, nor are they appropriately reported and valued in the scientific community. Both issues negatively impact scientific quality in the field.
Extended text descriptions do not boost annotation quality
The shortcomings in quality we found in common practice regarding labelling instructions called for an investigation on how this quality can be improved. As a first step in this direction, we sought to determine the impact of different types of labelling instructions on the quality of annotation of a particular dataset. The selected dataset44,45, which can be handled by crowdworkers, combines highest-quality reference annotations and 21 meta annotations per annotated image that reflect the annotation difficulty. We created three distinct types of labelling instructions with varying levels of information density, namely, (1) minimal text, (2) extended text and (3) extended text including pictures, as detailed in Methods and displayed in Supplementary Notes 6–8. Example instructions are provided in Fig. 3. The obtained 23,443 annotations on 14,040 images from the 864 annotators were analysed with a two-part zero-inflated beta mixed model (ZIBMM).
In contrast to the limited text labelling instructions, the extended text labelling instructions included more detailed descriptions, counter-examples and information on uncommon annotation cases that might appear. We define an annotation with a metric score equal to zero as a severe annotation error. For the 4,680 images annotated with the extended text labelling instructions, we observed a minor increase in the number of severe annotation errors compared with the limited text labelling instructions (median +0.4%, maximum +14.8%, minimum −31.7%). Furthermore, we observed no impact on the median Dice similarity coefficient (DSC)46, and only a minor increase in the interquartile range (IQR) (median +1.8%, maximum +33.3%, minimum −75.8%). These results contradict the initial assessment of the professional annotators. The absent effect of the extended text labelling instructions is reinforced by the results of the two-part ZIBMM, where we obtained no statistically significant difference for the extended text labelling instructions compared with minimal text from both the first and second part of the model, revealing that extended text descriptions do not boost annotation performance.
Exemplary images are crucial for high-quality annotations
Professional annotators claim that pictures help them understand labelling instructions (Fig. 2a). Therefore, the extended text including pictures labelling instructions were enriched by pictures including rare occurrences. In comparison with the extended text labelling instructions, the number of severe annotation errors was reduced for all five annotation providers (median −33.9%, maximum −13.6%, minimum −52.3%). Furthermore, their median DSC score increased (median +2.2%, maximum 20.0%, minimum +1.1%), and their IQR was reduced (median −58.3%, maximum −9.1%, minimum −84.2%) (Fig. 4a). This reinforces professional annotators’ initial assessment that pictures improve their understanding (Fig. 2a). The improvements occurred mainly on the difficult annotation cases (Supplementary Note 2). Based on the two-part ZIBMM, the odds of obtaining a severe annotation error with these labelling instructions are 0.37 times (credible interval (CI) 0.28–0.50) that with minimal text labelling instructions. From the second part of the two-part ZIBMM, we obtained no significant difference in the DSC score, once an object was identified. Thus, the improvements primarily stemmed from the additional reduction of severe annotation errors (Fig. 4b).
(1) Extended text descriptions (orange) do not necessarily boost annotation performance compared with minimal text descriptions (blue), while (2) including images (green) gives a clear benefit for all annotation providers. (3) Professional annotation companies (companies 1–4) provide substantially higher-quality annotations compared with the most popular crowdsourcing platform MTurk. a, The DSC score has been aggregated for each annotated image and is displayed aggregated for each pair of company and labelling instruction as dots and box plot (the band indicates the median, the box indicates the first (25th percentile) and third (75th percentile) quartiles and the whiskers indicate ±1.5 × IQR, the DSC score maximum is 1 and the minimum is 0 for each image). b, The absolute number of severe annotation errors, defined as annotations with a metric score equal to zero, is also shown. Metric scores for a and b were each processed from a total of 14,040 images annotated by 156 annotators from four professional annotation companies and 708 MTurk crowdworkers.
Professional annotation companies outperform crowdsourcing
In comparison with MTurk crowdworkers, professional annotators conduct labelling as their main source of income, label more often in a week and label for a higher number of weekly hours (Fig. 5a–c). In contrast, MTurk workers have a longer employment history in labelling than professional annotators, as displayed in detail in Fig. 5d. This observation is consistent with the historical development of the data annotation market, where general labour markets, including MTurk, preceded professional annotation companies. Given minimal text labelling instructions, professional annotators produced less severe annotation errors (companies median 307, MTurk median 549). Furthermore, the professional annotators’ annotations generated a higher median DSC score (companies median 0.93, MTurk median 0.67), and a smaller IQR of the DSC score (companies median 0.36, MTurk median 0.67). Both annotator types showed only minor or no improvement with the extended text labelling instructions. While both annotator types benefitted from added pictures, professional annotators displayed a stronger reduction of severe annotation errors (companies median −34.4%, MTurk median −13.6%) and of the IQR (companies median −63.00%, MTurk median −9.1%). In contrast, MTurk crowdworkers displayed a stronger improvement of the median DSC score (companies median +1.7%, MTurk median +20.0%). Under the same conditions, the odds of severe annotation errors for a professional annotator were 0.09 times (CI 0.06–0.12) that of a MTurk crowdworker, keeping all other factors constant. Similarly, once an object was identified, the odds for a professional annotator of achieving a perfect DSC score were 94.7% (1.947, CI 1.69–2.24) higher than those of a MTurk crowdworker.
a–c, In comparison with the MTurk crowdworkers, professional annotators conduct labelling as their main source of income (a), label more often in a week (b) and spend a higher number of hours per week on labelling images (c). d, In contrast, MTurk crowdworkers have a longer employment history with labelling than professional annotators. Answers were processed from 298 professional annotators from five annotation companies and 518 MTurk crowdworkers.
