Abstract
Ammonia production via glutamate dehydrogenase is inhibited by SIRT4, a sirtuin that displays both amidase and non-amidase activities. The processes underlying the regulation of ammonia removal by amino acids remain unclear. Here, we report that SIRT4 acts as a decarbamylase that responds to amino acid sufficiency and regulates ammonia removal. Amino acids promote lysine 307 carbamylation (OTCCP-K307) of ornithine transcarbamylase (OTC), which activates OTC and the urea cycle. Proteomic and interactome screening identified OTC as a substrate of SIRT4. SIRT4 decarbamylates OTCCP-K307 and inactivates OTC in an NAD+-dependent manner. SIRT4 expression was transcriptionally upregulated by the amino acid insufficiency-activated GCN2–eIF2α–ATF4 axis. SIRT4 knockout in cultured cells caused higher OTCCP-K307 levels, activated OTC, elevated urea cycle intermediates and urea production via amino acid catabolism. Sirt4 ablation decreased male mouse blood ammonia levels and ameliorated CCl4-induced hepatic encephalopathy phenotypes. We reveal that SIRT4 safeguards cellular ammonia toxicity during amino acid catabolism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data that support the findings of this study are available within source data and supplementary information files. Proteomics raw data are deposited in the PRIDE database (accession no. PXD037101). Source data are provided with this paper.
Code availability
No custom codes were used during this study.
References
Wijdicks, E. F. M. Hepatic encephalopathy. N. Engl. J. Med. 375, 1660–1670 (2016).
Keshet, R., Szlosarek, P., Carracedo, A. & Erez, A. Rewiring urea cycle metabolism in cancer to support anabolism. Nat. Rev. Cancer 18, 634–645 (2018).
Ogura, M. et al. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem. Biophys. Res. Commun. 393, 73–78 (2010).
Nakagawa, T. & Guarente, L. Urea cycle regulation by mitochondrial sirtuin, SIRT5. Aging 1, 578–581 (2009).
Yu, W. et al. Lysine 88 acetylation negatively regulates ornithine carbamoyltransferase activity in response to nutrient signals. J. Biol. Chem. 284, 13669–13675 (2009).
Zhao, S. et al. Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 (2010).
Ramponi, G. & Grisolia, S. Acylation of lysine and of arginine-rich histones with carbamyl phosphate and 1,3 diphosphoglycerate. Biochem Biophys. Res Commun. 38, 1056–1063 (1970).
Kollipara, L. & Zahedi, R. P. Protein carbamylation: In vivo modification or in vitro artefact. Proteomics 13, 941–944 (2013).
Stec, B. Structural mechanism of RuBisCO activation by carbamylation of the active site lysine. Proc. Natl Acad. Sci. USA 109, 18785–18790 (2012).
Wang, Z. et al. Protein carbamylation links inflammation, smoking, uremia and atherogenesis. Nat. Med. 13, 1176–1184 (2007).
Gorissea, L. et al. Protein carbamylation is a hallmark of aging. Proc. Natl Acad. Sci. USA 113, 1191–1196 (2016).
Anderson, K. A., Green, M. F., Huynh, F. K., Wagner, G. R. & Hirschey, M. D. SnapShot: mammalian sirtuins. Cell 159, 956 (2014).
Haigis, M. C. & Sinclair, D. A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. Mech. 5, 253–295 (2010).
Carrico, C., Meyer, J. G., He, W., Gibson, B. W. & Verdin, E. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metab. 27, 497–512 (2018).
van de Ven, R. A. H., Santos, D. & Haigis, M. C. Mitochondrial sirtuins and molecular mechanisms of aging. Trends Mol. Med. 23, 320–331 (2017).
He, X.-D. et al. Sensing and transmitting intracellular amino acid signals through reversible lysine aminoacylations. Cell Metab. 27, 151–166 (2018).
Hirschey, M. D. et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 (2010).
Du, J. T. et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011).
Peng, C. et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell Proteomics https://doi.org/10.1074/mcp.M111.012658 (2011).
Tan, M. et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 (2014).
Feldman, J. L., Baeza, J. & Denu, J. M. Activation of the Protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31350–31356 (2013).
Verdin, E. et al. Measurement of mammalian histone deacetylase activity. Methods Enzymol. 377, 180–196 (2004).
Haigis, M. C. et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 126, 941–954 (2006).
Mathias, R. A. et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 159, 1615–1625 (2014).
Anderson, K. A. et al. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. 25, 838–855 (2017).
Pannek, M. et al. Crystal structures of the mitochondrial deacylase sirtuin 4 reveal isoform-specific acyl recognition and regulation features. Nat. Commun. 8, 1513 (2017).
Laurent, G. et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol. Cell 50, 686–698 (2013).
