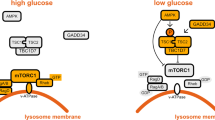
Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) kinase regulates cell growth by setting the balance between anabolic and catabolic processes. To be active, mTORC1 requires the environmental presence of amino acids and glucose. While a mechanistic understanding of amino acid sensing by mTORC1 is emerging, how glucose activates mTORC1 remains mysterious. Here, we used metabolically engineered human cells lacking the canonical energy sensor AMP-activated protein kinase to identify glucose-derived metabolites required to activate mTORC1 independent of energetic stress. We show that mTORC1 senses a metabolite downstream of the aldolase and upstream of the GAPDH-catalysed steps of glycolysis and pinpoint dihydroxyacetone phosphate (DHAP) as the key molecule. In cells expressing a triose kinase, the synthesis of DHAP from DHA is sufficient to activate mTORC1 even in the absence of glucose. DHAP is a precursor for lipid synthesis, a process under the control of mTORC1, which provides a potential rationale for the sensing of DHAP by mTORC1.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data supporting the plots within this paper are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism and disease. Cell 168, 960–976 (2017).
Kubrusly, M. S. et al. A role for mammalian target of rapamycin (mTOR) pathway in non-alcoholic steatohepatitis-related cirrhosis. Histol. Histopathol. 25, 1123–1131 (2010).
Guillén, C. & Benito, M. mTORC1 overactivation as a key aging factor in the progression to type 2 diabetes mellitus. Front. Endocrinol. 9, 621 (2018).
Ardestani, A., Lupse, B., Kido, Y., Leibowitz, G. & Maedler, K. mTORC1 signaling: a double-edged sword in diabetic β cells. Cell Metab. 27, 314–331 (2018).
Lee, P. L., Tang, Y., Li, H. & Guertin, D. A. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol. Metab. 5, 422–432 (2016).
Inoki, K., Zhu, T. & Guan, K.-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 (2003).
Shaw, R. J. et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6, 91–99 (2004).
Gwinn, D. M. & Shaw, R. J. AMPK control of mTOR signaling and growth. in The Enzymes Vol. 28 (eds Tamanoi, F. & Hall, M. N.) 49–75 (Academic Press, 2010).
Efeyan, A. et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683 (2013).
Wolfson, R. L. et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017).
Kalender, A. et al. Metformin, independent of AMPK, inhibits mTORC1 in a Rag GTPase-dependent manner. Cell Metab. 11, 390–401 (2010).
Buller, C. L., Heilig, C. W. & Brosius, F. C. GLUT1 enhances mTOR activity independently of TSC2 and AMPK. Am. J. Physiol. Renal Physiol. 301, F588–F596 (2011).
Almacellas, E. et al. Phosphofructokinases axis controls glucose-dependent mTORC1 activation driven by E2F1. iScience 20, 434–448 (2019).
Roberts, D. J., Tan-Sah, V. P., Ding, E. Y., Smith, J. M. & Miyamoto, S. Hexokinase-II positively regulates glucose starvation induced autophagy through TORC1 inhibition. Mol. Cell 53, 521–533 (2014).
Zhang, C.-S. et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 (2017).
Lee, M. N. et al. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol. Cell. Biol. 29, 3991–4001 (2009).
Sharma, S., Guthrie, P. H., Chan, S. S., Haq, S. & Taegtmeyer, H. Glucose phosphorylation is required for insulin-dependent mTOR signalling in the heart. Cardiovasc. Res. 76, 71–80 (2007).
Karlstaedt, A., Khanna, R., Thangam, M. & Taegtmeyer, H. Glucose 6-phosphate accumulates via phosphoglucose isomerase inhibition in heart muscle. Circ. Res. 126, 60–74 (2020).
Shiraj, S. et al. Glucose regulation of load‐induced mTOR signaling and ER stress in mammalian heart. J. Am. Heart Assoc. 2, e004796 (2013).
Hardie, D. G., Ross, F. A. & Hawley, S. A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251–262 (2012).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Sakai, K., Hasumi, K. & Endo, A. Two glyceraldehyde-3-phosphate dehydrogenase isozymes from the koningic acid producer Trichoderma koningii. Eur. J. Biochem. 193, 195–202 (1990).
Liberti, M. V. et al. A predictive model for selective targeting of the warburg effect through GAPDH inhibition with a natural product. Cell Metab. 26, 648–659 (2017).
Rieder, S. V. & Rose, I. A. The mechanism of the triosephosphate isomerase reaction. J. Biol. Chem. 234, 1007–1010 (1959).