Discussion
To our knowledge, this study is the first to quantitatively and critically examine the role of labelling instructions in professional and crowdsourced annotation work. We were able to uncover a major discrepancy between their importance and quality/availability, and determine which type of labelling instructions is the most effective. While labelling instructions play a crucial role in the creation of biomedical image analysis datasets, current common practice is insufficient and neither meets the requirements of industry (for example, in creating large-scale datasets) nor those of academia (for example, in hosting competitions) (Fig. 2c). Notably, professional annotators demand a higher time and resource commitment from the creators of labelling instructions. Furthermore, they identify current labelling instructions as a main cause for annotation delay and rework (Fig. 2a). Interestingly, we found that extended text descriptions do not necessarily boost annotation performance compared with minimal text descriptions (Fig. 4), although professional annotators expect them to improve the understanding of the labelling tasks (Fig. 2a). In contrast, the addition of pictures resulted in a clear improvement among all annotation providers (Fig. 4), which matches the assessment of the professional annotators (Fig. 2a). This improvement was mainly observed on ambiguous images with challenging conditions, such as poor illumination or intersecting objects, as depicted in Supplementary Note 2. Since ML models fail their prediction more often on ambiguous images than on clear images2, the correct annotation of such images in training datasets is particularly crucial for good performance. Lastly, annotators from professional annotation companies provide substantially higher-quality annotations compared with those from the most popular crowdsourcing platform in health research, MTurk, regardless of the type of labelling instruction.
One of the key implications of our findings is that there is a huge discrepancy between the potential impact of labelling instructions on generating the desired annotations and their role in current (research) practice. Our study shows that scope and specific design choices of labelling instructions play a major role in generating the desired annotations. Among other contributing factors such as prior annotation expertise or training, labelling instructions may thus determine the attention to detail that annotators pay to annotating challenging images. The importance of this is amplified in cases of datasets becoming increasingly complex and diversified over time, where more and more challenging data points are added due to initial prediction difficulties of the ML model. Consequently, researchers and practitioners alike should ensure proper representation of the necessary information in their labelling instructions, extend them if needed and invest the necessary time to produce informative annotated images. However, we observed that 76% of the recent MICCAI competitions did not report their labelling instructions (Fig. 2c). Since competitions are aimed at generating high publicity, it can be assumed that their current handling of labelling instructions represents the upper bound of quality regarding common practice, with the quality being substantially lower for research projects devoid of intense public scrutiny. This discrepancy is alarming and calls for a paradigm shift in common practice. Given our results, the MICCAI Special Interest Group47 for Biomedical Image Analysis Challenges is currently re-evaluating common competition practices and considering stricter rules for data quality for future challenges.
Another implication of our study is that generating and publishing labelling instructions is a pre-condition for enabling independent verification and reproduction of the created annotations. Similarly to how published code enables the verification of algorithmic results in research papers, access to labelling instructions is necessary to understand annotators’ decisions and potentially re-create annotations. Furthermore, the information provided in labelling instructions is important for ML practitioners. Based on the annotation decisions (for example, handling of occluded objects of interest) implemented in a dataset, ML practitioners need to define their own desired outcome for these occurrences and modify their ML model accordingly. Given their impact on the resulting annotations and the current poor state of datasets6,7,8,9,10, we argue that dataset creators and competition organizers should publish their labelling instructions, as proposed in ref. 39. To advance current practice, we recommend the dataset and labelling instruction creation to be an iterative process properly modelling the underlying distribution of the data space, as described in ref. 13. In earlier stages of the dataset creation process, the focus should be on common occurrences (for example, common surgical instruments in the case of laparoscopic surgery) to generate strong initial model performance. Throughout the process, special, conflicting or rare occurrences should be added to the dataset to maximize the model performance and reflect the real-world distribution.
A further recommendation motivated by our study is for annotation requesters to evaluate their annotator options more carefully and select their provider on the basis of suitability to their annotation requirements. While medical personnel alone may be too sparse and costly to satisfy the rising demand for annotated biomedical image data13, oftentimes, medical domain knowledge may be necessary only for the creation of the labelling instructions and not the annotation itself. Thus, crowdsourcing in combination with computer-assisted annotation strategies can be a valid and cost-effective approach. MTurk, the most commonly used crowdsourcing platform in health research17, follows a do-it-yourself model, where all components from annotator training to annotation tooling are provided by the annotation requester and the crowdworkers are employed on a freelance basis. In contrast, professional annotation companies assign a dedicated contact person that oversees the project, with annotators trained and directly employed by the annotation company. While MTurk can quickly scale up with a large number of people, professional annotation companies tend to scale up more slowly. However, professional annotators work on data annotation for a longer proportion of their workday and are usually assigned full-time to an annotation project (Fig. 5). Regarding location, the international annotation market leads to scenarios where requesters and annotators do not share the same (native) language. This reinforces the need for clear and concise labelling instructions with exemplary pictures as a fundamental requirement for scaling data annotation operations.
