Abstract
The brainstem dorsal vagal complex (DVC) is known to regulate energy balance and is the target of appetite-suppressing hormones, such as glucagon-like peptide 1 (GLP-1). Here we provide a comprehensive genetic map of the DVC and identify neuronal populations that control feeding. Combining bulk and single-nucleus gene expression and chromatin profiling of DVC cells, we reveal 25 neuronal populations with unique transcriptional and chromatin accessibility landscapes and peptide receptor expression profiles. GLP-1 receptor (GLP-1R) agonist administration induces gene expression alterations specific to two distinct sets of Glp1r neurons—one population in the area postrema and one in the nucleus of the solitary tract that also expresses calcitonin receptor (Calcr). Transcripts and regions of accessible chromatin near obesity-associated genetic variants are enriched in the area postrema and the nucleus of the solitary tract neurons that express Glp1r and/or Calcr, and activating several of these neuronal populations decreases feeding in rodents. Thus, DVC neuronal populations associated with obesity predisposition suppress feeding and may represent therapeutic targets for obesity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All genetic data generated in this study (bulk RNA-seq, snRNA-seq and snATAC–seq) are available in the GEO under SuperSeries accession number GSE166649. All other data are available from the authors upon reasonable request. Source data are provided with this paper.
Code availability
The source code used to analyse the data and produce the statistical figures is available at https://github.com/perslab/Ludwig-2021/.
References
Bray, G. A. Medical treatment of obesity: the past, the present and the future. Best Pract. Res. Clin. Gastroenterol. 28, 665–684 (2014).
Bray, G. A., Frühbeck, G., Ryan, D. H. & Wilding, J. P. H. Management of obesity. Lancet 387, 1947–1956 (2016).
Schwartz, M. W. et al. Obesity pathogenesis: an endocrine society scientific statement. Endocr. Rev. 38, 267–296 (2017).
O’Rahilly, S. & Farooqi, I. S. Genetics of obesity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1095–1105 (2006).
Srivastava, G. & Caroline, M. Current pharmacotherapy for obesity. Nat. Rev. Endocrinol. 14, 12–24 (2018).
Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Timshel, P. N., Thompson, J. J. & Pers, T. H. Genetic mapping of etiologic brain cell types for obesity. eLife 9, e55851 (2020).
Grill, H. J. & Hayes, M. R. Hindbrain neurons as an essential hub in the neuroanatomically distributed control of energy balance. Cell Metab. 16, 296–309 (2012).
Jensen, C. B. et al. Characterization of the glucagonlike peptide-1 receptor in male mouse brain using a novel antibody and in situ hybridization. Endocrinology 159, 665–675 (2018).
Cork, S. C. et al. Distribution and characterisation of glucagon-like peptide 1 receptor expressing cells in the mouse brain. Mol. Metab. 4, 718–731 (2015).
Mullican, S. E. et al. GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and non-human primates. Nat. Med. 23, 1150–1157 (2017).
Christopoulos, G. et al. Multiple amylin receptors arise from receptor activity-modifying protein interaction with the calcitonin receptor gene product. Mol. Pharmacol. 56, 235–242 (1999).
Adriaenssens, A. E. et al. Glucose-dependent insulinotropic polypeptide receptor-expressing cells in the hypothalamus regulate food intake. Cell Metab. 30, 987–996 (2019).
Braegger, F. E., Asarian, L., Dahl, K., Lutz, T. A. & Boyle, C. N. The role of the area postrema in the anorectic effects of amylin and salmon calcitonin: behavioral and neuronal phenotyping. Eur. J. Neurosci. 40, 3055–3066 (2014).
Richard, J. E. et al. Activation of the GLP-1 receptors in the nucleus of the solitary tract reduces food reward behavior and targets the mesolimbic system. PLoS ONE 10, e0119034 (2015).
Hayes, M. R. et al. Intracellular signals mediating the food intake suppressive effects of hindbrain glucagon-like-peptide-1 receptor activation. Cell Metab. 13, 320–330 (2011).
Cheng, W. et al. Calcitonin receptor neurons in the mouse nucleus tractus solitarius control energy balance via the non-aversive suppression of feeding. Cell Metab. 31, 301–312 (2020).
Coester, B., Foll, C. L. & Lutz, T. A. Viral depletion of calcitonin receptors in the area postrema: a proof-of-concept study. Physiol. Behav. 223, 112992 (2020).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Bentsen, M. A. et al. Transcriptomic analysis links diverse hypothalamic cell types to fibroblast growth factor 1-induced sustained diabetes remission. Nat. Commun. 11, 1–16 (2020).
