Abstract
White and beige adipocytes in subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT) are maintained by proliferation and differentiation of adipose progenitor cells (APCs). Here we use mice with tissue-specific telomerase reverse transcriptase (TERT) gene knockout (KO), which undergo premature telomere shortening and proliferative senescence in APCs, to investigate the effect of over-nutrition on APC exhaustion and metabolic dysfunction. We find that TERT KO in the Pdgfra+ cell lineage results in adipocyte hypertrophy, inflammation and fibrosis in SAT, while TERT KO in the Pdgfrb+ lineage leads to adipocyte hypertrophy in both SAT and VAT. Systemic insulin resistance is observed in both KO models and is aggravated by a high-fat diet. Analysis of human biopsies demonstrates that telomere shortening in SAT is associated with metabolic disease progression after bariatric surgery. Our data indicate that over-nutrition can promote APC senescence and provide a mechanistic link between ageing, obesity and diabetes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author on request. There are no restrictions on data availability. Raw scRNA-seq data are available from the Gene Expression Omnibus under accession GSE157815. Source data are provided with this paper.
References
Rosen, E. D. & Spiegelman, B. M. What we talk about when we talk about fat. Cell 156, 20–44 (2014).
Chau, Y. Y. et al. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol. 16, 367–375 (2014).
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Porter, C. et al. Human and mouse brown adipose tissue mitochondria have comparable UCP1 function. Cell Metab. 24, 246–255 (2016).
Wu, J. et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376 (2012).
Kajimura, S., Spiegelman, B. M. & Seale, P. Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 22, 546–559 (2015).
Gray, S. L. & Vidal-Puig, A. J. Adipose tissue expandability in the maintenance of metabolic homeostasis. Nutr. Rev. 65, S7–S12 (2007).
Ghaben, A. L. & Scherer, P. E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell. Biol. 20, 242–258 (2019).
Orci, L. et al. Rapid transformation of white adipocytes into fat-oxidizing machines. Proc. Natl Acad. Sci. USA 101, 2058–2063 (2004).
Kim, S. M. et al. Loss of white adipose hyperplastic potential is associated with enhanced susceptibility to insulin resistance. Cell Metab. 20, 1049–1058 (2014).
Hammarstedt, A., Gogg, S., Hedjazifar, S., Nerstedt, A. & Smith, U. Impaired adipogenesis and dysfunctional adipose tissue in human hypertrophic obesity. Physiol. Rev. 98, 1911–1941 (2018).
Kusminski, C. M. et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat. Med. 18, 1539–1549 (2012).
Frazier, T. et al. Gender and age-related cell compositional differences in C57BL/6 murine adipose tissue stromal vascular fraction. Adipocyte 7, 183–189 (2018).
Berry, D. C., Jiang, Y. & Graff, J. M. Emerging roles of adipose progenitor cells in tissue development, homeostasis, expansion and thermogenesis. Trends Endoc. Metab. 27, 574–585 (2016).
Wang, Q. A., Tao, C., Gupta, R. K. & Scherer, P. E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 19, 1338–1344 (2013).
Lee, Y. H., Petkova, A. P., Mottillo, E. P. & Granneman, J. G. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 15, 480–491 (2012).
Gawronska-Kozak, B., Staszkiewicz, J., Gimble, J. M. & Kirk-Ballard, H. Recruitment of fat cell precursors during high-fat diet in C57BL/6J mice is fat depot specific. Obesity 22, 1091–1102 (2014).
Klyde, B. J. & Hirsch, J. Increased cellular proliferation in adipose tissue of adult rats fed a high-fat diet. J. Lipid Res. 20, 705–715 (1979).
Maumus, M. et al. Evidence of in situ proliferation of adult adipose tissue-derived progenitor cells: influence of fat mass microenvironment and growth. J. Clin. Endocrinol. Metab. 93, 4098–4106 (2008).
Jeffery, E., Church, C. D., Holtrup, B., Colman, L. & Rodeheffer, M. S. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol. 17, 376–385 (2015).