Irrespective of the person chosen to conduct the annotations, the required domain knowledge to perform a given annotation task depends mainly on the underlying problem statement and the used imaging modality. Furthermore, the scope and form of high-quality labelling instructions may vary depending on the underlying problem. For simpler problems, such as eye colour classification, shorter labelling instructions may be sufficient. Complicated annotation tasks, on the other hand, may require elaborate labelling instructions up to 400 pages in length48 to properly communicate the necessary information. Regarding form, a labelling instruction is not restricted to being a document, and could be presented in video or app format as well. Which format or combination works best for the problem at hand should be determined individually. Regardless, the chosen format should be archivable to enable consistent onboarding of additional annotators and access for future users of the dataset to understand the data generation process. In case of doubt, sharing more information than necessary in the labelling instructions is preferable to sharing too little information, as evidenced by our work (Fig. 4).
Of note, with performance assessment of crowdworkers still conducted by examining their performance on reference standard datasets13,49 or measuring the inter-worker agreement50,51, the recommended iterative labelling instruction generation would potentially reduce errors resulting from lacking quality in labelling instructions and should thus result in a more precise assessment of crowdworkers’ actual performances.
Another implication of our findings is the effect of annotation errors on the life cycle of an ML system. Data critically impact the entire pipeline of an ML system, from the initial problem statement up to the final model deployment52. Annotation errors, often referred to as ‘label noise’, thus represent a long-standing challenge in the ML community, which has led to the development of pre-processing methods to clean data or the creation of models that are more robust to annotation errors53. This holds especially true for the safety-critical biomedical domain, in which high-quality test images are the foundation for the medical certification of new solutions54,55. It should be noted that robustness also plays a particularly important role in this domain. Hence, there is a risk of rare cases being mislabelled and not contributing sufficiently to the robustness assessment. Our experiments show that images with rare but relevant characteristics particularly benefit from labelling instructions with a higher density of information (Supplementary Note 2).
Furthermore, annotation errors in the test set have drastic consequences for ML competitions, which are often considered the gold standard for identifying the best algorithm for a specific research question. Competitions typically lead to winning algorithms becoming the new state-of-the-art method and being awarded tremendous monetary rewards and recognition40. Even for well-established datasets, such as ImageNet56, annotation errors in the test set falsify the selection of the best-performing model. For example, a trained ResNet-18 model was shown to outperform a trained ResNet-50 model if the prevalence of originally mislabelled test examples increases by just 6%, given the corrected ImageNet test set6. This issue is further aggravated by the fact that competition test sets are frequently inaccessible to the public after a competition ends, making auditing of the test sets impossible. A comprehensive overview of data issues and their cascading effect is provided in ref. 52. In summary, reducing annotation errors early on within the life cycle of an ML system by providing higher-quality labelling instructions positively impacts data pre-processing, model selection, model training, model validation and finally model deployment.
A limitation of our study could be seen in the fact that we included only one dataset. Our chosen dataset combines the advantages of high-quality reference annotations, representing the real-world complexity with gradually increasing stages of difficulty and high volume. We chose to include only one sample scenario since running several experiments with the same annotation companies and different datasets poses substantial risks of exposing the experimental setting. Annotation companies are typically well aware of the most common datasets and would additionally spot the known experimental structure of gradually increasing labelling instructions within the same layout. Awareness of the experimental setting would in turn lead to results being skewed in favour of high-quality performances, since these companies are inherently motivated to present their work as reliable. A further limitation of our study is that MTurk required a different annotation tooling than the annotation tooling used by the professional annotation companies. To mitigate a potential impact of the toolings, all participating annotators had no prior experience with their respective tooling and both toolings included best design practices to enable high-quality annotations.
Our study was subject to several design choices. For the performance measurement, we focused on the DSC as an overlap-based metric. Utilizing distance-based metrics, such as the normalized surface distance, yielded similar results. For the statistical analysis, we assumed that the non-zero DSC scores follow a beta distribution as it is the more natural distribution for this kind of data. We further assumed that the random effects in the two-part ZIBMM are normally distributed and correlated. The correlation assumption was reasonable in that, as the probability of severe annotation errors increases, the expected DSC score decreases and vice versa.
Even though this paper focuses on labelling instructions associated with biomedical image analysis, we believe that the findings can be translated to other research fields, and to crowdsourced data annotation in general. We expect the impact to go beyond academia because industrial production ML projects by nature depend on a pre-defined level of annotation quality to obtain the required algorithm performance level. Consequently, it stands to reason that more effort and monetary resources should especially be invested in developing labelling instructions in industry13.
