Abstract
The ten-eleven-translocation family of proteins (TET1/2/3) are epigenetic regulators of gene expression. They regulate genes by promoting DNA demethylation (i.e., catalytic activity) and by partnering with regulatory proteins (i.e., non-catalytic functions). Unlike Tet1 and Tet2, Tet3 is not expressed in mouse embryonic stem cells (ESCs) but is induced upon ESC differentiation. However, the significance of its dual roles in lineage specification is less defined. By generating TET3 catalytic-mutant (Tet3m/m) and knockout (Tet3–/–) mouse ESCs and differentiating them to neuroectoderm (NE), we identify distinct catalytic-dependent and independent roles of TET3 in NE specification. We find that the catalytic activity of TET3 is important for activation of neural genes while its non-catalytic functions are involved in suppressing mesodermal programs. Interestingly, the vast majority of differentially methylated regions (DMRs) in Tet3m/m and Tet3–/– NE cells are hypomethylated. The hypo-DMRs are associated to aberrantly upregulated genes while the hyper-DMRs are linked to downregulated neural genes. We find the maintenance methyltransferase Dnmt1 as a direct target of TET3, which is downregulated in TET3-deficient NE cells and may contribute to the increased DNA hypomethylation. Our findings establish that the catalytic-dependent and -independent roles of TET3 have distinct contributions to NE specification with potential implications in development.
Similar content being viewed by others
Introduction
The ten-eleven translocation family of enzymes (TET1/2/3) are DNA dioxygenases that are essential for embryonic development. They catalyze DNA demethylation through successive oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC)1,2,3. These bases can be excised by thymine DNA glycosylase (TDG) and removed through base excision repair pathways completing active DNA demethylation3,4,5. 5hmC is a stable base and is recognized by other proteins suggesting it may have independent regulatory roles6. It can evade recognition by UHRF1 and thus interfere with recruitment of DNMT1 to promote passive DNA demethylation during replication6,7.
TET enzymes are dynamically expressed during embryogenesis and play essential roles in gene regulation during development and in embryonic stem cell (ESC) biology8,9. Tet1 and Tet2 are highly expressed in ESCs, whereas Tet3 is not detectable10. While Tet1/2 levels decline during ESC differentiation, Tet3 is induced9,11. Loss of TET1 or TET2 in ESCs does not block pluripotency but compromises expression of lineage specific genes and ESC differentiation12,13,14. TET1 and TET2 deficient mice are viable, whereas TET3 loss results in perinatal lethality with no overt developmental defects13,15,16. TET1/3 double knockout embryos are embryonic lethal by E10.5 with impaired forebrain development and increased transcriptome variability17. TET1/2/3 triple knockout (TKO) ESCs have severe differentiation defects consistent with TET TKO embryos failing to complete gastrulation and die by E8.510,16. These finding support TET enzymes have both redundant and unique roles in embryogenesis.
Tet3 is uniquely expressed in the zygote where it is required for promoting DNA demethylation of the paternal genome16,18,19,20,21. Deletion of TET3 in oocytes causes developmental defects and lethality by mid-gestation16,22. Unlike Tet1/2, Tet3 is not expressed in the inner cell mass of the blastocyst nor in ESCs, but is induced during ESC differentiation and post-implantation development11. However, its roles in post-implantation development are less defined. TET3 has been shown to be important for development of some somatic cell types such as immune cells23,24, neural stem cells11,25,26,27 and cardiomyocytes28, and is also deregulated in some cancers29,30. TET3 mutations have also been implicated in human developmental disorders with all patients showing craniofacial abnormalities, and few with cardiovascular, motor, and speech deficits31,32. These studies underscore the need to understand the requirement of TET3 during development.
