Abstract
Alcohol is among the most widely used psychoactive substances worldwide. Ethanol metabolites such as acetate, thought to be primarily the result of ethanol breakdown by hepatic aldehyde dehydrogenase 2 (ALDH2), contribute to alcohol’s behavioural effects and alcoholism. Here, we show that ALDH2 is expressed in astrocytes in the mouse cerebellum and that ethanol metabolism by astrocytic ALDH2 mediates behavioural effects associated with ethanol intoxication. We show that ALDH2 is expressed in astrocytes in specific brain regions and that astrocytic, but not hepatocytic, ALDH2 is required to produce ethanol-derived acetate in the mouse cerebellum. Cerebellar astrocytic ALDH2 mediates low-dose ethanol-induced elevation of GABA levels, enhancement of tonic inhibition and impairment of balance and coordination skills. Thus, astrocytic ALDH2 controls the production, cellular and behavioural effects of alcohol metabolites in a brain-region-specific manner. Our data indicate that astrocytic ALDH2 is an important, but previously under-recognized, target in the brain to alter alcohol pharmacokinetics and potentially treat alcohol use disorder.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data from these studies are contained within this manuscript or are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Volkow, N. D., Koob, G. & Baler, R. Biomarkers in substance use disorders. ACS Chem. Neurosci. 6, 522–525 (2015).
Cui, C. & Koob, G. F. Titrating tipsy targets: the neurobiology of low-dose alcohol. Trends Pharmacol. Sci. 38, 556–568 (2017).
Abrahao, K. P., Salinas, A. G. & Lovinger, D. M. Alcohol and the brain: neuronal molecular targets, synapses and circuits. Neuron 96, 1223–1238 (2017).
McLaughlin, P. J., Chuck, T. L., Arizzi-LaFrance, M. N., Salamone, J. D. & Correa, M. Central versus peripheral administration of ethanol, acetaldehyde and acetate in rats: effects on lever pressing and response initiation. Pharmacol. Biochem. Behav. 89, 304–313 (2008).
Eng MY, L. S. & Wall, T. L. ALDH2, ADH1B and ADH1C genotypes in Asians: a literature review. Alcohol Res. Health 30, 22–27 (2007).
Edenberg, H. J. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health 30, 5–13 (2007).
Aragon, C. M., Rogan, F. & Amit, Z. Ethanol metabolism in rat brain homogenates by a catalase-H2O2 system. Biochem. Pharmacol. 44, 93–98 (1992).
Kim, J. I. et al. Aldehyde dehydrogenase 1a1 mediates a GABA synthesis pathway in midbrain dopaminergic neurons. Science 350, 102–106 (2015).
Chen, C. H., Ferreira, J. C., Gross, E. R. & Mochly-Rosen, D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol. Rev. 94, 1–34 (2014).
Deitrich, R., Zimatkin, S. & Pronko, S. Oxidation of ethanol in the brain and its consequences. Alcohol Res. Health 29, 266–273 (2006).
Jiang, L. et al. Increased brain uptake and oxidation of acetate in heavy drinkers. J. Clin. Invest. 123, 1605–1614 (2013).
Deelchand, D. K., Shestov, A. A., Koski, D. M., Ugurbil, K. & Henry, P. G. Acetate transport and utilization in the rat brain. J. Neurochem. 109, 46–54 (2009).
Carmichael, F. J. et al. Central nervous system effects of acetate: contribution to the central effects of ethanol. J. Pharmacol. Exp. Ther. 259, 403–408 (1991).
Volkow, N. D. et al. Acute alcohol intoxication decreases glucose metabolism but increases acetate uptake in the human brain. Neuroimage 64, 277–283 (2013).
Mews, P. et al. Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721 (2019).
Nuutinen, H., Lindros, K., Hekali, P. & Salaspuro, M. Elevated blood acetate as indicator of fast ethanol elimination in chronic alcoholics. Alcohol 2, 623–626 (1985).
Wagner, M. J., Kim, T. H., Savall, J., Schnitzer, M. J. & Luo, L. Cerebellar granule cells encode the expectation of reward. Nature 544, 96–100 (2017).
Carta, I., Chen, C. H., Schott, A. L., Dorizan, S. & Khodakhah, K. Cerebellar modulation of the reward circuitry and social behaviour. Science https://doi.org/10.1126/science.aav0581 (2019).