Laurent, G. et al. SIRT4 represses peroxisome proliferator-activated receptor-α activity to suppress hepatic fat oxidation. Mol. Cell. Biol. 33, 4552–4561 (2013).
Ho, L. et al. SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK. Aging 5, 835–849 (2013).
Nasrin, N. et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J. Biol. Chem. 285, 31995–32002 (2010).
Csibi, A. et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153, 840–854 (2013).
Wood, J. G. et al. Sirt4 is a mitochondrial regulator of metabolism and lifespan in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 115, 1564–1569 (2018).
Jeong, S. M. et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23, 450–463 (2013).
Wang, Y. S. et al. Sirtuin 4 depletion promotes hepatocellular carcinoma tumorigenesis through regulating adenosine‐monophosphate-activated protein kinase-α/mammalian target of rapamycin axis in mice. Hepatology 69, 1614–1631 (2019).
Guan, K. L., Yu, W., Lin, Y., Xiong, Y. & Zhao, S. Generation of acetyllysine antibodies and affinity enrichment of acetylated peptides. Nat. Protoc. 5, 1583–1595 (2010).
Roux, K. J., Kim, D. I., Burke, B. & May, D. G. BioID: a screen for protein–protein interactions. Curr. Protoc. Protein Sci. 91, 19.23.11–19.23.15 (2018).
Xie, C. Y., Yu, J. C., Huang, S., Gao, W. Q. & Tang, K. Q. A novel approach of matching mass-to-charge ratio for compound identification in gas chromatography-mass spectrometry. J. AOAC Int. 102, 638–645 (2019).
Hinnebusch, A. G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 (2005).
Sheikh, M. S. & Fornace, A. J. Regulation of translation initiation following stress. Oncogene 18, 6121–6128 (1999).
Pakos-Zebrucka, K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016).
Yamamoto, H. A. & Sugihara, N. Blood ammonia levels and hepatic-encephalopathy induced by Ccl4 in rats. Toxicol. Appl Pharm. 91, 461–468 (1987).
Hadjihambi, A. et al. Impaired brain glymphatic flow in experimental hepatic encephalopathy. J. Hepatol. 70, 40–49 (2019).
Kale, R. A. et al. Demonstration of interstitial cerebral edema with diffusion tensor MR imaging in type C hepatic encephalopathy. Hepatology 43, 698–706 (2006).
Kim, J. et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature 546, 168–172 (2017).
Herrero-Yraola, A. et al. Regulation of glutamate dehydrogenase by reversible ADP-ribosylation in mitochondria. EMBO J. 20, 2404–2412 (2001).
Komlos, D. et al. Glutamate dehydrogenase 1 and SIRT4 regulate glial development. Glia 61, 394–408 (2013).
Gorg, B., Karababa, A. & Haussinger, D. Hepatic encephalopathy and astrocyte senescence. J. Clin. Exp. Hepatol. 8, 294–300 (2018).
Gorg, B., Karababa, A., Shafigullina, A., Bidmon, H. J. & Haussinger, D. Ammonia-Induced senescence in cultured rat astrocytes and in human cerebral cortex in hepatic encephalopathy. Glia 63, 37–50 (2015).
Matsushita, M., Yamamoto, T. & Gemba, H. The role of astrocytes in the development of hepatic encephalopathy. Neurosci. Res. 34, 271–280 (1999).
Norenberg, M. D. The role of astrocytes in hepatic-encephalopathy. Neurochem. Pathol. 6, 13–33 (1987).
Wands, J. Hepatocellular carcinoma and sex. N. Engl. J. Med. 357, 1974–1976 (2007).
Wisniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, U359–U360 (2009).
Zhang, X. et al. Molecular basis for hierarchical histone de-β-hydroxybutyrylation by SIRT3. Cell Discov. https://doi.org/10.1038/s41421-019-0103-0 (2019).
Acknowledgements
This work was supported by grants from the State Key Development Programs of China (nos. 2018YFA0801300 (S.-M.Z.), 2018YFA0800300 (W.X.), 2020YFA0803601 (J.-Y.Z.) and 2019YFA0801900 (J.-Y.Z.)), the National Science Foundation of China (nos. 92253305 (S.-M.Z.), 31821002 (S.-M.Z.), 32230054 (S.-M.Z.), 31930062 (S.-M.Z.), 91857000 (S.-M.Z.), 92157001 (S.-M.Z.), 32171298 (W.X.), 81971449 (Y.-Y.Y.) and 82171672 (Y.-Y.Y.)) and the Program of Shanghai Academic Research Leader (21XD1423000) (W.X.).