Kras, K. M., Hausman, D. B., Hausman, G. J. & Martin, R. J. Adipocyte development is dependent upon stem cell recruitment and proliferation of preadipocytes. Obes. Res. 7, 491–447 (1999).
Merrick, D. et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364, eaav2501 (2019).
Tang, W. et al. White fat progenitor cells reside in the adipose vasculature. Science 322, 583–586 (2008).
Berry, R., Jeffery, E. & Rodeheffer, M. S. Weighing in on adipocyte precursors. Cell Metab. 19, 8–20 (2014).
Rodeheffer, M. S., Birsoy, K. & Friedman, J. M. Identification of white adipocyte progenitor cells in vivo. Cell 135, 240–249 (2008).
Hepler, C. et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal sub-populations in visceral adipose tissue of adult mice. eLife 7, e39636 (2018).
Traktuev, D. et al. A population of multipotent CD34-positive adipose stromal cells share pericyte and mesenchymal surface markers, reside in a periendothelial location, and stabilize endothelial networks. Circ. Res. 102, 77–85 (2008).
Vishvanath, L. et al. Pdgfrβ+ mural preadipocytes contribute to adipocyte hyperplasia induced by high-fat-diet feeding and prolonged cold exposure in adult mice. Cell Metab. 23, 350–359 (2016).
Berry, R. & Rodeheffer, M. S. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 15, 302–308 (2013).
Raajendiran, A. et al. Identification of metabolically distinct adipocyte progenitor cells in human adipose tissues. Cell Rep. 27, 1528–1540 (2019).
Daquinag, A. C., Salameh, A., Zhang, Y., Tong, Q. & Kolonin, M. G. Depletion of white adipocyte progenitors induces beige adipocyte differentiation and suppresses obesity development. Cell Death Differ. 22, 351–363 (2015).
Gao, Z., Daquinag, A. C., Su, F., Snyder, B. & Kolonin, M. G. PDGFRα/PDGFRβ signaling balance modulates progenitor cell differentiation into white and beige adipocytes. Development 145, 1–13 (2018).
Henninger, A. M., Eliasson, B., Jenndahl, L. E. & Hammarstedt, A. Adipocyte hypertrophy, inflammation and fibrosis characterize subcutaneous adipose tissue of healthy, non-obese subjects predisposed to type 2 diabetes. PLoS ONE 9, e105262 (2014).
Xu, M. et al. Targeting senescent cells enhances adipogenesis and metabolic function in old age. eLife 4, e12997 (2015).
Palmer, A. K. et al. Targeting senescent cells alleviates obesity-induced metabolic dysfunction. Aging Cell 18, e12950 (2019).
Berry, D. C. et al. Cellular aging contributes to failure of cold-induced beige adipocyte formation in old mice and humans. Cell Metab. 25, 166–181 (2017).
Gorgoulis, V. et al. Cellular senescence: defining a path forward. Cell 179, 813–827 (2019).
Whittemore, K., Vera, E., Martinez-Nevado, E., Sanpera, C. & Blasco, M. A. Telomere shortening rate predicts species life span. Proc. Natl Acad. Sci. USA 116, 15122–15127 (2019).
Calado, R. T. & Dumitriu, B. Telomere dynamics in mice and humans. Semin. Hematol. 50, 165–174 (2013).
Blasco, M. A. Telomere length, stem cells and aging. Nat. Chem. Biol. 3, 640–649 (2007).
Blackburn, E. H., Greider, C. W. & Szostak, J. W. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12, 1133–1138 (2006).
Welendorf, C. et al. Obesity, weight loss and influence on telomere length: new insights for personalized nutrition. Nutrition 66, 115–121 (2019).
Kipling, D. & Cooke, H. J. Hypervariable ultra-long telomeres in mice. Nature 347, 400–402 (1990).
Sacco, A. et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell 143, 1059–1071 (2010).
Liu, T. et al. Conditional knockout of telomerase reverse transcriptase in mesenchymal cells impairs mouse pulmonary fibrosis. PLoS ONE 10, e0142547 (2015).