Inoue, F. et al. Genomic and epigenomic mapping of leptin-responsive neuronal populations involved in body weight regulation. Nat. Metab. 1, 475–484 (2019).
Campbell, J. N. et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 20, 484–496 (2017).
O’Neil, P. M. et al. Efficacy and safety of semaglutide compared with liraglutide and placebo for weight loss in patients with obesity: a randomised, double-blind, placebo and active controlled, dose-ranging, phase 2 trial. Lancet 392, 637–649 (2018).
Adams, J. M. et al. Liraglutide modulates appetite and body weight through glucagon-like peptide 1 receptor-expressing glutamatergic neurons. Diabetes 67, 1538–1548 (2018).
Gabery, S. et al. Semaglutide lowers body weight in rodents via distributed neural pathways. JCI Insight 5, e133429 (2020).
Langlet, F., Mullier, A., Bouret, S. G., Prevot, V. & Dehouck, B. Tanycyte-like cells form a blood–cerebrospinal fluid barrier in the circumventricular organs of the mouse brain. J. Comp. Neurol. 521, 3389–3405 (2013).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Katsurada, K. et al. Central glucagon-like peptide 1 receptor signaling via brainstem catecholamine neurons counteracts hypertension in spontaneously hypertensive rats. Sci. Rep. 9, 1–13 (2019).
Holt, M. K. et al. Preproglucagon neurons in the nucleus of the solitary tract are the main source of brain GLP-1, mediate stress-induced hypophagia, and limit unusually large intakes of food. Diabetes 68, 21–33 (2019).
Cheng, W. et al. Leptin receptor–expressing nucleus tractus solitarius neurons suppress food intake independently of GLP1 in mice. JCI Insight 5, e134359 (2020).
Allison, M. B. et al. Defining the transcriptional targets of leptin reveals a role for Atf3 in leptin action. Diabetes 67, 1093–1104 (2018).
Yang, C. et al. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine beta-hydroxylase gene. J. Neurochem. 71, 1813–1826 (1998).
Bahrami, S. & Drabløs, F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 62, 37–49 (2016).
Frikke-Schmidt, H. et al. GDF15 acts synergistically with liraglutide but is not necessary for the weight loss induced by bariatric surgery in mice. Mol. Metab. 21, 13–21 (2019).
Barth, S. B., Riediger, T., Lutz, T. A. & Rechkemmer, G. Peripheral amylin activates circumventricular organs expressing calcitonin receptor a/b subtypes and receptor-activity modifying proteins in the rat. Brain Res. 997, 97–102 (2004).
Gross, P. M. Circumventricular organ capillaries. Prog. Brain Res. 91, 219–233 (1992).
Zhang, C. et al. Area postrema cell types that mediate nausea-associated behaviors. Neuron 109, 1–12 (2021).
Gaykema, R. P. et al. Activation of murine pre-proglucagon–producing neurons reduces food intake and body weight. J. Clin. Invest. 127, 1031–1045 (2017).
Berglund, E. D. et al. Melanocortin 4 receptors in autonomic neurons regulate thermogenesis and glycemia. Nat. Neurosci. 17, 911–913 (2014).
Resch, J. M. et al. Aldosterone-sensing neurons in the NTS exhibit state-dependent pacemaker activity and drive sodium appetite via synergy with angiotensin II signaling. Neuron 96, 190–206 (2017).
Do, J., Chang, Z., Sekerková, G., McGrimmon, D. R. & Martina, M. A leptin-mediated neural mechanism linking breathing to metabolism. Cell Rep. 33, 108358 (2020).
Brierley, D. I. et al. Central and peripheral GLP-1 systems independently suppress eating. Nat. Metab. 3, 258–273 (2021).
Ranadive, S. A. & Vaisse, C. Lessons from extreme human obesity: monogenic disorders. Endocrinol. Metab. Clin. North Am. 37, 733–751 (2008).
Philippe, J. et al. A nonsense loss-of-function mutation in PCSK1 contributes to dominantly inherited human obesity. Int. J. Obes. 39, 295–302 (2015).
Bariohay, B., Lebrun, B., Moyse, E. & Jean, A. Brain-derived neurotrophic factor plays a role as an anorexigenic factor in the dorsal vagal complex. Endocrinology 146, 5612–5620 (2005).