Fang, Y., Vilella-Bach, M., Bachmann, R., Flanigan, A. & Chen, J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294, 1942–1945 (2001).
Menon, D. et al. Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J. Biol. Chem. 292, 6303–6311 (2017).
Ye, J. et al. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 29, 2331–2336 (2015).
Wolfson, R. L. et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 (2016).
Chantranupong, L. et al. The CASTOR proteins are arginine sensors for the mTORC1 Pathway. Cell 165, 153–164 (2016).
Huttlin, E. L. et al. The BioPlex Network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Düvel, K. et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010).
Peterson, T. R. et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 (2011).
Herman, M. A., She, P., Peroni, O. D., Lynch, C. J. & Kahn, B. B. Adipose tissue branched chain amino acid metabolism (BCAA) modulates circulating BCAA levels. J. Biol. Chem. 285, 11348–11356 (2010).
Nelson, D. L. & Cox, M. M. Lehninger Principles of Biochemistry (W. H. Freeman, 2012).
Jang, C. et al. The small intestine converts dietary fructose into glucose and organic acids. Cell Metab. 27, 351–361 (2018).
Herman, M. A. & Samuel, V. T. The sweet path to metabolic demise: fructose and lipid synthesis. Trends Endocrinol. Metab. 27, 719–730 (2016).
Frias, M. A. et al. Phosphatidic acid drives mTORC1 lysosomal translocation in the absence of amino acids. J. Biol. Chem. 295, 263–274 (2020).
Joy, J. M. et al. Phosphatidic acid enhances mTOR signaling and resistance exercise induced hypertrophy. Nutr. Metab. 11, 29 (2014).
Dennis, P. B. et al. Mammalian TOR: a homeostatic ATP sensor. Science 294, 1102–1105 (2001).
Yang, W.-C. et al. Simultaneous quantification of metabolites involved in central carbon and energy metabolism using reversed-phase liquid chromatography−mass spectrometry and in vitro 13C labeling. Anal. Chem. 80, 9508–9516 (2008).
Jannasch, A., Sedlak, M. & Adamec, J. Quantification of pentose phosphate pathway (PPP) metabolites by LC–MS. in Metabolic Profiling: Methods and Protocols (ed. Metz, T. O.) 159–171 (Humana Press, 2011).
Acknowledgements
The authors would like to thank M. Valenstein, J. Spinelli and all the current members of the Sabatini laboratory for helpful insights. This work was supported by grants from the the National Institutes of Health (NIH; R01 CA103866, R01 CA129105 and R37 AI047389). J.M.O. was supported by a fellowship grant (no. F30CA210373) from the National Cancer Institute and the Harvard–MIT MSTP training grant (no. T32GM007753) from the National Institute of General Medical Sciences. P.A.K. was supported by a scholarship from Santander Universidades mobility fund granted by A. Mickiewicz University in Poznan. A.L.C. was supported by a fellowship grant (no. F31DK113665) from the National Institute of Diabetes and Digestive and Kidney Diseases. D.M.S. is an investigator of the Howard Hughes Medical Institute and an American Cancer Society research professor.
Author information
Authors and Affiliations
Contributions
J.M.O. and D.M.S. initiated the project and designed the research plan with input from P.A.K. J.M.O. and P.A.K. conducted experiments and analysed data with technical assistance from S.M.S. A.L.C. assisted with experiments. S.H.C., T.K. and C.A.L. operated the LC–MS platform, analysed the metabolomics data and assisted in the method development to measure GAP–aniline. J.M.O. and D.M.S. wrote the manuscript with assistance from P.A.K. and C.A.L.
Corresponding author
Ethics declarations
Competing interests
D.M.S. is a founder, shareholder and member of the scientific advisory board for Navitor Pharmaceuticals, which is targeting the mTORC1 pathway for therapeutic benefit. J.M.O. is a shareholder of Navitor Pharmaceuticals. The other authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Christoph Schmitt.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Glucose metabolism via the pentose phosphate pathway (PPP) is not required for the activation of mTORC1.
a, AMPKα DKO cells were incubated for 1 hour without glucose, then either maintained without an added sugar or 10 mM of the indicated sugar or sugar analog was added for 15 minutes prior to cell lysis. Whole cell lysates were analyzed for the phosphorylation state or level of S6K1 by immunoblotting. b, 2-Deoxyglucose inhibits mTORC1 activity in a glucose-competitive manner. Wildtype HEK-293T cells were incubated in the given concentration of glucose for 1 hour and the indicated amount of 2-DG was added to them for 15 minutes. Whole cell lysates were analyzed for the phosphorylation state or level of S6K1 by immunoblotting. c, Loss of G6PD, the rate limiting enzyme for entry into the PPP, does not prevent the mTORC1 pathway from sensing glucose. Cells targeted with CRISPR/Cas9 using a control guide (AAVS1) or one against G6PD were incubated in the given glucose condition for 2 hours. Whole cell lysates were analyzed as in (a). d, Loss of G6PD causes a predictable decrease in pentose phosphate intermediates but had no impact on most glycolytic intermediates. Cells were treated the same as in (c) and metabolite extracts were analyzed by LC/MS for the indicated metabolites. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. An asterisk denoted The peak used to quantify fructose 6-phosphate (F6P) levels may also contain glucose 1-phosphate.