Over recent years, our lab and others have shown that TET enzymes can regulate gene expression independent of their enzymatic activity, which entails partnering with or recruitment of other transcription factors and epigenetic modifiers to chromatin5,12,14,33,34,35,36. Specifically, we and others have shown that in ESCs, TET1 recruits PRC2 and SIN3A to promoters of bivalent genes for H3K27 trimethylation and deacetylation, respectively12,33,34,37. Additionally, in ESCs, we have shown that TET2 recruits SIN3A to active enhancers of its target genes independent of its catalytic activity14. Further, in hematopoietic stem cells, TET2 non-catalytic functions are important for regulating the lymphoid lineage and preventing lymphoid malignancies35. The non-catalytic functions of TET3 are less defined and studies have been limited to lineage committed cell types. In immune cells, TET3 partnership with HDAC1 is critical for eliciting an immune response following viral infection24. Likewise, TET3 non-catalytic functions are important in astrocyte specification from neural stem cells (NSCs)25,26. However, the catalytic and non-catalytic functions of TET3 during early lineage specification have not been investigated. Here, we have used TET3 catalytic deficient and knockout mouse ESCs to study how TET3 regulates genes during ESC differentiation to neuroectoderm (NE), a lineage where Tet3 is highly expressed. We have identified distinct catalytic dependent and independent roles of TET3 in activation of neural and suppression of mesodermal programs as ESCs commit to NE. These findings not only define the roles of TET3 in lineage specification but may also have potential implications in neurodevelopment where TET3 is mutated or dysregulated.
Results
Generation of TET3 catalytic mutant (Tet3 m/m) and knockout (Tet3 –/–) mouse ESCs and their differentiation to neuroectoderm (NE)
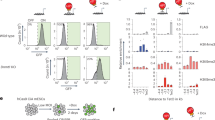
To define the catalytic and non-catalytic functions of TET3, we generated TET3 catalytic deficient (Tet3m/m) and knockout (Tet3–/–) mouse ESCs using CRISPR/Cas9 mediated gene editing. To generate TET3 catalytic deficient ESCs, we used a guide RNA (gRNA) targeting exon 9 and a donor oligo coding for amino acid substitutions H950Y and D952A in the iron binding site of TET3 as well as a HaeIII restriction site to allow for screening of targeted clones by restriction fragment length polymorphism (RFLP) (Fig. 1a). The amino acids H950 and D952 are conserved among all TET enzymes and their respective substitutions to H950Y and D952A is shown to abrogate the catalytic activity of TET31,24,26,38. To generate Tet3–/– ESCs, two gRNA flanking exon 4 were used to delete the exon (Fig. 1a), which leads to an out of frame transcript with an immediate stop codon resulting in complete loss of TET3 as shown before10. Properly targeted Tet3m/m ESC clones (n = 3) were validated by RFLP and Sanger sequencing (Fig. 1b, c) and deletion of exon 4 in Tet3–/– ESC clones (n = 3) was confirmed by PCR (Fig. 1b). Tet3m/m and Tet3–/– ESCs maintained normal morphology in culture (Fig. 1d) and remained pluripotent forming teratomas with structures of the three germ layers (Fig. 1e) consistent with previous reports that loss of TET3 does not overtly effect pluripotency.
a Schematic of the gene targeting strategy to generate catalytic mutant (Tet3m) and knockout (Tet3–) mouse ESCs. b Genotyping of Tet3m/m clones by RFLP (restriction fragment length polymorphism) using HaeIII enzyme (top). Correctly targeted (mutated) allele bands are 138 bp and 116 bp. Allele without mutation is 354 bp. (3 independent clones were generated A12, B2, and H1). Genotyping of Tet3–/– clones by PCR (bottom). Amplification of a shorter fragment (~500 bps) confirms deletion of exon 4 (3 independent clones were generated E10, H4, and H5). c Sanger sequencing confirming mutations for amino acid substitutions HKD to YKA in Tet3m/m ESCs. d Representative brightfield images of ESCs cultured on feeder cells. Scale bar = 100μm. e Hematoxylin and Eosin (H&E) staining of sections of teratomas derived from ESCs of indicated genotypes. Scale bars = 400 μm. f Representative brightfield images of Tet3+/+, Tet3m/m, and Tet3–/– ESCs on gelatin (top) and after differentiation to NE by LIF withdrawal and RA treatment for 72 h (bottom). Scale bar = 100 μm. g Quantification of Tet1/2/3 mRNA levels at day 0 and 3 of –LIF + RA treatment. Data normalized to Gapdh expression. h Growth curve of ESCs differentiated to NE. Viable cells were counted during each day of differentiation. i Cell cycle analysis of NE cells at day 3 of –LIF + RA treatment. Note Tet3–/–, but not Tet3m/m, cells show a reduction of cells in S phase. For all panels error bars represent SD and * indicates statistical significance (p < 0.05).