Chen, C. H., Fremont, R., Arteaga-Bracho, E. E. & Khodakhah, K. Short latency cerebellar modulation of the basal ganglia. Nat. Neurosci. 17, 1767–1775 (2014).
Sathyanesan, A. et al. Emerging connections between cerebellar development, behaviour and complex brain disorders. Nat. Rev. Neurosci. 20, 298–313 (2019).
Lackey, E. P., Heck, D. H. & Sillitoe, R. V. Recent advances in understanding the mechanisms of cerebellar granule cell development and function and their contribution to behavior. F1000Res https://doi.org/10.12688/f1000research.15021.1 (2018).
Reeber, S. L., Otis, T. S. & Sillitoe, R. V. New roles for the cerebellum in health and disease. Front Syst. Neurosci. 7, 83 (2013).
Langevin, F., Crossan, G. P., Rosado, I. V., Arends, M. J. & Patel, K. J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475, 53–58 (2011).
Song, A. J. & Palmiter, R. D. Detecting and avoiding problems when using the Cre-lox system. Trends Genet. 34, 333–340 (2018).
Sun, Q., Zhang, W., Zhong, W., Sun, X. & Zhou, Z. Dietary fisetin supplementation protects against alcohol-induced liver injury in mice. Alcohol Clin. Exp. Res. 40, 2076–2084 (2016).
Xiang, Y. & Shen, J. In vivo detection of intermediate metabolic products of [1-13C]ethanol in the brain using 13C magnetic resonance spectroscopy. NMR Biomed. 24, 1054–1062 (2011).
Guillot, A. et al. Targeting liver aldehyde dehydrogenase 2 prevents heavy but not moderate alcohol drinking. Proc. Natl Acad. Sci. USA 116, 25974–25981 (2019).
Wang, W. T., Lee, P., Hui, D., Michaelis, E. K. & Choi, I. Y. Effects of ethanol exposure on the neurochemical profile of a transgenic mouse model with enhanced glutamate release using in vivo 1H MRS. Neurochem. Res. 44, 133–146 (2019).
Carta, M., Mameli, M. & Valenzuela, C. F. Alcohol enhances GABAergic transmission to cerebellar granule cells via an increase in Golgi cell excitability. J. Neurosci. 24, 3746–3751 (2004).
Diaz, M. R. & Valenzuela, C. F. Sensitivity of GABAergic tonic currents to acute ethanol in cerebellar granule neurons is not age- or delta subunit-dependent in developing rats. Alcohol Clin. Exp. Res. 40, 83–92 (2016).
Lee, S. et al. Channel-mediated tonic GABA release from glia. Science 330, 790–796 (2010).
Woo, J. et al. Control of motor coordination by astrocytic tonic GABA release through modulation of excitation/inhibition balance in cerebellum. Proc. Natl Acad. Sci. USA 115, 5004–5009 (2018).
Cinar, R. et al. Hybrid inhibitor of peripheral cannabinoid-1 receptors and inducible nitric oxide synthase mitigates liver fibrosis. JCI Insight https://doi.org/10.1172/jci.insight.87336 (2016).
Liang, J. et al. Chronic intermittent ethanol-induced switch of ethanol actions from extrasynaptic to synaptic hippocampal GABAA receptors. J. Neurosci. 26, 1749–1758 (2006).
Valenzuela, C. F. & Jotty, K. Mini-review: effects of ethanol on GABAA receptor-mediated neurotransmission in the cerebellar cortex–recent advances. Cerebellum 14, 438–446 (2015).
Zhu, H. et al. Moderate UV exposure enhances learning and memory by promoting a novel glutamate biosynthetic pathway in the brain. Cell 173, 1716–1727 (2018).
Sonnewald, U. et al. NMR spectroscopic studies of [13C]acetate and [13C]glucose metabolism in neocortical astrocytes: evidence for mitochondrial heterogeneity. Dev. Neurosci. 15, 351–358 (1993).
Sonnewald, U. et al. Direct demonstration by [13C]NMR spectroscopy that glutamine from astrocytes is a precursor for GABA synthesis in neurons. Neurochem. Int. 22, 19–29 (1993).