The present work opens up several future research avenues: First, a structured competition submission system has improved the quality of biomedical image analysis competitions in recent years39. By collecting the submitted labelling instructions and the corresponding feedback over time, a model could be trained to provide an automated quality feedback mechanism for labelling instructions in the biomedical domain. Second, professional annotation companies usually conduct quality assurance checks by experienced annotators or team leads before providing their created annotations to the annotation requester. Although we obtained substantial quality improvements in the obtained annotations by optimizing the labelling instructions, it would be of interest to analyse potential further impact of such quality assurance checks. Additionally, the interaction effects between experienced annotators working on quality assurance checks and instructions with varying levels of information density could be of interest to the scientific community. Third, data pipelines with a long-term focus face the risk of concept drift, where the initially captured distribution of input data changes. How labelling instructions should evolve along ever-changing data and its distribution remains an open question to be tackled. Finally, with an increasing educational shift towards digitalization and data science, involving medical students in medical image annotation as part of their study programme could potentially become a new source of crowdsourcing. Future research should examine this promising symbiotic relationship, where medical students obtain hands-on ML-related skills highly relevant to their profession while at the same time easing the annotation bottleneck for the scientific community.
In summary, our study is the first to examine the impact of the quality of labelling instructions on annotation work performed both by professional annotators and crowdworkers. We uncovered a substantial discrepancy between the demand of professional annotators for better labelling instructions and current common practice. Given the rapidly increasing complexity and diversity of datasets, we envision the establishment and widespread adoption of quality standards for labelling instructions to become imperative in the future.
Methods
Following the definition of terms used throughout this study, we will describe the selected data, the annotation providers, the labelling instructions, the experimental setup and the statistical analysis in detail throughout this section.
Definitions
We use the following terms throughout the paper:
Annotation provider: An entity that provides annotations performed by human workers. They can be categorized into two types of annotation providers: (1) crowdsourcing platforms, such as MTurk, and (2) professional annotation companies (see the following definitions).
Annotation requester: An entity that wants a dataset annotated by an external annotation provider.
Challenge/competition: We follow the challenge definition of the BIAS statement, which defines a challenge as an “open competition on a dedicated scientific problem in the field of biomedical image analysis. A challenge is typically organized by a consortium that issues a dedicated call for participation. A challenge may deal with multiple different tasks for which separate assessment results are provided. For example, a challenge may target the problem of segmentation of human organs in computed tomography (CT) images. It may include several tasks corresponding to the different organs of interest”40.
Challenge/competition task: The BIAS statement defines a challenge task as a “subproblem to be solved in the scope of a challenge for which a dedicated ranking/leaderboard is provided (if any)”40.
Labelling instruction: A document or tool that specifies how to correctly label (imaging) data. The different types of labelling instructions are defined below and presented in greater detail in Supplementary Notes 6–8.
MTurk: A two-sided crowdsourcing marketplace that enables annotation requesters to hire freelance workers, referred to as MTurk crowdworkers, to perform discrete tasks on-demand.
MTurk crowdworker: A remotely located person performing discrete on-demand tasks on MTurk on their own hardware. They are employed on a freelance basis.
Professional annotation company: A company focusing mainly on generating annotations for (imaging) data. Their workers are located in regular office space and mainly employed full-time.
Professional annotator: An on-site located and full-time employed person performing annotations for a professional annotation company in a provided office space with according hardware.
International professional annotator survey
To obtain a comprehensive understanding of the current issues professional annotators face with respect to labelling instructions and their work characteristics, we developed a 26-item questionnaire (provided in Supplementary Note 4). The survey was distributed among five internationally operating annotation companies in best-cost countries that exclusively employ professional annotators. To increase the statistical validity of the submitted entries (n = 363), we employed a twofold filtering strategy of the entries: (1) a control question pair, which consists of the positive and negative formulation of a question, and (2) an instructional manipulation check, as recommended in ref. 57. The check asks the participant to answer an open-ended question with a specific set of words that can only be answered by carefully reading the question text. The filtering resulted in 298 remaining entries.
Competition analysis
Our goal was to capture the current handling of labelling instructions in biomedical image analysis. Thus, we included all MICCAI registered competitions that were published until the end of 2021. We retrieved the registered competitions from the MICCAI website, where all MICCAI registered competitions are published. This resulted in a list of 53 competitions with 96 competition tasks. Two engineers with a proven history in reviewing competitions rated all submitted standardized competition design documents of the individual competition tasks as to whether a labelling instruction was provided by the competition task. In addition, ambiguous cases were marked as such. This resulted in an inter-rater agreement of 90.6%. Contradictory ratings were mainly cases where both raters marked the competition task as ambiguous and were solved by an independent third engineer with a proven history in reviewing competitions. Twenty competition tasks were excluded as not applicable in the process, as they provided a valid reasoning why labelling instructions cannot be provided. An example is the Medical Out-of-Distribution Analysis Challenge58, where the publication of the labelling instruction would enable cheating, because it contains information about the placement of out-of-distribution objects in the competition data. The result of the competition tasks analysis is provided in Supplementary Note 3.
Dataset selection
The Heidelberg Colorectal (HeiCo) segmentation44,45 dataset, comprising medical instrument segmentations in laparoscopic video data, served as the basis for this study. Each image was enhanced by 21 meta annotations representing relevant image characteristics or artefacts (for example, whether overlapping instruments or motion blur are present on the image), where each image characteristic is a binary annotation decision59. The meta annotations were implemented by a trained engineer with extensive annotation experience, who was involved in the original creation process of the dataset. On the basis of the meta annotations, the images were categorized into nine different image categories, which represented the potential annotation difficulty and served for the subsequent image selection:
-
(1)
Simple category: Images do not contain any artefact on the instruments.
-
(2)
Chaos category: Images contain at least three different artefacts on the instruments. Moreover, images containing a higher number of instruments are preferred.