Williams, D. L., Kaplan, J. M. & Grill, H. J. The role of the dorsal vagal complex and the vagus nerve in feeding effects of melanocortin-3/4 receptor stimulation. Endocrinology 141, 1332–1337 (2000).
Fortin, S. M., Chen, J. & Hayes, M. R. Hindbrain melanocortin 3/4 receptors modulate the food intake and body weight suppressive effects of the GLP-1 receptor agonist, liraglutide. Physiol. Behav. 220, 112870 (2020).
Alhadeff, A. L. et al. Endogenous glucagon-like peptide 1 receptor signaling in the nucleus tractus solitarius is required for food intake control. Neuropsychopharmacology 42, 1471–1479 (2017).
Hayes, M. R., Bradley, L. & Grill, H. J. Endogenous hindbrain glucagon-like peptide 1 receptor activation contributes to the control of food intake by mediating gastric satiation signaling. Endocrinology 150, 2654–2659 (2009).
Borner, T. et al. GDF15 induces anorexia through nausea and emesis. Cell Metab. 31, 351–362 (2020).
Ahrén, B. et al. Semaglutide induces weight loss in subjects with type 2 diabetes regardless of baseline BMI or gastrointestinal adverse events in the SUSTAIN 1 to 5 trials. Diabetes Obes. Metab. 20, 2210–2219 (2018).
Georgescu, T. et al. Neurochemical characterization of brainstem pro-opiomelanocortin cells. Endocrinology 161, 1–13 (2020).
Secher, A. et al. The arcuate nucleus mediates GLP-1 receptor agonist liraglutide-dependent weight loss. J. Clin. Invest. 124, 4473–4488 (2014).
Baraboi, E. D., Smith, P., Ferguson, A. V. & Richard, D. Lesions of area postrema and subfornical organ Alter exendin-4-induced brain activation without preventing the hypophagic effect of the GLP-1 receptor agonist. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R1098–R1110 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wang, R., Nambiar, R., Zheng, D. & Tian, B. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res. 46, D315–D319 (2018).
Haeussler, M. et al. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 47, D853–D858 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA-sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 (2019).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Chen, R., Wu, X., Jiang, L. & Zhang, Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 18, 3227–3241 (2017).
Romanov, R. A. et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 20, 176–188 (2017).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists. Nucleic Acids Res. 47, W191–W198 (2019).
Fang, R. et al. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 12, 1337 (2021).
Zhang, Y. et al. Model-based analysis of ChIP–Seq (MACS). Genome Biol. 9, 1–9 (2008).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Loh, P. R., Kichaev, G., Gazal, S., Schoech, A. P. & Price, A. L. Mixed-model association for biobank-scale datasets. Nat. Genet. 50, 906–908 (2018).
Rai, V. et al. Single-cell ATAC–seq in human pancreatic islets and deep learning upscaling of rare cells reveals cell-specific type 2 diabetes regulatory signatures. Mol. Metab. 32, 109–121 (2020).
Pyke, C. Automated ISH for validated histological mapping of lowly expressed genes. Methods Mol. Biol. 2148, 219–228 (2020).
Pan, W. et al. Essential role for hypothalamic calcitonin receptor-expressing neurons in the control of food intake by leptin. Endocrinology 159, 1860–1872 (2018).
Krashes, M. J. et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011).
Acknowledgements
Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research centre, based at the University of Copenhagen, and partially funded by an unconditional donation from the Novo Nordisk Foundation (https://www.cbmr.ku.dk/; grant no. NNF18CC0034900). We acknowledge the Novo Nordisk Foundation (grant no. NNF16OC0021496 to T.H.P.) and the Lundbeck Foundation (grant no. R190-2014-3904 to T.H.P.). We acknowledge J. J. Thompson from the Single-Cell Omics Platform at the Novo Nordisk Foundation Center for Basic Metabolic Research for help with uploading the data to the GEO database. Furthermore, this work was supported by a research grant from the Danish Diabetes Academy, which is funded by the Novo Nordisk Foundation (grant no. NNF17SA0031406). Studies at the University of Michigan were funded by the NIH (P01DK117821 to M.G.M.), BioPharmaceuticals R&D, AstraZeneca (to M.G.M.), the American Diabetes Association (1-16-PDF-021 to W.C.) and supported by the Michigan Diabetes Research Center Molecular Genetics Core (P30DK020572).