Tiwari, V., Veeraiah, P., Subramaniam, V. & Patel, A. B. Differential effects of ethanol on regional glutamatergic and GABAergic neurotransmitter pathways in mouse brain. J. Neurochem. 128, 628–640 (2014).
Rustay, N. R. et al. Sensitivity and tolerance to ethanol-induced incoordination and hypothermia in HAFT and LAFT mice. Pharmacol. Biochem. Behav. 70, 167–174 (2001).
Rustay, N. R., Wahlsten, D. & Crabbe, J. C. Assessment of genetic susceptibility to ethanol intoxication in mice. Proc. Natl Acad. Sci. USA 100, 2917–2922 (2003).
Saeed Dar, M. Co-modulation of acute ethanol-induced motor impairment by mouse cerebellar adenosinergic A1 and GABAA receptor systems. Brain Res. Bull. 71, 287–295 (2006).
Hipolito, L., Sanchez, M. J., Polache, A. & Granero, L. Brain metabolism of ethanol and alcoholism: an update. Curr. Drug Metab. 8, 716–727 (2007).
Wilson, D. F. & Matschinsky, F. M. Ethanol metabolism: The good, the bad, and the ugly. Med. Hypotheses 140, 109638 (2020).
Deelchand, D. K., Nelson, C., Shestov, A. A., Ugurbil, K. & Henry, P. G. Simultaneous measurement of neuronal and glial metabolism in rat brain in vivo using co-infusion of [1,6-13C2]glucose and [1,2-13C2]acetate. J. Magn. Reson. 196, 157–163 (2009).
Zimatkin, S. M., Pronko, S. P., Vasiliou, V., Gonzalez, F. J. & Deitrich, R. A. Enzymatic mechanisms of ethanol oxidation in the brain. Alcohol Clin. Exp. Res. 30, 1500–1505 (2006).
Isse, T., Matsuno, K., Oyama, T., Kitagawa, K. & Kawamoto, T. Aldehyde dehydrogenase 2 gene targeting mouse lacking enzyme activity shows high acetaldehyde level in blood, brain and liver after ethanol gavages. Alcohol Clin. Exp. Res. 29, 1959–1964 (2005).
Martinez-Rodriguez, R. et al. Synaptic and non-synaptic immunolocalization of GABA and glutamate acid decarboxylase in cerebellar cortex of rat. Cell Mol. Biol. 39, 115–123 (1993).
Le Meur, K., Mendizabal-Zubiaga, J., Grandes, P. & Audinat, E. GABA release by hippocampal astrocytes. Front Comput Neurosci. 6, 59 (2012).
Joshi, A. U. et al. Aldehyde dehydrogenase 2 activity and aldehydic load contribute to neuroinflammation and Alzheimer’s disease related pathology. Acta Neuropathol. Commun. 7, 190 (2019).
Overstreet, D. H., Knapp, D. J., Breese, G. R. & Diamond, I. A selective ALDH-2 inhibitor reduces anxiety in rats. Pharmacol. Biochem. Behav. 94, 255–261 (2009).
Zambelli, V. O. et al. Aldehyde dehydrogenase 2 regulates nociception in rodent models of acute inflammatory pain. Sci. Transl. Med. 6, 251ra118 (2014).
Yao, L. et al. Inhibition of aldehyde dehydrogenase 2 suppresses cocaine seeking by generating THP, a cocaine use-dependent inhibitor of dopamine synthesis. Nat. Med. 16, 1024–1028 (2010).
Mitew, S., Kirkcaldie, M. T., Dickson, T. C. & Vickers, J. C. Altered synapses and gliotransmission in Alzheimer’s disease and AD model mice. Neurobiol. Aging 34, 2341–2351 (2013).
Jo, S. et al. GABA from reactive astrocytes impairs memory in mouse models of Alzheimer’s disease. Nat. Med. 20, 886–896 (2014).
Wu, Z., Guo, Z., Gearing, M. & Chen, G. Tonic inhibition in dentate gyrus impairs long-term potentiation and memory in an Alzheimer’s disease model. Nat. Commun. 5, 4159 (2014).
Marek, S. et al. Spatial and temporal organization of the individual human cerebellum. Neuron 100, 977–993 (2018).