-
(3)
Trocar category: At least one trocar is present on the image.
-
(4)
Intersection category: At least two medical instruments are intersecting on the image.
-
(5)
Motion blur category: A minimum of one medical instrument with the motion blur artefact is present on the image.
-
(6)
Underexposure category: At least one medical instrument on the image is underexposed.
-
(7)
Text overlay category: Text overlay is present and obstructs the view of the image.
-
(8)
Image overlay category: An image overlay is present and obstructs the view of the image.
-
(9)
Random category: Images are randomly selected from the remaining images in the test set.
We selected 234 unique frames corresponding to the defined categories. Each unique image was annotated four times per labelling instruction and per annotation provider, resulting in 60 annotations per image (generating a total of 14,040 annotated images). Fifteen unique images from category 1 to category 8 were selected by hand, accounting for roughly half of the images. The only exception was category 8, because there existed only 11 unique images that matched the definition of the category. The other half was selected randomly (category 9).
Annotation providers
The study was conducted on the basis of five annotation providers, consisting of four professional annotation companies and the crowdsourcing platform MTurk. Each annotator participated exclusively with one of the three labelling instructions. We selected high-quality representatives for the professional annotation companies and the crowdsourcing platform. The professional annotation companies operate internationally, and their annotators are located in best-cost countries. The selected companies had a proven track record in large-scale industry annotation projects. Participating crowdworkers on MTurk had to fulfil the following quality requirements: (1) 98% accepted human intelligence tasks (HITs) and (2) a minimum of 5,000 accepted HITs. Our quality requirements thus far surpassed the quality requirements of researchers working with MTurk, which normally require 95% accepted HITs with a minimum of 100 accepted HITs20,21,25,35. To capture a representative sample of the population on MTurk, we spread our HITs across 40 days and all times of day. We followed Litman et al. to ensure a fair worker compensation23.
Labelling instructions
As a labelling instruction specifies how to correctly label the (imaging) data, we defined a set of design rules that are shared across all labelling instructions:
-
(1)
The information of a labelling instruction is provided in a slide layout for better human information processing.
-
(2)
Each slide represents a chunk, an encapsulated unit of information. Chunking reduces the demand on the working memory.
-
(3)
Related information chunks are positioned near each other.
-
(4)
A consistent layout with defined fonts, symbols and colours is applied.
Minimal text labelling instructions
The minimal text labelling instructions consist of a limited textual description, including positive examples and the most common annotation occurrences (Supplementary Note 6). This represents a situation where only little effort was put into creating the labelling instruction. For example, text overlay references the uncommon occurrence of text that is visible in the image. Because it is an uncommon occurrence, it is not mentioned in the minimal text labelling instruction. As a baseline, the minimal text labelling instructions consist of seven slides with 168 words.
Extended text labelling instructions
The extended text labelling instructions extend the minimal text labelling instructions with a comprehensive text description, which is supported by both positive examples and counterexamples in text form. Furthermore, both common and uncommon cases are included (Supplementary Note 7). In this labelling instruction for example, the uncommon occurrence of text overlay is described in detail. This resulted in ten slides with 446 words.
Extended text including pictures labelling instructions
The extended text including pictures labelling instructions complement the extended text labelling instructions with pictures (Supplementary Note 8). The pictures include textual descriptions, symbols, markings and the usage of colour to convey the information on the slides. In addition, rare annotation occurrences are included as well. This represents a situation where extensive (domain) knowledge about the labelling process is present and documented in detail in the labelling instruction. Hence, in our example, the uncommon occurrence of text overlay is described in detail with text and pictures. These labelling instructions consist of 16 slides with 961 words.
Setup for the labelling instruction experiments
Each of the five labelling providers annotated the same images subsequently with each labelling instruction, using separate annotators. We started with the minimal text labelling instructions, followed by the extended text labelling instructions, and provided the extended text including pictures labelling instructions last, to prevent information leakage. As an additional security measure, we added a minimum break of 10 days between two labelling instructions. No individual questions from the annotators regarding the labelling instruction content were answered to prevent a potential information advantage for an annotation provider that could impact the statistical analysis. Each annotator was only allowed to participate for a single labelling instruction. All participating annotators had no prior experience with their respective annotation tooling. Furthermore, no worker selection tasks were used for the annotation providers. Each professional annotator annotated a total of 72 images. To properly simulate the parallelization of crowdsourcing, each MTurk crowdworker annotated four images. After the submission of their annotations, each MTurk crowdworker could submit a short optional survey about their work characteristics.