Tet3 is not expressed in ESCs but is induced upon differentiation and is highly expressed in the neural lineage. Therefore, we differentiated Tet3+/+, Tet3m/m, and Tet3–/– ESCs to neuroectoderm (NE) by LIF withdrawal and retinoic acid (RA) treatment for three days39. Tet3m/m and Tet3–/– NE cells were morphologically indistinguishable from their wildtype counterparts (Fig. 1f). Expression of Tet3 mutant mRNA in Tet3m/m and loss of exon 4 containing transcripts in Tet3–/– NE cells were confirmed by RT-qPCR (Fig. 1g). We note that there was a reduction in mRNA levels of mutant Tet3 suggesting some instability of this transcript. We also confirmed that Tet1 and Tet2 transcript levels were not affected in Tet3m/m or Tet3–/– ESCs, and both were downregulated upon ESC differentiation (Fig. 1g). While growth of Tet3m/m or Tet3–/– ESCs were unaffected, upon differentiation to NE, we found a subtle trend in reduced proliferation of NE cells which reached significance at day 2 (Fig. 1h). Cell cycle analysis of NE cells confirmed a mild increase in G1 and decrease in S phase cells in Tet3–/–, but not in Tet3m/m, cultures (Fig. 1i) suggesting that Tet3 non-catalytic functions may subtly influence cell cycle progression of NE cells. Since Tet3m/m cells only lack the catalytic activity of TET3, their comparisons to Tet3–/– cells allows for dissecting the catalytic and non-catalytic functions of TET3 in NE specification.
TET3 promotes activation of neural genes and suppression of mesodermal genes through catalytic and non-catalytic functions, respectively
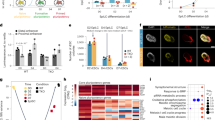
To identify catalytic and non-catalytic target genes of TET3, we compared the transcriptomes of Tet3+/+, Tet3m/m, and Tet3–/– NE cells (harvested 72 h after LIF withdrawal and RA treatment) by RNAseq (Supplementary Fig. 1a). Sample clustering by Euclidean distance revealed that the biological replicates clustered with each other (Supplementary Fig. 1b), and principal component analysis (PCA) confirmed they were separated by genotype (Supplementary Fig. 1c). We identified a total of 3,184 differentially expressed genes (DEGs) between Tet3+/+, Tet3m/m, and Tet3–/– cells showing distinct expression profiles in each genotype (Fig. 2a). The expression of Tet1 and Tet2 was not affected in Tet3m/m and Tet3–/– cells (Supplementary Fig. 1d) which is consistent with our earlier quantification of Tet mRNA levels by RT-qPCR in NE cells (Fig. 1g). Comparison of gene expression of Tet3m/m vs. Tet3+/+ cells identified 2402 DEGs, with nearly equal number of DEGs up- (1306, 54%) and down- (1096, 46%) regulated (Fig. 2b, c). In contrast, comparison of gene expression of Tet3–/– vs. Tet3+/+ revealed 1071 DEGs where 68% of the DEGs (727) were upregulated and 32% (344) were downregulated, indicating a role for TET3 in gene silencing (Fig. 2b, c). To distinguish the DEGs impacted by the catalytic vs. non-catalytic functions of TET3, we overlapped DEGs in Tet3m/m and Tet3–/– cells (Fig. 2d). Genes deregulated in Tet3m/m