Extended Data Fig. 2 Metabolite profiling of cells lacking GAPDH activity.
a, In a dose-dependent manner, Koningic Acid (KA) leads to an increase in the metabolites upstream of GAPDH despite the removal of glucose from the cell media. Metabolites downstream of GAPDH are predictably depleted upon KA treatment. NAD levels fall slightly upon glucose starvation and are not affected by KA treatment. NADH became undetectable in the absence of glucose, consistent with the role of glucose metabolism in NADH production. The effect of glucose deprivation on ATP/ADP/AMP levels was not reversed by KA treatment. HEK-293T cells were treated with the indicated concentrations of KA at the beginning of a glucose starvation period, and metabolites were extracted and analyzed by LC/MS. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. * The peak used to quantify fructose 6-phosphate (F6P) levels may also contain glucose 1-phosphate. b, Overexpression of TK-GAPDH prevents all the effects on glycolytic metabolites caused by KA. Cells stably expressing FLAG-metap2 or FLAG-TK-GAPDH were incubated under the indicated conditions, and metabolites were extracted and analyzed by LC/MS. KA was added at the beginning of starvation. This panel is an extension of the data shown in Fig. 3e. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. * The peak used to quantify fructose 6-phosphate (F6P) levels may also contain glucose 1-phosphate. c, Consistent with pharmacological inhibition of GAPDH, loss of GAPDH expression increases levels of GAP and DHAP and decreases those of phosphoenolpyruvate (PEP). GAPDH Dox-off cells were exposed to the indicated glucose concentrations in the absence or presence of dox. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test.
Extended Data Fig. 3 GAP can be measured as its aniline adduct as it has a unique mass.
a, In cells with normal TPI activity, the DHAP and GAP metabolites elute at similar retention times (11 mins vs 11.5 mins, respectively) but the peaks are separate enough to integrate independently. However, when cells lose TPI expression, the DHAP levels increase by an order of magnitude, and the peak normally integrated to measure GAP is obscured by the broader DHAP peak, which has a shoulder that extends into the GAP peak at 11.5 mins. Because the mass of DHAP and GAP are the same, quantifying them accurately in the context of TPI loss is an analytical challenge. HEK-293T AMPKα DKO TPI dox-off cells were treated with (+Dox) or without dox (-Dox) for 10 days. The media was then replaced with fresh RPMI media containing 5 mM glucose for thirty minutes. Metabolites were extracted and analyzed by HILIC LC/MS. Peak traces are shown for the +TPI (-Dox) condition in black and -TPI (+Dox) in blue, both separately with different y-axis scales, or overlaid to highlight the observed changes in DHAP. b, DHAP and GAP can form aniline adducts that are singly labeled with an m/z value of 244.03803 but only GAP can form an aniline adduct that is doubly labeled with an m/z value of 319.08532. c, As expected, only GAP forms a product with m/z value of 319.08532 when reacted with aniline. 10 μM GAP or DHAP standards were reacted with aniline in the presence of EDC in 80% methanol to yield their respective aniline adducts. The reaction was analyzed by reverse phase LC/MS. d, Metabolites upstream of TPI increase and metabolites downstream of TPI decrease in the absence of TPI expression. HEK-293T AMPKα DKO TPI dox-off cells were incubated in full media (5mM glucose) following 10 days of doxycycline. Half of the extract was used in an aniline labeling reaction and analyzed by reverse phase LC/MS and the other half was analyzed by HILIC LC/MS. These data are an extension of the data shown in Fig. 4b, showing only the unstarved samples. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates.