Statistical analysis
To quantify the impact of labelling instructions and the two annotator types, the following statistical methods were used:
To ensure compatibility with prior work on the data, we utilized the same metrics as suggested in ref. 45, in which the dataset was originally introduced as part of the MICCAI Robust Medical Instrument Segmentation Challenge 2019. We analysed the annotation results based on the DSC scores with a two-part ZIBMM60: (1) the first part included a logistic mixed model analysing the probability of severe annotation errors (at least one instrument with DSC of 0 in one frame) and (2) the second part consisted of a beta mixed model analysing the non-zero DSC values of an image when valid annotations occurred. The image variable and the annotation worker variable were modelled as random effects while the type of labelling instructions, annotator type, image category and access to context video were modelled as fixed effects with two-side hypothesis tests. The model was implemented in the brms package in R (ref. 61), where vague Gaussian priors centred on 0 were used for the fixed effects and half-Cauchy priors were assigned for the standard deviation of the random effects. A total of 4,000 Markov chain Monte Carlo samples were generated across four chains. The obtained estimates of the covariates are on the log-odds scale and were exponentiated to obtain the odds ratio for each covariate. Software: R version 4.0.2 (package brms version 2.16.0).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Six datasets were utilized during the current study: DS1: Captured biomedical competition design documents from publicly available sources (2020–2021). DS2: Reference annotations for the HeiCo44,45 dataset. DS3: Captured annotations from professional annotators and MTurk crowdworkers. DS4: Individual professional annotator responses to the survey ‘Labeling Instructions Survey’. DS5: Individual MTurk crowdworker responses to optional work characteristics survey. These questions are a subset of the DS4 questions. DS6: DSC scores between DS2, DS3 and the existing output of six algorithms from a recent medical instrument instance segmentation challenge. For DS1, the individual challenge design documents are freely available at MICCAI62. A reporting summary for the evaluation is available as Supplementary Note 3. DS2 is freely available from Synapse63. DS3, DS4, DS5 and DS6 are available from the corresponding author L.M.-H. upon reasonable request.
Code availability
The repository with the statistical code (excluding the raw data) is publicly available at https://github.com/IMSY-DKFZ/labeling_instructions_matter (ref. 64).
References
Benjamens, S., Dhunnoo, P. & Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. NPJ Digit. Med. 3, 118 (2020).
Shad, R., Cunningham, J. P., Ashley, E. A., Langlotz, C. P. & Hiesinger, W. Designing clinically translatable artificial intelligence systems for high-dimensional medical imaging. Nat. Mach. Intell. 3, 929–935 (2021).
Peiffer-Smadja, N. et al. Machine learning for COVID-19 needs global collaboration and data-sharing. Nat. Mach. Intell. 2, 293–294 (2020).
Hu, Y. et al. The challenges of deploying artificial intelligence models in a rapidly evolving pandemic. Nat. Mach. Intell. 2, 298–300 (2020).
Willemink, M. J. et al. Preparing medical imaging data for machine learning. Radiology 295, 4–15 (2020).
Northcutt, C. G., Athalye, A. & Mueller, J. Pervasive label errors in test sets destabilize machine learning benchmarks. In Proc. 35th Conference on Neural Information Processing Systems Track on Datasets and Benchmarks (eds Vanschoren, J. & Yeung, S.) (NeurIPS, 2021).
Rädsch, T. et al. What your radiologist might be missing: using machine learning to identify mislabeled instances of X-ray images. In Proc. 54th Hawaii International Conference on System Sciences (HICSS) (ed. Bui, T. X.) (HICSS, 2021).
Paullada, A., Raji, I. D., Bender, E. M., Denton, E. & Hanna, A. Data and its (dis)contents: a survey of dataset development and use in machine learning research. Patterns 2, 100336 (2021).
Peng, K., Mathur, A. & Narayanan, A. Mitigating dataset harms requires stewardship: lessons from 1000 papers. In Proc. 35th Conference on Neural Information Processing Systems Track on Datasets and Benchmarks (eds Vanschoren, J. & Yeung, S.) (NeurIPS, 2021).
The rise and fall (and rise) of datasets. Nat. Mach. Intell. 4, 1–2 (2022).
Maier-Hein, L. et al. Surgical data science—from concepts toward clinical translation. Med. Image Anal. 76, 102306 (2022).
Joskowicz, L., Cohen, D., Caplan, N. & Sosna, J. Inter-observer variability of manual contour delineation of structures in CT. Eur. Radiol. 29, 1391–1399 (2019).
Freeman, B. et al. Iterative quality control strategies for expert medical image labeling. Proc. AAAI Conference on Human Computation and Crowdsourcing 9, 60–71 (2021).
Kohli, M. D., Summers, R. M. & Geis, J. R. Medical image data and datasets in the era of machine learning—whitepaper from the 2016 C-MIMI meeting dataset session. J. Digit. Imaging 30, 392–399 (2017).
Balagopal, A. et al. PSA-Net: deep learning-based physician style-aware segmentation network for postoperative prostate cancer clinical target volumes. Artif. Intell. Med. 121, 102195 (2021).
Ørting, S. N. et al. A survey of crowdsourcing in medical image analysis. Hum. Comput. 7, 1–26 (2020).
Créquit, P., Mansouri, G., Benchoufi, M., Vivot, A. & Ravaud, P. Mapping of crowdsourcing in health: systematic review. J. Med. Internet Res. 20, e187 (2018).
Amazon Mechanical Turk (Amazon Mechanical Turk, 2022); https://www.mturk.com/
Budd, S. et al. in Domain Adaptation and Representation Transfer, and Affordable Healthcare and AI for Resource Diverse Global Health (eds Albarqouni, S. et al.) 251–262 (Springer, 2021).
Heim, E. et al. Large-scale medical image annotation with crowd-powered algorithms. J. Med. Imaging 5, 034002 (2018).