cells were influenced by the catalytic activity of Tet3 (i.e., catalytic target genes), while the 749 uniquely deregulated genes in Tet3–/– cells were genes impacted by non-catalytic roles of TET3 (i.e., non-catalytic target genes) with most of them being upregulated (Fig. 2d). Gene ontology (GO) analysis revealed that the catalytic DEGs were enriched in neural pathways while the non-catalytic DEGs were enriched in mesodermal pathways (Fig. 2e), suggesting that the catalytic and non-catalytic activities of TET3 in NE are involved in controlling the neural and mesoderm programs, respectively. Consistently, key neural transcription factors including Sox1, Neurog2, and Hes5 were downregulated in both Tet3m/m and Tet3–/– cells while key mesodermal genes such as Gata6, Mcam, and Bin1 were aberrantly upregulated in Tet3–/–, but not in Tet3m/m, cells (Fig. 2f and Supplementary Fig. 1e). We also found an increase in expression of cyclin dependent kinase inhibitors p15, p16, and p21 only in Tet3–/– cells (Supplementary Fig. 1f) which is in line with the mild delay in cell cycle progression of these cells (Fig. 1i). Together, these findings support that the catalytic and non-catalytic roles of TET3 activate neural and suppress mesodermal programs, respectively, during specification of NE from ESCs. To confirm that the gene expression changes observed in Tet3m/m and Tet3–/– NE cells are due to loss of TET3, we re-expressed Tet3 in Tet3m/m and Tet3–/– cells during differentiation to NE. This ameliorated the expression of both aberrantly upregulated and downregulated genes in these cells (Supplementary Fig. 1g), including key mesodermal genes such as Hand1 and neural genes such as Sox5, supporting that TET3 is responsible for activation and suppression of these genes during NE specification.
a Heatmap of all 3184 differentially expressed genes (DEGs, fold changes > 1.5, FDR < 0.05) in Tet3m/m and Tet3–/– cells on day 3 of –LIF + RA. Color represents relative expression extracted from normalized counts. (Three independent Tet3+/+ and Tet3m/m clones and two independent Tet3–/– clones were used for this analysis). b Up and downregulated DEGs between Tet3+/+ vs Tet3m/m (top) and Tet3+/+ vs Tet3–/– (bottom). c Volcano plots showing DEGs between Tet3+/+ vs Tet3m/m (top) and Tet3+/+ vs Tet3–/– (bottom). Some downregulated neural genes and upregulated mesodermal genes are noted. d Venn diagram (left) showing the overlap of DEGs between Tet3m/m and Tet3–/–. The percent of up and downregulated genes in each category is plotted (right). Genes deregulated in Tet3m/m are catalytic targets of TET3. Genes uniquely deregulated in Tet3–/–are TET3 non-catalytic targets. e Gene Ontology (GO) analysis of all catalytic target genes (top) and non-catalytic target genes (bottom). Line represents significance (p < 0.05). f Heatmap of selected catalytic target neural genes that are downregulated in both Tet3m/m and Tet3–/– and selected non-catalytic target mesodermal genes uniquely upregulated in Tet3–/– cells.