Lin, C. Y., Chen, C. H., Tom, S. E. & Kuo, S. H., Alzheimer’s Disease Neuroimaging Initiative. Cerebellar volume is associated with cognitive decline in mild cognitive impairment: results from ADNI. Cerebellum 19, 217–225 (2020).
Garcia, A. D., Doan, N. B., Imura, T., Bush, T. G. & Sofroniew, M. V. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat. Neurosci. 7, 1233–1241 (2004).
Maeda, M., Hasumura, Y. & Takeuchi, J. Localization of cytoplasmic and mitochondrial aldehyde dehydrogenase isozymes in human liver. Lab. Invest. 59, 75–81 (1988).
Kitagawa, K. et al. Aldehyde dehydrogenase 2 associates with oxidation of methoxyacetaldehyde; in vitro analysis with liver subcellular fraction derived from human and Aldh2 gene targeting mouse. FEBS Lett. 476, 306–311 (2000).
Koivisto, T., Carr, L. G., Li, T. K. & Eriksson, C. J. Mitochondrial aldehyde dehydrogenase polymorphism in AA and ANA rats: lack of genotype and phenotype line differences. Pharmacol. Biochem. Behav. 45, 215–220 (1993).
Gao, Y. et al. Alcohol inhibits T cell glucose metabolism and hepatitis in ALDH2-deficient mice and humans: roles of acetaldehyde and glucocorticoids. Gut 68, 1311–1322 (2019).
Zhu, H. et al. Single-neuron identification of chemical constituents, physiological changes and metabolism using mass spectrometry. Proc. Natl Acad. Sci. USA 114, 2586–2591 (2017).
Ende, G. Proton magnetic resonance spectroscopy: relevance of glutamate and GABA to neuropsychology. Neuropsychol. Rev. 25, 315–325 (2015).
Xu, S. et al. Decreased taurine and creatine in the thalamus may relate to behavioral impairments in ethanol-fed mice: a pilot study of proton magnetic resonance spectroscopy. Mol Imaging 17, 1536012117749051 (2018).
Pawlosky, R. J. et al. Alterations in brain glucose utilization accompanying elevations in blood ethanol and acetate concentrations in the rat. Alcohol Clin. Exp. Res. 34, 375–381 (2010).
Xiong, W. et al. Cannabinoids suppress inflammatory and neuropathic pain by targeting α3 glycine receptors. J. Exp. Med. 209, 1121–1134 (2012).
Acknowledgements
We thank F. Langevin and K. J. Patel (University of Cambridge) for Aldh2 flox mice. We especially thank M. Chen, X. Sun and X. Li (Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland) for their technical support for in vivo MRS experiments and data analysis. We thank A. Salinas, G. Luo, T. Ren, A. Guillot and H.-J. White (NIAAA, NIH) for technical assistance and comments on the manuscript. We thank the Human Brain Collection Core at the National Institute of Mental Health for providing human cerebellar tissues. This work was supported by grants 1UL1TR003098 (to Q. Cao) from the University of Maryland, Baltimore, Institute for Clinical & Translational Research (ICTR) and the National Center for Advancing Translational Sciences (NCATS) Clinical Translational Science Award (CTSA), K08AA024895-01A1 to (Q. Cao) from the National Institue on Alcohol Abuse and Alcoholism (NIAAA), National Institutes of Health (NIH), chairman seed grant award (to Q. Cao) of the Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland, Baltimore and 81801938 (to S.J.) from the National Natural Science Foundation of China.
Author information
Authors and Affiliations
Contributions
L.Z. conceived, designed and supervised the study. S.J., L.Z., F.Y., Y.L., R.C. and R.J.P. contributed to initial data collection and analysis. Q. Cao and S.X. conducted the MRS experiment and analysed the data. Q. Chen, Z.W., H.Z. and W.X. conducted high-resolution single-cell GC–MS analysis and analysed the data. S.J. and L.Z. performed the final data analysis. S.J. made the figures and tables. L.Z. wrote the manuscript. Y.Z., D.M.L., B.G., Q. Cao and G.F.K. helped with data interpretation and revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editors: Pooja Jha; Isabella Samuelson. Nature Metabolism thanks Juan Bolaños and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
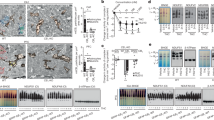
Extended Data Fig. 1 Astrocyte-specific distribution of ALDH2 in the cerebellum.