Lee, H. W. et al. Essential role of mouse telomerase in highly proliferative organs. Nature 392, 569–574 (1998).
Sahin, E. & Depinho, R. A. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature 464, 520–528 (2010).
Sahin, E. et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365 (2011).
Mender, I. & Shay, J. W. Telomere restriction fragment (TRF) analysis. Bio. Protoc. 5, e1658 (2015).
O’Callaghan, N. J. & Fenech, M. A quantitative PCR method for measuring absolute telomere length. Biol. Proced. Online 13, 3 (2011).
Randle, D. H., Zindy, F., Sherr, C. J. & Roussel, M. F. Differential effects of p19Arf and p16Ink4a loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc. Natl Acad. Sci. USA 98, 9654–9659 (2001).
Capparelli, C. et al. CDK inhibitors (p16/p19/p21) induce senescence and autophagy in cancer-associated fibroblasts, ‘fueling’ tumor growth via paracrine interactions, without an increase in neo-angiogenesis. Cell Cycle 11, 3599–3610 (2012).
Demaria, M. et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014).
Burl, R. B. et al. Deconstructing adipogenesis induced by β3-adrenergic receptor activation with single-cell expression profiling. Cell Metab. 28, 300–309 (2018).
Daquinag, A. C., Zhang, Y., Amaya-Manzanares, F., Simmons, P. J. & Kolonin, M. G. An isoform of decorin is a resistin receptor on the surface of adipose progenitor cells. Cell Stem Cell 9, 74–86 (2011).
Patel, P. & Abate, N. Role of subcutaneous adipose tissue in the pathogenesis of insulin resistance. J. Obes. 2013, 489187 (2013).
Iwayama, T. et al. PDGFRα signaling drives adipose tissue fibrosis by targeting progenitor cell plasticity. Genes Dev. 29, 1106–1119 (2015).
Sun, K., Gao, Z. & Kolonin, M. G. Transient inflammatory signaling promotes beige adipogenesis. Sci. Signal. 11, eaat3192 (2018).
Romaniuk, A. et al. The non-canonical functions of telomerase: to turn off or not to turn off. Mol. Biol. Rep. 46, 1401–1411 (2019).
Stewart, S. A. et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl Acad. Sci. USA 99, 12606–12611 (2002).
Muñoz-Lorente, M. A., Cano-Martin, A. C. & Blasco, M. A. Mice with hyper-long telomeres show less metabolic aging and longer lifespans. Nat. Commun. 10, 4723 (2019).
Karastergiou, K. & Fried, S. K. Cellular mechanisms driving sex differences in adipose tissue biology and body shape in humans and mouse models. Adv. Exp. Med. Biol. 1043, 29–51 (2017).
Nordstrom, A., Hadrevi, J., Olsson, T., Franks, P. W. & Nordstrom, P. Higher prevalence of type 2 diabetes in men than in women is associated with differences in visceral fat mass. J. Clin. Endoc. Metab. 101, 3740–3746 (2016).
Cherif, H., Tarry, J. L., Ozanne, S. E. & Hales, C. N. Ageing and telomeres: a study into organ- and gender-specific telomere shortening. Nuc. Acids Res. 31, 1576–1583 (2003).
Wang, J. et al. Association between telomere length and diabetes mellitus: a meta-analysis. J. Int. Med. Res. 44, 1156–1173 (2016).
Elks, C. E. & Scott, R. A. The long and short of telomere length and diabetes. Diabetes 63, 65–67 (2014).
Mundstock, E. et al. Effect of obesity on telomere length: systematic review and meta-analysis. Obesity 23, 2165–2174 (2015).
Kirchner, H. et al. The telomeric complex and metabolic disease. Genes 8, 176 (2017).
Gielen, M. et al. Body mass index is negatively associated with telomere length: a collaborative cross-sectional meta-analysis of 87 observational studies. Am. J. Clin. Nutr. 108, 453–475 (2018).
Lakowa, N. et al. Telomere length differences between subcutaneous and visceral adipose tissue in humans. Biochem. Biophys. Res. Commun. 457, 426–432 (2015).