Hyper-DMRs are associated with downregulated neural genes while hypo-DMRs are associated with aberrantly upregulated genes in Tet3 –/– NE cells
TET3 promotes DNA demethylation, therefore, we analyzed the genome-wide DNA methylation landscape of NE cells (day 3 of –LIF + RA) by whole genome bisulfite sequencing (WGBS) (Supplementary Fig. 2a). As expected, global DNA methylation levels were mildly increased in both Tet3m/m (73.7%) and Tet3–/– (73.4%) compared to Tet3+/+ (72.7%) cells at CpG dinucleotides (Fig. 3a). Consistently, we identified ~9000 DMRs in both cell types. However, to our surprise, the vast majority of the DMRs were hypomethylated. Of the 9,222 DMRs in Tet3m/m cells 626 were hyper- and 8596 were hypo-methylated. Likewise of the 9,335 DMRs in Tet3–/– cells 571 were hyper- and 8,764 were hypo-methylated (Fig. 3b). Methylation levels were sharply affected at the center of the DMRs (Fig. 3c) and there was a substantial overlap of hyper- or hypo- DMRs between Tet3m/m and Tet3–/– cells (Supplementary Fig. 2b, c). The hyper-DMRs were associated to 544 genes in Tet3m/m and 485 genes in Tet3–/– cells, with 270 genes common between them (Supplementary Fig. 2b). The hypo-DMRs were associated to 5,989 genes in Tet3m/m and 5,828 genes in Tet3–/– cells, with 4283 genes common between them (Supplementary Fig. 2c). Both hyper- and hypo-DMRs were comparably represented throughout the genome at promoters, gene bodies and distal intergenic regions (Fig. 3d). Hyper-DMRs were significantly enriched for downregulated catalytic target genes (Tet3m/m DEGs) consistent with the role of TET enzymes in DNA demethylation and gene activation (Fig. 3e). Additionally, hypo-DMR associated genes were significantly enriched for upregulated catalytic and non-catalytic (Tet3–/– only) DEGs (Fig. 3e). These findings suggest that both Tet3 catalytic and non-catalytic mechanisms may regulate DNA methylation dynamics during ESC differentiation.
a Percent methylated CpG sites genome-wide in NE cells of indicated genotypes by Whole Genome Bisulfite Sequencing (WGBS). b Hyper- and hypo- differentially methylated regions (DMRs = >5 CpGs, methylation difference >20%, and FDR < 0.05) in Tet3m/m and Tet3–/– (compared to Tet3+/+) NE cells. c DNA methylation levels at all hyper-DMRs (n = 924) and all hypo-DMRs (n = 12,050) in Tet3m/m and Tet3–/– cells. d Annotation of hyper- and hypo-DMRs to genomic regions. e Percent of catalytic and non-catalytic DEGs that overlap with all hyper- and hypo-DMRs in both Tet3m/m and Tet3–/– NE cells. * Hypergeometric test p-value < 0.05.
To investigate how DNA hypermethylation impacted gene expression, we overlapped all hyper-DMRs (n = 924) with all TET3 catalytic target genes (Tet3m/m DEGs). We identified 104 hyper-DMR containing catalytic DEGs (Fig. 4a) that were associated with developmental processes including neural differentiation, pattern specification, and transcription (Fig. 4b). This included key neural genes such as Neurog2 and Sox1 (Fig. 4c) that are normally upregulated during differentiation of ESCs to NE but were significantly downregulated in both Tet3m/m and Tet3–/– cells (Fig. 2f). This is consistent with Tet3 activating neural genes through its catalytic activity of DNA demethylation.
a Heatmap of all TET3 catalytic target genes (n = 104) associated with hyper-DMRs. b GO analysis of hyper-DMR containing TET3 catalytic target genes revealing enrichment for neural terms. Line represents significance (p < 0.05). c IGV tracks of DNA methylation levels at Neurog2 and Sox1 loci (both downregulated in Tet3m/m and Tet3–/–). Hyper-DMRs are highlighted in orange. d Heatmap of all TET3 non-catalytic target genes (n = 239) associated with hypo-DMRs. e GO analysis of hypo-DMR-containing TET3 non-catalytic target genes revealing enrichment for mesodermal signaling pathways. Line represents significance (p < 0.05). f IGV tracks of DNA methylation levels at Wnt10a and Myl9 loci (both upregulated in Tet3–/– but not in Tet3m/m NEs). Hypo-DMRs are highlighted in orange.