Cheplygina, V., Perez-Rovira, A., Kuo, W., Tiddens, H. A. W. M. & de Bruijne, M. in Deep Learning and Data Labeling for Medical Applications (Carneiro, G. et al.) 209–218 (Springer, 2016).
Maier-Hein, L. et al. Can masses of non-experts train highly accurate image classifiers? In International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Golland, P. et al.) 438–445 (Springer, 2014).
Litman, L., Robinson, J. & Rosenzweig, C. The relationship between motivation, monetary compensation, and data quality among US- and India-based workers on Mechanical Turk. Behav. Res. Methods 47, 519–528 (2015).
Denton, E., Díaz, M., Kivlichan, I., Prabhakaran, V. & Rosen, R. Whose ground truth? Accounting for individual and collective identities underlying dataset annotation. NeurIPS Data-Centric AI Workshop (NeurIPS, 2021).
Kennedy, R. et al. The shape of and solutions to the MTurk quality crisis. Polit. Sci. Res. Methods 8, 614–629 (2020).
Hossfeld, T., Keimel, C. & Timmerer, C. Crowdsourcing quality-of-experience assessments. Computer 47, 98–102 (2014).
Tokarchuk, O., Cuel, R. & Zamarian, M. Analyzing crowd labor and designing incentives for humans in the loop. IEEE Internet Comput. 16, 45–51 (2012).
Clark, H. H. & Brennan, S. E. in Perspectives on Socially Shared Cognition (eds Resnick, L. et al.) 127–149 (American Psychological Association, 1991).
Sullivan, D. P. et al. Deep learning is combined with massive-scale citizen science to improve large-scale image classification. Nat. Biotechnol. 36, 820–828 (2018).
Albarqouni, S., Matl, S., Baust, M., Navab, N. & Demirci, S. in Deep Learning and Data Labeling for Medical Applications (eds Carneiro, G. et al.) 269–277 (Springer, 2016).
Mavandadi, S. et al. Distributed medical image analysis and diagnosis through crowd-sourced games: a malaria case study. PLoS ONE 7, e37245 (2012).
Luengo-Oroz, M. A., Arranz, A. & Frean, J. Crowdsourcing malaria parasite quantification: an online game for analyzing images of infected thick blood smears. J. Med. Internet Res. 14, e2338 (2012).
Ning, Q. et al. Easy, reproducible and quality-controlled data collection with CROWDAQ. In Proc. 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations (eds Liu, Q. & Schlangen, D.) 127–134 (ACL, 2020).
Chaithanya Manam, V. K., Jampani, D., Zaim, M., Wu, M.-H. & J. Quinn, A. TaskMate: a mechanism to improve the quality of instructions in crowdsourcing. In Companion Proc. 2019 World Wide Web Conference (Liu, L. & White, R.) 1121–1130 (ACM, 2019).
Bragg, J., Mausam & Weld, D. S. Sprout: crowd-powered task design for crowdsourcing. In Proc. 31st Annual ACM Symposium on User Interface Software and Technology (eds Baudisch, P. et al.) 165–176 (ACM, 2018).
Manam, V. C. & Quinn, A. Wingit: efficient refinement of unclear task instructions. Proc. AAAI Conference on Human Computation and Crowdsourcing 6, 108–116 (2018).
Chang, J. C., Amershi, S. & Kamar, E. Revolt: collaborative crowdsourcing for labeling machine learning datasets. In Proc. 2017 CHI Conference on Human Factors in Computing Systems (eds Mark, G. et al.) 2334–2346 (ACM, 2017).
Gebru, T. et al. Datasheets for datasets. Commun. Assoc. Comput. Mach. 64, 86–92 (2021).
Maier-Hein, L. et al. BIAS: transparent reporting of biomedical image analysis challenges. Med. Image Anal. 66, 101796 (2020).
Maier-Hein, L. et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 9, 5217 (2018).
Call for challenges. The Medical Image Computing and Computer Assisted Intervention Society http://www.miccai.org/news/2021/10/25/call-for-challenges (2021).
Reinke, A. et al. How to exploit weaknesses in biomedical challenge design and organization. In International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Frangi, A. F. et al.) 388–395 (Springer, 2018).
Page, M. J. et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Int. J. Surg. 88, 105906 (2021).
Maier-Hein, L. et al. Heidelberg colorectal data set for surgical data science in the sensor operating room. Sci. Data 8, 101 (2021).
Roß, T. et al. Comparative validation of multi-instance instrument segmentation in endoscopy: results of the ROBUST-MIS 2019 challenge. Med. Image Anal. 70, 101920 (2021).
Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
MICCAI special interest group for biomedical image analysis challenges. The Medical Image Computing and Computer Assisted Intervention Society https://miccai.org/index.php/special-interest-groups/challenges/ (2022).
Shankar, V. et al. Evaluating machine accuracy on ImageNet. In Proc. 37th International Conference on Machine Learning (eds Daumé III, H. and Singh, A.) 8634–8644 (PMLR, 2020).
Lampert, T. A., Stumpf, A. & Gançarski, P. An empirical study into annotator agreement, ground truth estimation, and algorithm evaluation. IEEE Trans. Image Process. 25, 2557–2572 (2016).
Lendvay, T. S., White, L. & Kowalewski, T. Crowdsourcing to assess surgical skill. JAMA Surg. 150, 1086–1087 (2015).