Gustafson, B., Nerstedt, A. & Smith, U. Reduced subcutaneous adipogenesis in human hypertrophic obesity is linked to senescent precursor cells. Nat. Commun. 10, 2757 (2019).
Joe, A. W., Yi, L., Even, Y., Vogl, A. W. & Rossi, F. M. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells 27, 2563–2570 (2009).
Cuttler, A. S. et al. Characterization of Pdgfrb-Cre transgenic mice reveals reduction of ROSA26 reporter activity in remodeling arteries. Genesis 49, 673–680 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies and species. Nat. Biotechnol. 36, 411–420 (2018).
Acknowledgements
We thank S. H. Pan for the TERT floxed mice and V. Lindner for Pdgfrb-Cre mice. We thank Z. Mao for help with microscopy. We also thank A. Ribas-Latre and E. Sahin for expert advice. This work was supported by the Harry E. Bovay, Jr. Foundation. Z.Z. and Y.D. were partially supported by the Cancer Genomics Core funded by the Cancer Prevention and Research Institute of Texas (CPRIT) (RP180734) and National Institutes of Health grant (R01LM012806).
Author information
Authors and Affiliations
Contributions
M.G.K. and Z.G. conceived/designed experiments; Z.G., A.C.D., C.F., K.M., Z.Z. and Y.D. performed experiments and analysed data. M.G.K., Z.G. and K.L.E.-M. wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary handling editor: Christoph Schmitt. Nature Metabolism thanks Thomas von Zglinicki and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
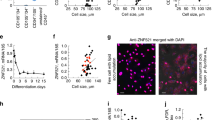
Extended Data Fig. 1 Telomere attrition / cell senescence in WAT of a-KO mice.
a, TRF assay reveals advanced telomere shortening in SAT of both a-KO males (M) and females (F) at 8 months of age. b, Telo-FISH staining (red) on sections from Pdgfra-Cre;mTmG;TERTfl/fl (WT) and Pdgfra-Cre;mTmG;TERTfl/fl male mice reveals normal telomeres in mG+ cells of brain and muscle. c, Telo-FISH staining on culture-plated cells from VAT and SAT of control Pdgfra-Cre; mTmG and Pdgfra-Cre; mTmG; TERTfl/fl mice reveals advanced telomere shortening (less red signal) in SAT mG+ cells of a-KO mice. d, Telo-FISH staining on SAT sections from mice in c reveals telomere shortening and nuclear p16 IF signal in mG+ cells of a-KO mice. e, q-PCR reveals telomere shortening in SAT of 8-month old WT mice compared to 3-month old WT mice. Evident is more advanced telomere shortening in SAT of 8-month old α-KO males (M) and females (F), compared to 8-month old WT males and females, respectively. f, Quantitative RT-PCR analysis reveals higher p16 and p21 mRNA expression in VAT and SAT of a-KO mice, compared to WT mice, at 6 months of age. Data are normalized to 18S RNA. N = 3 mice / group. Scale bar=50 µm in all panels. P: Student’s t-test (2-sided).
Extended Data Fig. 2 Transient AT browning in young Pdgfra-TERT-KO (a-KO) mice.
a, Quantitative RT-PCR demonstrates higher Pdgfa mRNA expression in VAT and SAT of 6 month old KO males, compared to 6 month old WT males. Data are normalized to 18S RNA. b, Quantitative RT-PCR demonstrates higher Pdgfa mRNA expression in SVF from SAT of 3 month-old KO males, which is not accompanied by increased p16 and p21 expression at that age. In contrast, in SVF from SAT of 7 month-old KO males, higher Pdgfa expression is accompanied by increased p16 and p21 mRNA expression. Data are normalized to 18S RNA. N = 3 mice / group. P: Student’s t-test (2-sided).