To determine how DNA hypomethylation influenced gene expression, we overlapped all hypo-DMRs identified with TET3 non-catalytic target genes (DEGs unique to Tet3–/–). We identified 239 hypo-DMR containing genes that were mostly upregulated in Tet3–/– cells (Fig. 4d). They were associated with signaling pathways such as Wnt and mesodermal processes such as heart development (Fig. 4e) and included key mesoderm genes like Wnt10a and Myl9 (Fig. 4f), which were upregulated in Tet3–/– cells. This suggests that TET3 influences DNA methylation dynamics leading to aberrant upregulation of Wnt signaling and mesodermal programs.
To examine changes in 5hmC levels in Tet3m/m and Tet3–/– NE cells, we mapped the genomic distribution of 5hmC by hmeDIP in these cells. Majority of 5hmC was at gene bodies and distal intergenic regions followed by promoters. 5hmC distribution was slightly shifted in Tet3m/m and Tet3–/– NE cells with less 5hmC being at promoters (Supplementary Fig. 3a), which is in agreement with the majority of DMRs being at promoters (Fig. 3d). Importantly, consistent with a role for TET3 in DNA hydroxylation the overall 5hmC levels at 5hmC-enriched regions was reduced in Tet3m/m and Tet3–/– cells (Supplementary Fig. 3b). This was very notable at gene bodies and distal intergenic regions (Supplementary Fig. 3c, d). We also found that downregulated genes in TET3 deficient NE cells had remarkably reduced levels of 5hmC (Supplementary Fig. 3e). These included several direct targets of TET3 such as Axin2 and Pdik1l, which were not only bound by TET3 in wild type cells but also had reduced levels of 5hmC and were downregulated in Tet3m/m and Tet3–/– cells (Supplementary Fig. 3e, f). This data supports that TET3 drives deposition of 5hmC and activation of its direct target genes.
The maintenance DNA methyltransferase Dnmt1 is a direct target of TET3 in NE cells
To determine genes directly bound by TET3 in NE cells, we mapped the genomic occupancy of TET3 by CUT&Tag. We identified 1167 peaks genome wide and the majority (92.7%) of them were associated to promoters (+/– 2 kb of TSS; Fig. 5a). The peaks identified annotated to 1,134 genes which were enriched for regulators of transcription, chromatin organization, and cell cycle (Fig. 5b). Because of the low number of peaks identified, we observed only a small overlap of TET3-bound genes to DEGs and hyper- or hypo-DMRs in Tet3m/m or Tet3–/– cells (Supplementary Fig. 4a–c). Consistent with a role for TET3 in DNA hydroxylation, we also found that 5hmC levels were reduced at all TET3 bound regions, especially at TET3 bound promoters (Supplementary Fig. 4d, Supplementary Fig. 3f). Among the genes bound by TET3, we found the maintenance methyltransferase Dnmt1 as a direct target of TET3. Specifically, we found that TET3 binds at the promoter of Dnmt1 at a region that does not have changes in DNA methylation and hydroxymethylation, suggesting that TET3 might regulate Dnmt1 independent of DNA methylation (Fig. 5c). We validated TET3 binding at the Dnmt1 promoter by ChIP-qPCR where TET3 was enriched in Tet3+/+, but not in in the negative control Tet3–/– cells, confirming the specificity of this enrichment (Fig. 5d). Dnmt1 was downregulated in Tet3m/m and Tet3–/– NE cells (Fig. 5e) while the levels of the de novo methyltransferases Dnmt3a and Dnmt3b were unaffected (Supplementary Fig. 4e). Reduced Dnmt1 levels can influence DNA methylation maintenance dynamics during ESC differentiation and may be responsible for the increased occurrence of hypo-DMRs at the aberrantly upregulated genes in Tet3m/m or Tet3–/– cells. To examine if Dnmt1 can restore proper expression of upregulated genes, we expressed Dnmt1 in Tet3m/m and Tet3–/– NE cells (Fig. 5f) and analyzed gene expression by RNA-seq. We found that Dnmt1 overexpression ameliorated the aberrant upregulation of genes in Tet3m/m and Tet3–/– NE cells (Fig. 5g). This included expression of several mesodermal genes such as Hand1 and Hrc (Fig. 5g), which were also corrected by re-expression of Tet3 (Supplementary Fig. 1g). This supports that Dnmt1, as a direct target of TET3, contributes to repressing some mesodermal programs during NE specification.