Nowak, S. & Rüger, S. How reliable are annotations via crowdsourcing: a study about inter-annotator agreement for multi-label image annotation. In Proc. International Conference on Multimedia Information Retrieval (eds Wang, J. Z. et al.) 557–566 (ACM 2010).
Sambasivan, N. et al. “Everyone wants to do the model work, not the data work”: data cascades in high-stakes AI. In Proc. 2021 CHI Conference on Human Factors in Computing Systems (eds Kitamura, Y. et al.) 1–15 (ACM, 2021).
Karimi, D., Dou, H., Warfield, S. K. & Gholipour, A. Deep learning with noisy labels: exploring techniques and remedies in medical image analysis. Med. Image Anal. 65, 101759 (2020).
Maier-Hein, L. et al. Metrics reloaded: pitfalls and recommendations for image analysis validation. Preprint at https://arxiv.org/abs/2206.01653 (2022).
Reinke, A. et al. Common limitations of image processing metrics: a picture story. Preprint at https://arxiv.org/abs/2104.05642 (2021).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017).
Oppenheimer, D. M., Meyvis, T. & Davidenko, N. Instructional manipulation checks: detecting satisficing to increase statistical power. J. Exp. Soc. Psychol. 45, 867–872 (2009).
Zimmerer, D. et al. MOOD 2020: A public benchmark for out-of-distribution detection and localization on medical images. IEEE Trans. Med. Imaging 41, 2728–2738 (2022).
Roß, T. et al. How can we learn (more) from challenges? A statistical approach to driving future algorithm development. Preprint at https://arxiv.org/abs/2106.09302 (2021).
Chen, E. Z. & Li, H. A two-part mixed-effects model for analyzing longitudinal microbiome compositional data. Bioinformatics 32, 2611–2617 (2016).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020).
MICCAI registered challenges. The Medical Image Computing and Computer Assisted Intervention Society https://miccai.org/index.php/special-interest-groups/challenges/miccai-registered-challenges/ (2021).
Roß, T. & Reinke, A. Robust Medical Instrument Segmentation (ROBUST-MIS) Challenge 2019 - syn18779624 - Wiki. SYNAPSE https://www.synapse.org/#!Synapse:syn18779624/wiki/592660 (2019).
Rädsch, T. Labeling instructions matter code repository. GitHub https://github.com/IMSY-DKFZ/labeling_instructions_matter (2023).
Acknowledgements
understand.ai, Karlsruhe provided the annotations for this work and funded a part of this work, namely, M. Mengler and S. Funke. A part of this work was funded by Helmholtz Imaging, a platform of the Helmholtz Incubator on Information and Data Science, and the Surgical Oncology Program of the National Center for Tumor Diseases (NCT) Heidelberg. We thank M. Eisenmann for his continuous feedback, and M. Gelz, S. Strzysch and T. Klocke for their continuous support. Furthermore, we thank K. D. Pandl, A. Sunyaev and the Karlsruhe Institute of Technology (KIT), where a part of this research was conducted. We thank the participants and organizers of the Robust Medical Instrument Segmentation Challenge 2019 for enabling the experiments presented in Supplementary Note 4.
Funding
Open access funding provided by Deutsches Krebsforschungszentrum (DKFZ).
Author information
Authors and Affiliations
Contributions
Ti.R., A.R., N.S., M.D.T., A.K.-S. and L.M.-H. designed the study. Ti.R. prepared the annotation experiments with the help of A.R and M.D.T. Ti.R. conducted the annotation experiments and implemented the code. Ti.R. and A.E.K. prepared and conducted the competition analysis with the help of A.R. B.P. and To.R. created the meta annotations for the HeiCo dataset. Ti.R., V.W., N.S., A.R., A.K.-S. and L.M.-H. analysed the results. Ti.R. designed and created the figures with the help of A.R. and L.M.-H. Ti.R., A.R., M.D.T. and L.M.-H. wrote the paper with substantial contributions from V.W. and A.K.-S., and feedback from all co-authors. Ti.R. initiated and managed the contact with the involved companies and annotators. Ti.R. acquired the funding for the annotations. L.M.-H. managed and coordinated the overall project with substantial input from A.K.-S.
Corresponding authors
Ethics declarations
Competing interests
Ti.R. was an employee of the company understand.ai, which sponsored the creation of the annotations. After his research, To.R. was employed by Quality Match GmbH. The other authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Bernhard Kainz and Wei Shao for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rädsch, T., Reinke, A., Weru, V. et al. Labelling instructions matter in biomedical image analysis. Nat Mach Intell 5, 273–283 (2023). https://doi.org/10.1038/s42256-023-00625-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-023-00625-5
This article is cited by
-
Real-time near infrared artificial intelligence using scalable non-expert crowdsourcing in colorectal surgery
npj Digital Medicine (2024)
-
Accelerated evidence synthesis in orthopaedics—the roles of natural language processing, expert annotation and large language models
Journal of Experimental Orthopaedics (2023)
-
Deep learning-enabled segmentation of ambiguous bioimages with deepflash2
Nature Communications (2023)
-
Improving image labelling quality
Nature Machine Intelligence (2023)
-
Online citizen science with the Zooniverse for analysis of biological volumetric data
Histochemistry and Cell Biology (2023)