Author information
Authors and Affiliations
Contributions
M.Q.L., W.C., D.G., J.L., S.J.P., S.N.H., K.L.E., P.B., C.J.R., A.S., L.B.K., C.P., M.G.M. and T.H.P. designed the experiments. W.C., S.J.P., S.N.H. and P.B. performed the mouse experiments. D.G. performed the rat experiments. J.L., S.J.P. and K.L.E. performed the single-nucleus experiments. M.Q.L. analysed the data. M.Q.L., M.G.M. and T.H.P. wrote the initial draft of the manuscript, and all authors edited, read and approved the final version. T.H.P. is the guarantor of the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.B. is employed by Gubra. S.J.P., S.N.H., A.S., L.B.K. and C.P. are employed by Novo Nordisk A/S. C.J.R. is employed by AstraZeneca and holds stock in the company. All other authors declare no competing interests.
Additional information
Peer review information Nature Metabolism thanks Zachary Knight, Karolina P. Skibicka and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Christoph Schmitt.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Body weight and food intake for the single-nucleus RNA- and ATAC-seq in vivo study.
a,b, Daily body weight (a) and food intake (b) in semaglutide-administered (n=9), ad libitum-fed vehicle-administered (n=9) and weight-matched control mice (n=8). Values are mean ± s.e.m. *P<0.05, **P<0.01, ***P<0.001 versus vehicle and #P<0.05, ##P<0.01, ###P<0.001 semaglutide versus weight-matched, linear mixed effects model, Bonferroni-adjusted least-squares means two-tailed t-test.
Extended Data Fig. 2 Expression of tanycyte-like cell marker genes.
a, Expression of marker genes for tanycyte-like cells. b-e, ISH of sagittal brain sections (Allen Mouse Brain Atlas) of Wt1 (n=1 mouse) (b), Wif1 (n=2 mice) (c), Slc22a3 (n=1 mouse) (d) and Cdon (n=1 mouse) (e). Scale bar for panel b is representative for panels c-e, 100 μm. Data for reconstruction of a are available in Supplementary Data 2.
Extended Data Fig. 3 Expression of DVC neuronal marker genes.
a, Expression of marker genes for different neuronal populations. From top to bottom, dendrogram illustrating the similarity of the neuronal populations computed based on their gene expression levels, heatmap depicting the gene expression specificity values (ESμ) of the neuronal marker genes, the most likely DVC origin of the neuronal populations. b-z, ISH of coronal brain sections (Allen Mouse Brain Atlas). N=1 mouse for all hybridizations except panels m and o (n=2 mice) and panel p (n=3 mice). Scale bar for panel b is representative for panels c-z, 100 μm. Data for reconstruction of a are available in Supplementary Data 2.
Extended Data Fig. 4 Expression of appetite-supressing receptors in mice and non-human primates.
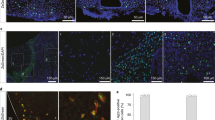
a, Representative image showing db-ISH of GFRALASR (blue) and GLP1R (red) in non-human primates (n=2). Scale bar, 250 μm. b, Representative image showing db-ISH of CALCR (blue) and GLP1R (red) in non-human primates (n=2). Scale bar, 100 μm. c, Representative image showing db-ISH of CASR (blue) and GLP1R (red) in non-human primates (n=2). Scale bar, 250 μm. d, Representative image showing db-ISH of Grpr (blue) and Calcr (red) in mice (n=4). Scale bar, 100 μm. e, Representative image showing db-ISH of Casr (blue) and Bdnf (red) in mice (n=4). Scale bar, 100 μm. f, Representative image showing db-ISH of Grpr (blue) and Bdnf (red) in mice (n=4). Scale bar, 100 μm. g, Representative image showing db-ISH of Casr (blue) and Mc4r (red) in mice (n=4). Scale bar, 100 μm. h, Representative image showing db-ISH of Grpr (blue) and Mc4r (red) in mice (n=4). Scale bar, 100 μm. i-l, Representative images showing TH (purple) or DDC immunoreactivity (purple) and GFP immunoreactivity (green) in Glp1r-Cre;GFP or Calcr-Cre;GFP mice (n=3). Scale bar, 150 μm.
Extended Data Fig. 5 Expression of appetite-supressing receptors and peptides in mice and non-human primates.
a, Representative image showing db-ISH of CALCR (blue) and RAMP3 (red) in non-human primates (n=2). Scale bar, 100 μm. b, Representative image showing db-ISH of Ccbe1 (blue) and Gipr (red) in mice (n=4). Scale bar, 100 μm. c, Representative image showing db-ISH of Pax5 (blue) and Gipr (red) in mice (n=4). Scale bar, 100 μm. d, Representative image showing db-ISH of Gipr (blue) and Glp1r (red) in mice (n=4). Scale bar, 100 μm. e, Representative image showing db-ISH of Asb4 (blue) and Gcg (red) in mice (n=4). Scale bar, 100 μm.