a Genomic distribution of 1167 peaks identified by CUT&Tag in Tet3+/+ NE cells. Note, vast majority of the peaks are at promoters. b GO analysis of TET3-bound genes. c IGV tracks showing TET3 CUT&Tag peaks and DNA methylation levels at Dnmt1 promoter region. d TET3 enrichment at the Dnmt1 promoter quantified by ChIPqPCR. Data normalized to IgG. e Quantification of Dnmt1 mRNA levels in NE cells (day 3 of –LIF + RA) by RT-qPCR. f Quantification of Dnmt1 by RT-qPCR in NE cells expressing either empty vector (EV) or Dnmt1. g Heatmap of upregulated genes in Tet3m/m and Tet3–/– NE cells where their expression is corrected by overexpression of Dnmt1. For all panels error bars represent SD and * p < 0.05 **p < 0.005.
Discussion
Tet3, unlike Tet1 and Tet2, is not expressed in ESCs but is induced upon differentiation. While the catalytic and non-catalytic functions of TET1 and TET2 in ESC biology have been investigated by us and others recently12,14,35,37, these dual roles of TET3 in lineage specification have not been well defined. Here, we provide evidence in support of catalytic dependent and independent roles of TET3 in activation of neural and silencing of mesodermal programs, respectively, during differentiation of ESCs to NE. (1) Neural genes were downregulated in both Tet3m/m and Tet3–/– NE cells while mesoderm gene were aberrantly upregulated in Tet3–/– NE cells only. (2) The majority of DMRs in Tet3m/m and Tet3–/– NE cells were hypomethylated, which correlated with gene upregulation, while the few hyper-DMRs were linked to downregulated neural genes. (3) Dnmt1 is directly bound by TET3 in NE cells and is downregulated in Tet3m/m and Tet3–/– NE cells, which may be in part responsible for the increased region-specific DNA hypomethylation. Our findings establish that the catalytic dependent and independent roles of TET3 have distinct contributions to NE specification with implications in development.
We identified ~2,400 genes regulated by TET3 catalytic activity and ~750 genes regulated by TET3 non-catalytic roles. In agreement with a prior study, we find that TET3 maintains a balance between neural and mesoderm gene expression programs11. Our data further expands upon this study by revealing that the catalytic functions of TET3 activate neural genes while its non-catalytic functions suppress mesodermal genes. Interestingly, it was previously shown that TET3 catalytic activity promotes repression of mesodermal genes via demethylation of the Wnt inhibitor Sfrp4, where promoter hypermethylation of Sfrp4 in TET3 null neural progenitor cells resulted in aberrant activation of Wnt signaling and mesodermal programs11. We do not observe hypermethylation nor deregulation of Sfrp4 in Tet3m/m and Tet3–/– NE cells. However, we find robust aberrant upregulation of Wnt pathway and mesoderm genes uniquely in Tet3–/–, but not in Tet3m/m, NE cells supporting that silencing of Wnt signaling during NE specification depends on TET3 non-catalytic functions. Non-catalytic functions of other TETs, in particular TET1, have also been implicated in silencing of Wnt signaling and mesodermal genes suggesting TET catalytic independent roles curtails mesodermal programs as ESCs commit to NE37.