Author information
Authors and Affiliations
Contributions
S.-M.Z., W.X. and J.-Y.Z. conceived the concept and designed and supervised the experiments. S.-H.H., Y.-Y.F., Y.-X.Y., H.-D.M., S.-X.Z., K.-H.Z. and Y.-N.Q. performed the biological experiments. L.Z. performed LC–MS/MS-based target metabolites measurement and metabolomics experiments. L.H. performed proteomics experiments. Y.-Y.Y., Y. Lin, X.-Y.Z., Y. Li and H.-T.L. participated in the discussions. S.-M.Z., W.X. and J.-Y.Z. wrote the manuscript. All authors read and discussed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks René Zahedi and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editor: Alfredo Giménez-Cassina, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
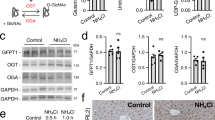
Extended Data Fig. 1 Characterizing specificity of pan CP-K antibody and Sirt4-/- mice.
a-c. CP-K antibody specifically recognizes CP-K. The reactivity of the CP-K antibody used in the current study toward synthetic acetylated and carbamylated peptides (a), HepG2 cell lysate heated or not heated with 8 M urea (b) and acetylated proteins in HepG2 cell lysate (b, c). d. SIRT4 deletion had mild impact on amino acids levels. The amino acids levels were compared between wild-type (set as 100%) and Sirt4-/- mice hepatocytes (n = 3 biologically independent samples, P = 0.0093, 0.0499, 0.0152, 0.0280, 0.0047, 0.0068,0.0017).
Extended Data Fig. 2 SIRT4 decreases CP-K levels.
a. Schematic diagram of CP-K peptides screening. CP-K antibody was employed to affinity-enrich CP-K modified tryptic peptides from mice livers, followed by MS detection. b-d. SIRT4 decreases mitochondrial enzymes CP-K levels in vitro and in cells. The ability of purified SIRT4 to decrease CP-K levels of its potential substrates, namely purified GDH, GOT2 and PDHA from HEK293T cells (b) and Sirt4-/- mice liver mitochondrial lysate proteins (c) in vitro and the ability of SIRT3, SIRT4 and SIRT5 overexpression to decrease CP-K levels of recombinant GDH, GOT2 and PDHA from cells (d) were tested.
Extended Data Fig. 3 SIRT4 regulates urea cycle.
a. Heatmap of metabolite changes in the livers of wild-type (n = 5) and Sirt4-/- (n = 5) mice. Significance threshold was set as VIP (Variable Importance in the Projection) > 1, p < 0.05 (Supplementary Table 4). b-g. Loss of SIRT4 elevates urea cycle metabolites. Levels of CP (b), citrulline (c), arginino-succinate (d), arginine (e), urea (f) and ornithine (g) in wild-type and Sirt4-/- mouse kidneys were measured using target LC–MS. (n = 5 biologically independent samples, b: P = 0.0004; c: P = 0.0075; d: P = 0.0044; e: P = 0.0313; f: P = 0.0268; g: P = 0.0018). h-k. Carbohydrates metabolism has limited impact on SIRT4 deletion-induced urea cycle activation. 13C-Aspartate production from 13C-glucose (h) were compared between primary hepatocytes of wild-type and Sirt4-/- mice (i); glucose limitation impact on 13C-Aspartate production (j), aspartate (k) were determined. The chasing time for 13C-glucose was 6 h (n = 3 biologically independent samples, j: P = 0.00014; k: P = 0.0025).
Extended Data Fig. 4 OTCK307 was carbamylated.a. MS/MS spectrum generated tryptic peptide libraries of mouse liver.
b. Formation of CP-K307 of synthetic OTC K307 peptide, OTC K307R peptide after reacting with 5 mM CP was detected using MS. c-d. Validation of custom-made OTCCP-K307 antibody (abclonal, China). Employing dot-blot assays, the OTCCP-K307 antibody was tested for its reactivity to acetylated and carbamylated OTC K307 peptide (c) and to OTC K307 and OTC K307R peptides after reacting with 5 mM CP (d). e. CP increases intracellular OTCCP-K307 levels. OTCCP-K307 levels of ectopically expressed OTC in HEK293T after treatment with 5 mM CP for 4 h.
Extended Data Fig. 5 SIRT4 decarbamylates OTCCP-K307.
a. SIRT4 has decarbamylase activity. The formation of decarbamylated OTC peptide from synthetic OTC CP-K307 peptide were detected by MS after the synthetic peptide was incubated each 5 μM recombinant SIRT3, SIRT4 and SIRT5. b. HPLC analysis for the decarbamylation of OTC CP-K307 peptide by SIRT4. Time-dependent production of decarbamylated OTC K307 peptide was monitored (upper) and confirmed by MS/MS analysis (bottom). c-d. OTC interacts with SIRT4. Co-immunoprecipitation analysis was employed to analyze ectopically expressed OTC and SIRT4 in HEK293T (c) and endogenous OTC and SIRT4 in mice hepatocytes (d).