Extended Data Fig. 3 Metabolism impairment in old Pdgfra-TERT-KO (a-KO) mice raised on chow.
a, qPCR analysis of DNA from whole tissue reveals telomere shortening in SAT but not in VAT of a-KO mice at 8 months of age, compared to WT mice at 8 months of age. Real-time PCR data are normalized to data for a single copy gene. b, Quantitative RT-PCR demonstrates higher p16 and TNFa mRNA expression, normalized to 18S RNA, in cultured MEFs of a-KO mice, compared to WT mice. c, EdU incorporation measurement by flow cytometry in control mTmG;TERTfl/fl and a-KO Pdgfra-Cre;mTmG; TERTfl/fl males (2-month-old, HFD-fed) and females (20-month-old, chow-fed). 4 h after EdU injection, mG+ and mT+ SVF cells were gated and EdU incorporation was compared for SAT. EdU fluorescence: 647 nm channel, side scatter (SSC) is used for separation. d, IF analysis of SAT from 12-month-old mice reveals increased infiltration of CD68+ CD206- (M1) macrophages and increased phosphorylation of NFkB subunit p65 in PDGFRα+ cells of a-KO mice, compared to WT mice. Adipocytes are positive for perilipin1 (PLN1). Nuclei are blue. e, Masson’s trichrome staining reveals increased fibrosis in livers of 12-month-old a-KO males compared to WT males. In all panels, N = 3 mice / group; P: Student’s t-test (2-sided). Scale bar=50 µm in all panels.
Extended Data Fig. 4 HFD exacerbates metabolism impairment in male Pdgfra-TERT-KO (a-KO) mice.
a, Echo MRI data demonstrating comparable fat mass in a-KO and WT males raised on HFD for indicated number of months. N = 5-6 mice / group. b, Comparable food intake by a-KO and WT males raised on HFD. N = 5-6 mice / group. c, Comparable spontaneous locomotor activity of a-KO and WT males raised on HFD. N = 5-6 mice / group. d, Comparable physical endurance, reflected by Joules of work performed, by a-KO and WT males. N = 5-6 mice / group. e, Indirect calorimetry based on oxygen consumption (VO2) and carbon dioxide production (VCO2) indicates decreased energy expenditure in a-KO males pre-fed HFD for 8 months. N = 5-6 mice / group. f, Masson’s trichrome staining of VAT from a-KO and WT males raised on HFD for 8 months. g, Telo-FISH staining (red) on SAT and VAT sections from Pdgfra-Cre;mTmG mice raised on chow or HFD revealing a higher degree of telomere shortening (less red signal) induced by DIO in mG+ cells. a-g, N = 5-6 mice / group. Scale bar=50 µm in all panels. P: Student’s t-test (2-sided).
Extended Data Fig. 5 Pdgfra-TERT-KO (a-KO) females are protected from metabolism impairment.
a, Echo MRI data demonstrating lower fat mass in a-KO females raised on HFD for indicated number of months. N = 5-6 mice / group. b, H&E staining demonstrates smaller BAT adipocytes and normal liver and muscle anatomy in a-KO females raised on HFD. Scale bar=50 µm. c, Quantitative RT-PCR demonstrates a lack of p16 induction, despite higher Pdgfa mRNA expression, in SAT of 4-month old a-KO females, compared to 4 month old WT females. Data normalized to 18S RNA. N = 5-6 mice / group. P: Student’s t-test (2-sided).
Extended Data Fig. 6 Senescence and metabolism in Pdgfrb-TERT-KO (b-KO) mice.
a, Quantitative RT-PCR demonstrates higher senescence marker and pdgfa induction in VAT than in SAT of 8-month old b-KO males compared to WT males. Data normalized to 18S RNA. N = 5 mice / group. b, Comparable food intake by b-KO and WT males. c, Comparable spontaneous locomotor activity of b-KO and WT males. d, RER in males calculated based on oxygen consumption (VO2) and carbon dioxide production (VCO2). e, Indirect calorimetry-based measurement of energy expenditure in b-KO and WT males at 8 months of age. f, Comparable cold tolerance in b-KO and WT males placed at 4 °C. g, H&E staining does not reveal abnormality in BAT and liver of b-KO males at 8 months of age. Scale bar=50 µm. h, Echo MRI data: no body composition abnormality in b-KO females at 8 months of age. i, ITT: no insulin resistance in 8-month-old b-KO females. j, Comparable cold tolerance in b-KO and WT females placed at 4 °C. N = 6 mice / group. P: Student’s t-test (2-sided).