a, Bar graphs of ALDH2 activity in cerebellar anterior, vermis and posterior regions. (n=6 mice/group). Data are presented as means±s.e.m. Analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey’s test. b, Epifluorescence images of ALDH2 (green), GFAP (violet) and L7/PC (red) gene in a cerebellar sagittal slice. Red scale bar, 200 μm; White scale bars, 50 μm. c, Epifluorescence images of ALDH2 (green), MAP2 (violet) and L7/PC (red) gene. No colocalization across these genes. Red scale bar, 200 μm; White scale bars, 50 μm. d, Images showing a colocalization of ALDH2 (green) with GFAP (red) but not MAP2 (violet) gene in cerebellum. Red scale bar, 200 μm; White scale bars, 50 μm. For b, c and d, Experiments were repeated four times for each biologically independent mouse, with similar results obtained. e, Quantification of cluster colocalization between ALDH2 and ALDH1L1 in cerebellum. The experiment was repeated four times for each biologically independent mouse, with similar results obtained (n=3 mice). Quantitative data was provided in Supplementary Table 2.YES: colocalization; NO: no colocalization; White scale bars, 50 μm.
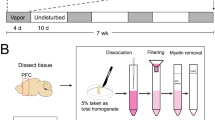
Extended Data Fig. 2 ALDH2 expression in different brain regions of Aldh2Gfap-/- mice.
a, the schematic diagram shows the generation of astrocytic specific Aldh2 deficient mice. b, A representative image of PCR genotyping result. The image of agar gel migration shows flox (homozygous: one band, 365 bp; heterozygous: two bands, 365 and 300 bp) and Cre (negative: one band, 350 bp; positive: two bands, 350 and 200 bp) gene. c, RNAscope images of ALDH2 signals in cerebellum and liver slices from conditional and global Aldh2-/- mice. Noted that ALDH2 was selectively depleted from microdomains surrounding PC signal in cerebellar slice from Aldh2Gfap-/- mice. Liver ALDH2 mRNA was intact in Aldh2Gfap-/- mice, whereas liver ALDH2 signal was abolished in Aldh2-/- mice. Each experiment was repeated four times with similar results obtained. White scale bars, 100 μm. d, e, Representative peaks from electrophoresis densitometry of cerebellar (n=7, 7 and 3 mice, respectively) and liver (n=5, 5 and 3 mice, respectively) ALDH2 proteins from Aldh2Gfap+/+ (black line or rectangle), Aldh2Gfap-/- and Aldh2-/- mice. f, Summary statistics of cerebellar ALDH2 enzymatic activity detected in Aldh2L7-/- and Aldh2Camk2a-/- mice (n=6, 6, 7 and 7 mice, respectively). g, Representative electropherogram traces of ALDH2 protein samples collected from different brain areas of Aldh2Gfap+/+ and Aldh2Gfap-/- mice. h, Summary statistics of brain ALDH2 protein detected in difference brain areas from Aldh2Gfap+/+ (n=5, 4, 6, 4, 4 and 7 mice, respectively) and Aldh2Gfap-/- (n=5, 4, 6, 4, 4 and 7 mice, respectively) mice. i, Representative electropherogram traces of protein samples collected from different brain areas from Aldh2Camk2a+/+ and Aldh2Camk2a-/- mice. j, Summary statistics of brain ALDH2 protein detected in difference brain areas from Aldh2Camk2a+/+ and Aldh2Camk2a-/- mice (n=4 mice/group). k, l, Representative electropherogram traces and summary statistics of brain ALDH2 protein detected in different brain areas from astrocytic ALDH2 deficient mice generated by crossing Aldh2 flox with GFAPCre (Jax, 024098), Aldh2Gfap-/- mice and their wildtype littermates, Aldh2Gfap+/+ (n=3 mice/group). Data are presented as means ±s.e.m. Analysis was performed using unpaired two tailed student’s t test (h, j, f, l) or one-way analysis of variance (ANOVA) followed by Tukey’s test (d, e). PFC, Prefrontal cortex; CE, Cerebellum; HIPP, Hippocampus; CeA, Central nucleus of the amygdala; VTA, Ventral tegmental area; BLA, Basolateral amygdala; NAc, Nucleus accumbent.