Extended Data Fig. 6 GCN2–eIF2α-ATF4 axis regulates SIRT4 and the urea cycle.
a. Diagram of how 15N-labeled ammonia incorporates into the urea cycle intermediates. b. Sirt4 knockout increases ammonia incorporation into urea cycle metabolites. The incorporation of 15 N into CP, citrulline, arginino-succinate, arginine and urea were determined in wild-type and Sirt4-/- mice hepatocytes that were chased with 50 μM 15NH4Cl for 6 h (n = 4 biologically independent samples, P = 0.0257, 0.0124, 0.0154, 0.0015, 0.0178). c-d. mRNA levels of Sirt4 were determined in Hepa1-6, Eif2α knockdown Hepa1-6 (c) and Atf4 knockdown Hepa1-6 cells (d) cultured in medium with or without glutamine or amino acids for 12 h (n = 3 biologically independent samples, c: P = 0.0003, 0.0001, > 0.05, > 0.05, > 0.05, > 0.05; d: P = 0.0002, 0.0019, > 0.05, > 0.05, > 0.05, > 0.05). e-g. Food intake (e), weight (f), and water intake (g) were determined by metabolic cave for wild-type and Sirt4-/- C57 mice (n = 5 biologically independent samples, e: P> 0.05; f: P> 0.05; g: P = 0.0090).
Extended Data Fig. 7 Deletion of Sirt4 ameliorates hepatic encephalopathy phenotypes.
a. Schematic depicting the experimental setup of HE mice. Wild-type and Sirt4-/- C57 mice were intraperitoneally injected with 30% CCl4 (5 mL/kg body weight, twice a week) for 10 weeks to build the mouse model of HE before assessment. b-c. Food intake (b) and weight (c) were determined for wild-type and Sirt4-/- C57 mice treated with or without CCl4; number of mice = 10. Data are presented as mean ± SEM. d-e. Sirt4 ablation has no impact on hepatic damage induced by CCl4. Total blood bilirubin (d) and blood AST activity (e) were determined in wild-type and Sirt4-/- C57 mice treated or not treated with CCl4 (n = 10 biologically independent samples, d: P = < 0.0001, > 0.05, > 0.05; e: P > 0.05; g: P = < 0.0001, > 0.05, > 0.05).
Extended Data Fig. 8 Deletion of Sirt4 prevented brain edema induced by CCl4.
Brain images were obtained from wild-type and Sirt4-/- C57 mice after CCl4 treatments for 10 weeks; number of mice were 10 for each experimental group. Brain edema indicated by asymmetry hyper-intensity was pointed out by red arrows.
Extended Data Fig. 9 Deletion of Sirt4 prevented mouse exploratory and locomotor activity loss induced by CCl4.
a-c. Mobile time (a), immobile time (b), and mean speed (c) were determined via the open field test in wild-type and Sirt4-/- C57 mice treated with our without CCl4 (n = 10 biologically independent samples, a: P = > 0.05, 0.0025; b: P = > 0.05, 0.0071; c:P = > 0.05, 0.0269). d-g. Total distance (d), novel arm exploration time (e), old arm distance (f), and old arm exploration time (g) were determined via the Y-maze test in wild-type and Sirt4-/- C57 mice treated with or without CCl4 (n = 10 biologically independent samples, d: P = > 0.05, 0.0060; e: P = > 0.05, 0.0145; f:P = > 0.05, > 0.05; g: P = > 0.05, 0.0142).
Extended Data Fig. 10 Schematic diagram of amino acids-regulated SIRT4 coordinates ammonia detoxification and urea cycle.
GCN2 senses amino acids to downregulate SIRT4 expression, which suppresses ammonia detoxification by inhibiting CP incorporation into the urea cycle by inhibiting OTCCP-K307 that activates OTC.
Supplementary information
Supplementary Tables 1–5
Supplementary Table 1: Identification of substrate of K-CP in mice liver. Supplementary Table 2: SIRT4 interactome in mice hepatoma cell by BioID. Supplementary Table 3: Co-identified proteins in both K-CP substrates and SIRT4 interactome. Supplementary Table 4: Untargeted metabolomic analysis of liver tissues of WT and Sirt4−/− mice. Supplementary Table 5: Oligonucleotides.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, SH., Feng, YY., Yang, YX. et al. Amino acids downregulate SIRT4 to detoxify ammonia through the urea cycle. Nat Metab 5, 626–641 (2023). https://doi.org/10.1038/s42255-023-00784-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-023-00784-0