Extended Data Fig. 7 Gene expression in ASC of Pdgfra-TERT-KO (a-KO) mice.
a, ScRNAseq data violin plots showing that Pdgfra is expressed in both Dpp4+ and Cd142+ cells, while Pdgfrb is mainly expressed in Cd142+ cells. b, UMAPs of WT, a-KO and b-KO SAT CD45- SVF cells from Fig. 7c were overlapped to identify cells expressing genes assigned in Fig. 7d. N = 4 mice / group (combined). c, Expression of Cdkn1a and Pdgfa determined by scRNAseq for combined Dpp4+ and Cd142+ ASC compared for WT, a-KO, and b-KO mice.
Extended Data Fig. 8 Changes in AT stroma resulting from senescence of Pdgfra and Pdgfrb lineages.
a, IF analysis of SAT reveals increased infiltration of PDGFRα+ (orange, anti-rat Cy5 secondary antibody) and PDGFRb+ (red, anti-rabbit Cy3 secondary antibody) cells in a-KO and b-KO 1-year-old males raised on chow compared to WT male littermates. Isolectin B4 (green) stains endothelium. Nuclei are blue. Scale bar=50 µm. Graph: the ratio of positive cells / total nucleated cells in 4-5 view fields / sample. Plotted are mean +/- SEM. P: Student’s t-test (2-sided). Analysis of tissues from 3 mice / genotype showed similar results. b, Changes in AT stroma resulting from senescence of Pdgfra and Pdgfrb lineages in VAT. UMAP clusters of cells identified by scRNAseq in VAT of TERT a-KO and b-KO mice. Shown are ASC, EC and PA only, CD45+ leukocytes are not shown. Note selective accumulation of Cd142+ ASC in VAT of b-KO mice. Note high expression of Cdkn1a and IL-6 and low expression on Ki67 in ASC.
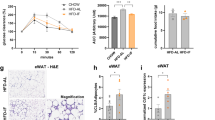
Extended Data Fig. 9 Consequences of pharmacological adipocyte progenitor depletion in mice treated with D-WAT.
WT males pre-treated with D-WAT or PBS (control) at 4 months of age were subsequently maintained on HFD and analyzed 9 months later. a, Echo MRI data reveals higher adiposity in old mice pre-treated with D-WAT. b, H&E staining reveals larger adipocytes and fibrosis (arrow) in SAT of old mice pre-treated with D-WAT. Scale bar=50 µm. c, GTT reveals lower glucose tolerance of old mice pre-treated with D-WAT. N = 4 mice / group. *P < 0.05 (Student’s t-test, 2-sided).
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Supplementary Fig. 1.
Source data
Source Data Fig. 1
Uncropped gel for Fig. 1c.
Source Data Fig. 3
Uncropped immunoblots for Fig. 3h.
Rights and permissions
About this article
Cite this article
Gao, Z., Daquinag, A.C., Fussell, C. et al. Age-associated telomere attrition in adipocyte progenitors predisposes to metabolic disease. Nat Metab 2, 1482–1497 (2020). https://doi.org/10.1038/s42255-020-00320-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-00320-4
This article is cited by
-
WRN loss accelerates abnormal adipocyte metabolism in Werner syndrome
Cell & Bioscience (2024)
-
The mediating role of telomere length in multi-pollutant exposure associated with metabolic syndrome in adults
Environmental Science and Pollution Research (2023)
-
Secretion of miRNA-326-3p by senescent adipose exacerbates myocardial metabolism in diabetic mice
Journal of Translational Medicine (2022)
-
Telomere dysfunction in ageing and age-related diseases
Nature Cell Biology (2022)
-
Distinct functional properties of murine perinatal and adult adipose progenitor subpopulations
Nature Metabolism (2022)