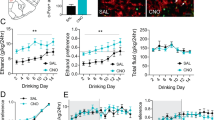
Extended Data Fig. 3 Astrocytic Aldh2 deficiency does not alter serum and brain (cerebellar cortex) ethanol and acetaldehyde contents.
a, b, Serum and cerebellum ethanol concentrations measured 50 min after ethanol (2 g/kg, i.p.) in WT and Aldh2*2KI mice (n=4 mice/group). c, d, Graphs showing ethanol concentrations in the serum and cerebellum 50 min after ethanol in Aldh2Gfap+/+ and Aldh2Gfap-/- mice (0 g/kg ethanol: n=4 mice/group; 1 g/kg ethanol: n=12 and 13 from Aldh2Gfap+/+ and Aldh2Gfap-/- mice respectively (serum), n=7 and 8 from Aldh2Gfap+/+ and Aldh2Gfap-/- mice respectively (cerebellum); 2 g/kg ethanol: n=5 mice/group; 3.6 g/kg ethanol: n=5 mice/group). e, f, Serum and cerebellum acetaldehyde contents measured after ethanol (1 g/kg, i.p.) in Aldh2Gfap+/+ and Aldh2Gfap-/- mice (0 min: n=4 mice/group; 10 min: n=9 mice/group (serum), n=4 mice/group (cerebellum); 50 min: n=11 mice/group (serum), n=7 and 8 from Aldh2Gfap+/+ and Aldh2Gfap-/- mice respectively (cerebellum)). g, h, Serum and cerebellum ethanol contents measured after ethanol (1 g/kg, i.p.) in Aldh2Gfap+/+ and Aldh2Gfap-/- mice (0 min: n=4 mice/group; 10 min: n=9 mice/group (serum), n=4 mice/group (cerebellum); 50 min: n=12 and 13 from Aldh2Gfap+/+ and Aldh2Gfap-/- mice respectively (serum), n=7 and 8 from Aldh2Gfap+/+ and Aldh2Gfap-/- mice respectively (cerebellum)). Data are presented as mean±s.e.m. Analysis was performed using unpaired two tailed student’s t test (a, b) or two-way analysis of variance (ANOVA) (c-h).
Extended Data Fig. 4 In vivo MRS quantitative measurement of brain ethanol metabolites and neurochemicals.
a, b, Representative peaks and Summary statistics of brain ALDH2 protein in cerebellum and liver from Aldh2Hep+/+ and Aldh2Hep-/-mice (n=4 mice/group).c, d, In vivo MRS measurement of cerebellar acetate and GABA contents after systemic ethanol (1-2 g/kg, i.p.) in Aldh2Gfap+/+ mice (n=5, 3 and 5 mice, respectively). e, Ethanol enhancement of cerebellar GABA, Glutamine, Glutamate and NAA in Aldh2 flox mice (n=9 mice/group). f, g, Statistic summaries of cerebellar ethanol by MRS after systemic administration of ethanol (1-2 g/kg, i.p.) in astrocytic ALDH2 deficient mice (f: n=3 mice/group; g: n=5 mice/group), hepatocytic ALDH2 deficient mice (Aldh2Hep+/+: n=4 mice and Aldh2Hep-/-: n=6 mice) and their wild type littermates mice. g, Bar graphs showing cerebellar ethanol concentrations after ethanol (2 g/kg, i.p.) by in vivo MRS measurement in Aldh2Hep+/+ (n=4) and Aldh2Hep-/- (n=4) mice. h, Bar graph showing the basal levels of GABA, Glutamine, Glutamate and NAA in cerebellum in Aldh2Gfap+/+ and Aldh2Gfap-/- mice (n=5 mice/group). i, Bar graph showing the basal levels of GABA, Glutamine, Glutamate and NAA in cerebellum in Aldh2Hep+/+ (n=4) and Aldh2Hep-/- (n=6) mice. Data are presented as mean±s.e.m. Groups were compared by unpaired two tailed student’s t test (b, e-i), or one-way analysis of variance (ANOVA) followed by Tukey’s test (c, d).