Extended Data Fig. 4 Glycerol Kinase-mediated glycerol 3-phosphate (G3P) synthesis partially activates mTORC1 in glucose starved cells.
a, Diagram detailing the metabolism of glycerol, and its connection to the G3P-shuttle and the glycolytic metabolite dihydroxyacetone phosphate (DHAP). The role of GPD1 in coupling DHAP reduction to NADH oxidation is emphasized. b, HEK-293T AMPKα DKO cells stably overexpressing either a FLAG-tagged control cDNA (metap2) or glycerol kinase (GK) were incubated with the indicated glucose or glycerol concentrations for 1 hour. Whole cell protein lysates were analyzed by immunoblotting for the phosphorylation state and levels of the indicated proteins. Dashed line in panel B marks the removal of irrelevant intervening lanes in the original western blot. c, Cells were treated as in (B) and metabolite extracts were analyzed by LC/MS. Notably, glycerol addition to cells expressing GK, but not the control protein, generated supraphysiological levels of G3P but only partially restored DHAP levels. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. d, Diagram describing the position of phosphatidic acid (PA) in relation to its precursors DHAP and G3P. e, Addition of 100 uM PA does not activate mTORC1 in cells starved of either glucose or leucine. HEK-293T AMPKα DKO cells were starved of either glucose or leucine for 2.5 h and were restimulated for 30 mins with either glucose or leucine in the absence or presence of 100 uM PA.
Extended Data Fig. 5 DHAP plays a key role in the activation of mTORC1 by glucose.
a, GPD1 overexpression decreases the levels of several glycolytic intermediates (G6P and DHAP are shown) but increases those of glycerol 3-phosphate. HEK-293T AMPKα DKO cells expressing the indicated cDNAs were incubated with the indicated glucose concentration for 15 mins following a 3-hour glucose starvation. Metabolite extracts were analyzed by HILIC LC/MS and by reverse phase LC/MS for GAP-aniline. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. b, Dihydroxyacetone (DHA) makes DHAP only in TKFC-expressing cells, restoring levels to nearly those seen upon glucose restimulation. DHA treatment has little to no effect on glucose 6-phosphate (G6P) or fructose 1,6-bisphosphate (FBP) levels and only partially rescues glyceraldehyde 3-phosphate (GAP) levels. HEK-293T AMPKα DKO cells expressing a FLAG-tagged control cDNA (metap2) or triose kinase (TKFC) were incubated with the indicated concentrations of glucose or DHA for 1 hour. Metabolite extracts were analyzed by LC/MS. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. c, Glyceraldehyde (GA partially rescues mTORC1 signaling in the absence of glucose. HEK-293T AMPKα DKO cells expressing the indicated cDNAs were incubated with the indicated concentrations of glucose or GA for one hour. d, The concentrations required to obtain partial rescue of mTORC1 signaling with GA addition are also capable of leading to generation of DHAP along with GAP, here measured as the derivative GAP-aniline. Cells were treated as in (c). Metabolite extracts were analyzed by HILIC LC/MS for DHAP or reacted with aniline and analyzed by reverse phase LC/MS for GAP-aniline. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. e, DHA does not require TPI or ALDO expression in order to activate mTORC1. HEK-293T AMPKα DKO TPI Dox-off and AMPKα DKO ALDO Dox-off cells were treated with either 10mM glucose or 0.2 mM DHA in the absence or presence of doxycycline, and cell lysates were analyzed by immunoblotting for the phosphorylation state and levels of the indicated proteins.
Extended Data Fig. 6 Glucose sensing does not require canonical stress pathway components.
a, GCN2 loss is associated with increased mTORC1 activity but the pathway remains capable of sensing glucose and leucine in GCN2 KO cells. HEK-293T AMPKα DKO or AMPKα/GNC2 triple KOs (TKO) were starved of glucose or leucine for 3 hours or starved and restimulated with glucose or leucine for 15 minutes. Whole cell lysates were analyzed for the phosphorylation states and levels of the indicated proteins by immunoblot. b, ATF4 loss is associated with increased mTORC1 activity but the pathway remains capable of sensing glucose and leucine in ATF4 KO cells. HEK-293T AMPKα DKO stably expressing Cas9 with either a control guide (AAVS1) or guides against ATF4 were starved of glucose or leucine for 3 hours or starved and restimulated with glucose or leucine for 15 minutes. Whole cell lysates were analyzed for the phosphorylation states and levels of the indicated proteins by immunoblotting. c, Loss of Axin1 does not affect glucose sensing by the mTORC1 pathway or that of AMPK, measured by phosphorylation of Raptor and ACC. Wildtype or AMPKα DKO HEK-293T cells stably expressing Cas9 and either a guide against a control locus (AAVS1) or Axin1 were starved of glucose for 1 hour or starved and restimulated with glucose for 10 minutes. Whole cell lysates were analyzed for the phosphorylation states and levels of the indicated proteins by immunoblotting. d, The RagC-RagB heterodimer but not Axin1 is able to co-immunoprecipitate the Ragulator components c7orf59 and p18. None of the tested proteins were able to co-immunoprecipitate AMPK. Wildtype HEK-293T cells transfected with cDNAs for either HA-tagged metap2, Axin1(N-terminal or C-terminal tags), or HA-RagB/Flag-RagC were starved of glucose for 1 hour or starved and restimulated with glucose for 15 minutes. HA-immunopreciptates and whole cell lysates were analyzed for the levels of the indicated proteins by immunoblotting.