Extended Data Fig. 6 Additional DVC neuronal populations with previously-defined functions.
Expression of genes defining DVC neurons previously implicated in metabolic control. Top, expression specificity (ESμ) of selected genes. Bottom, the most likely DVC origin of the neuronal populations. Mc4r cholinergic (Chat3) DMV neurons regulate circulating insulin39, Hsd11b2 and Nr3c2 glutamatergic (Glu2) NTS neurons drive sodium appetite40 and Lepr and Gal glutamatergic (Glu3) NTS neurons module breathing41. Data for reconstruction of figure are available in Supplementary Data 2.
Extended Data Fig. 8 Expression of Bdnf and Mc4r following GLP-1RA administration.
a, Representative ic of Casr (blue) and Mc4r (red) in mice administered with Ex4 (i.v., 200 μg kg−1; n=2). Scale bar, 100 μm. b, Representative image showing db-ISH of Casr (blue) and Bdnf (red) in mice administered with Ex4 (i.v., 200 μg kg−1; n=2). Scale bar, 100 μm. i.v., intravenous.
Extended Data Fig. 9 FOS immunoreactivity in Calcr AP and NTS cells following GLP-1RA or salmon calcitonin administration.
a, Representative image showing FOS immunoreactivity (purple) and GFP immunoreactivity (green) in Calcr-Cre;GFP mice administered with Ex4 (i.p., 150 μg kg−1; n=3) or vehicle (n=3). Scale bar, 150 μm. b, Representative image showing FOS immunoreactivity (purple) and GFP immunoreactivity (green) in Calcr-Cre;GFP mice administered with sCT (i.p., 150 μg kg−1; n=3) or vehicle (n=3). Scale bar, 150 μm.
Extended Data Fig. 10 Activation of Calcr NTS neurons suppresses feeding.
a, Representative image showing mCherry immunoreactivity (pseudo-colored green) and FOS immunoreactivity (purple) after CNO treatment in Calcr-Cre mice injected with hM3Dq-mCherry in the NTS (n=7). Scale bar, 150 μm. b,c, Long-term chow food intake (b) and body weight (c) in control (n=6) and Calcr-Cre mice injected with hM3Dq-mCherry in the NTS (n=7) measured during 1 day of saline, 2 days of CNO (i.p., 1 mg kg−1) followed by 1 day of saline treatment. d, Short-term HFD food intake in Calcr-Cre mice injected with hM3Dq-mCherry in the NTS and treated with saline (n=7) or CNO (i.p., 1 mg kg−1; n=7). e,f, Long-term HFD food intake (e) and body weight (f) in control (n=5) or Calcr-Cre mice injected with hM3Dq-mCherry in the NTS (n=7) measured during 3 days of saline, 2 days of CNO (i.p., 1 mg kg-1) followed by 2 days of saline treatment. g, Short-term chow food intake in control mice injected with hM3Dq-mCherry in the NTS and treated with saline (n=6) or CNO (i.p., 1 mg kg-1; n=6). Values are the mean ± s.e.m. P<0.05 are specified, linear mixed effects model, Bonferroni-adjusted least squares means two-tailed t-test. h, Long-term chow food intake in control mice injected with hM3Dq-mCherry in the NTS and treated with saline (n=6) or CNO (i.p., 1 mg kg−1; n=6). Values are the mean ± s.e.m. Linear model, least squares means two-tailed t-test.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3
Supplementary Data 1
Differentially expressed genes in bulk RNA-seq data.
Supplementary Data 2
CELLEX scores for cell populations identified in the snRNA-seq data.
Supplementary Data 3
AP and NTS marker genes.
Supplementary Data 4
Gene modules and their association with treatment.
Supplementary Data 5
Motif enrichment for cell populations identified in the snATAC–seq data.
Supplementary Data 6
Meta-analysis of CELLECT results.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Ludwig, M.Q., Cheng, W., Gordian, D. et al. A genetic map of the mouse dorsal vagal complex and its role in obesity. Nat Metab 3, 530–545 (2021). https://doi.org/10.1038/s42255-021-00363-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-021-00363-1