Extended Data Fig. 5 Ethanol does not significantly alter GABAergic mIPSCs and GABA-activated current in GCs.
a, b, Trace records and summary statistics of tonic current recorded using whole cell recording in cerebellar GCs from Aldh2Gfap+/+ and Aldh2Gfap-/- mice (ACSF: n=8 cells, 6 mice). c, Summary statistics of the cell membrane capacitance with and without ethanol and acetate (n=8, 8, 7, 7, 7, 8, 9, 9, 9 and 10 cells, respectively). d, e, Left, Trace records of GABA (20 µM) activated currents in cerebellar GCs from Aldh2Gfap+/+ and Aldh2Gfap-/- mice. Right, Summary of the average amplitudes of GABA-activated currents (n=4 cells/group, 4 mice). f, Statistic summary of the average levels of cerebellar GABA contents measured by LC-MS/MS in Aldh2Gfap+/+ and Aldh2Gfap-/- mice (n=8 mice/group). g-j, Trace records and summary statistics of mIPSCs in GCs from Aldh2Gfap+/+ and Aldh2Gfap-/- mice (n=8 cells/group). k-n, The effect of ethanol on GABAergic IPSCs (n=9 cells/group, 6 mice). Data are presented as mean±s.e.m. Groups were compared by one-way analysis of variance (ANOVA) (c), or unpaired two tailed student’s t test (b, c, e, f, h, i, l, m).
Extended Data Fig. 6 Characterization of unilateral conditional deletion of cerebellar astrocytic ALDH2.
a, Bar graph showing the rotarod performance of Aldh2Camk2a+/+ and Aldh2Camk2a-/- mice (n=10 mice/group) after 10 min 1 g/kg ethanol i.p. injection. b, Time course of the basal levels of the rotarod performance in consecutive trails in Aldh2Gfap+/+ (n=9) and Aldh2Gfap-/- (n=10) mice. c, Time courses of core-body temperature after ethanol (2 g/kg, i.p.) in Aldh2Gfap+/+ (n=9) and Aldh2Gfap-/- (n=13) mice. d,e, Florescent microscopy images of ALDH2 signal (green) in cerebellar slices from Aldh2 flox mice previously injected with AAV-GFAPCre and AAV-Synt1Cre virus (4 weeks after injection). Experiments were repeated four times for each biologically independent mouse, with similar results obtained (n=3 mice). Red scale bar, 500 μm; White scale bars, 50 μm. f, Bar graph showing the baseline levels of rotarod performance in cerebellar virus injected mice (n=15 mice for AAV-Synt1Cre; n=16 mice for AAV-GFAPCre). g, Time courses of the levels of rotarod performance after i.c.v. injection of CGP (200 ng, n=7 mice) or Bicuculline (200 ng, n=5 mice). Data are presented as mean±s.e.m. Analysis was performed using unpaired two tailed student’s t test (f) or two-way analysis of variance (ANOVA) followed by Turkey’s test (a), or two-way repeated-measures analysis of variance (ANOVA) (b,c,g).
Extended Data Fig. 7 A hypothetical mechanism for astrocytic ALDH2 control of alcohol metabolism and action in the brain.
Simplified diagram of ALDH2-dependent pathways that mediate ethanol-induced elevation in cerebellar GABA levels and discoordination.
Supplementary information
Supplementary Information
Supplementary Tables 1–3
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Jin, S., Cao, Q., Yang, F. et al. Brain ethanol metabolism by astrocytic ALDH2 drives the behavioural effects of ethanol intoxication. Nat Metab 3, 337–351 (2021). https://doi.org/10.1038/s42255-021-00357-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-021-00357-z
This article is cited by
-
Physiological acetic acid concentrations from ethanol metabolism stimulate accumbens shell medium spiny neurons via NMDAR activation in a sex-dependent manner
Neuropsychopharmacology (2024)
-
Potentiation of the depressant effect of alcohol by flunitrazepam in rats: an electrocorticographic, respiratory and electrocardiographic study
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Alcohol spiked with zolpidem and midazolam potentiates inflammation, oxidative stress and organ damage in a mouse model
Forensic Toxicology (2024)
-
Pathogenic mechanisms and regulatory factors involved in alcoholic liver disease
Journal of Translational Medicine (2023)
-
Identifying novel proteins underlying loneliness by integrating GWAS summary data with human brain proteomes
Neuropsychopharmacology (2023)