Extended Data Fig. 7 Glucose sensing by mTORC1 requires an intact GATOR/KICSTOR pathway.
a, mTORC1 is insensitive to glucose starvation in Nprl3 KO but not AMPK DKO cells. HEK-293T cells of the indicated genotypes were tested starved of or restimulated with glucose. Protein lysates were analyzed for the phosphorylation states or levels of the indicated proteins. b-c, KICSTOR is required for mTORC1 to sense glucose and leucine. Wildtype HEK-293T cells or with the indicated genotypes were starved of either glucose or leucine for one hour or starved and restimulated with glucose or leucine. d, FLCN is not required for mTORC1 sensing of glucose or leucine starvation. HEK-293T AMPKα DKO or AMPKα/FLCN TKO cells were starved of either glucose or leucine for one hour or starved and restimulated with glucose or leucine for 15 minutes. e, A decrease in Mios expression leads to a concomitant decrease in the activation by glucose of mTORC1. HEK-293T AMPKα DKO stably expressing Cas9 and either a control guide or one targeting Mios were starved of glucose for one hour or starved and restimulated with glucose for 15 minutes. f, Overexpression of the components of GATOR2 prevents glucose or leucine, but not amino acid, starvation from regulating mTORC1. HEK-293T AMPKα DKO co-transfected with cDNAs for FLAG-S6K and either metap2 or all five components of the GATOR2 complex (Mios, WDR24, WDR59, SEC13, SEH1L) were starved of glucose, leucine, or amino acids or were starved and restimulated with glucose, leucine, or amino acids. FLAG-IP were analyzed for the phosphorylation state or levels of the indicated proteins. g-h, Glucose and amino acids regulate mTOR localization to the lysosome consistent with glucose regulating the Rag-GTPase pathway upstream of mTORC1. HEK-293T AMPK DKO cells were starved of amino acids or glucose for 50 minutes or starved for 50 minutes and restimulated with amino acids or glucose for 10 minutes. Cells were fixed and permeabilized and analyzed in an immunofluorescence assay with mTOR and LAMP2 antibodies. Representative images are shown in (g) and quantification of mTOR/LAMP2 colocalization is shown in (h), mean ± s.d. are represented for the indicated number of lysosomes obtained from 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test.
Extended Data Fig. 8 Testing of the role of GPD1/1L/2 in glucose sensing and the reported GAPDH-Rheb and ALDOA-v-ATPase interactions.
a, Loss of GPD1, GPD1L, or GPD2 does not affect glucose sensing by mTORC1. HEK-293T AMPKα DKO stably expressing Cas9 with either a control guide (AAVS1) or guides against GPD1, GPD1L, or GPD2 were starved of glucose for 3 hours or starved for 3 hours and restimulated with glucose for 15 minutes. Whole cell lysates were analyzed for the phosphorylation state and levels of the indicated proteins by immunoblot. b, ALDOA does not co-immunopreciptate the v-ATPase component ATP6V1B2 nor did GAPDH co-immunopreciptate RHEB. HEK-293T AMPKα DKO stably expressing FLAG-metap2, FLAG-ALDOA, or FLAG-GAPDH were starved of glucose or starved and restimulated with glucose. FLAG-IPs or whole cell lysates were analyzed for the levels of the indicated proteins by immunoblotting.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Orozco, J.M., Krawczyk, P.A., Scaria, S.M. et al. Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat Metab 2, 893–901 (2020). https://doi.org/10.1038/s42255-020-0250-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-0250-5
This article is cited by
-
A multidimensional atlas of human glioblastoma-like organoids reveals highly coordinated molecular networks and effective drugs
npj Precision Oncology (2024)
-
Accumulation of G3P and pEtN drives cellular senescence via altered lipid metabolism
Nature Metabolism (2024)
-
Newly discovered roles of triosephosphate isomerase including functions within the nucleus
Molecular Medicine (2023)
-
Multi-omics quantitative data of tomato fruit unveils regulation modes of least variable metabolites
BMC Plant Biology (2023)
-
The molecular basis of nutrient sensing and signalling by mTORC1 in metabolism regulation and disease
Nature Reviews Molecular Cell Biology (2023)