While the slight increase in the overall 5mC levels and reduction of 5hmC in Tet3m/m and Tet3–/– NE cells is consistent with a role for TET3 in DNA demethylation, the identification of a large number of hypo-DMRs (~8000) in contrast to hyper-DMRs (~1000) in both genotypes was surprising. This has not been observed in TET1 and TET2 deficient ESCs or progenitor cell types12,14. The association of hyper-DMRs with neural genes, including Neurog2 and Sox1, supports a catalytic role for TET3 in activating neural programs. The hypo-DMRs were associated to 239 non-catalytic DEGs, of which most were aberrantly upregulated in Tet3–/–, but not in Tet3m/m cells, and were associated to mesoderm pathways. This suggests that TET3 regulates DNA methylation through catalytic and non-catalytic functions which influences neural and mesodermal programs. Our finding that the maintenance methyltransferase Dnmt1 is a direct target of TET3 and is downregulated in Tet3m/m and Tet3–/– cells may provide a possible explanation to the increased number of hypo-DMRs. The reduced levels of Dnmt1 alone is sufficient to affect DNA methylation maintenance leading to hypomethylation and gene upregulation irrespective of any mild cell cycle or proliferation defects in Tet3-deficient cells. This is further supported by the fact that overexpression of Dnmt1 in Tet3m/m and Tet3–/– cells corrected the aberrant upregulation of genes. The expression of de novo methyltransferases Dnmt3a and Dnm3b, were unaffected in Tet3m/m and Tet3–/– NE cells suggesting they are unlikely involved. However, we cannot rule out any changes in their subcellular localization and intra nuclear redistribution potentially impacting DNA methylation, a phenomenon that has been reported for DNMT3A in some cell types40. Future studies mapping the genome-wide distribution of DNMTs and TETs in NE cells may provide better molecular insights underpinning the hypo-DMRs of TET-deficient cells.
The dual roles of TET3 in balancing neuroectoderm and mesoderm fates may have some developmental ramifications during early lineage specification and embryogenesis. Deficiency of TET3 in mice allows overtly normal early and mid-gestation development, but TET3 null embryos die perinatally for reasons not yet defined16. While gross organs, and in particular the nervous system components, are well developed in TET3 null embryos, histopathological and molecular studies are limited16. Future generation and comparison of TET3 catalytic deficient and knockout embryos at cellular and molecular levels during early development can provide better in vivo insights into how TET3 regulates neuroectoderm fate and neurodevelopment. Indeed, similar studies focused on TET1 and TET2 comparing knockout vs catalytic mutant mice have revealed their distinct biological contributions to embryogenesis and hematopoiesis12,35,41.
This study defines the dual functions of TET3 in NE specification. Our transcriptomic and epigenomic comparisons of TET3 catalytic mutant and TET3 knockout ESCs has provided key insights on how TET3 regulates neural and mesodermal gene programs via its catalytic and non-catalytic functions, respectively. In contrast, the few studies in the literature that have investigated the role of TET3 in the neural lineage11,25,27 have only used knockout or knock down approaches which abrogates the entire TET3 protein and fails to dissect its dual catalytic and noncatalytic functions. One of these studies reports a requirement for TET3 in differentiation of neural progenitors to neurons and preventing apoptosis27. While this study does not provide mechanistic insights into how TET3 regulates these two processes, it suggests that TET3 does not influence cell death in early neuroectoderm commitment but rather prevents apoptosis of established neural progenitor cells. Consistently, we do not find increased apoptosis or deregulation of apoptotic programs in Tet3m/m and Tet3–/– neuroectoderm cells. This is not surprising because neuroectoderm specified from ESCs is a different cell type than establish neural progenitors or terminally differentiated neurons, and therefore TET3 may have different functions in each. Future studies aimed at dissecting the catalytic and noncatalytic roles of TET3 in formation of neurons may elaborate which TET3 functions contribute to neurogenesis. Future studies are also warranted to further understand the mechanisms of how TET3 represses mesodermal genes, perhaps either through directly binding at its target genes and recruiting repressive factors to silence them, as we have reported for TET1 and TET2 in ESCs12,14, or indirectly through modulation of signaling pathways37. It will also be important to better define the relationship between TETs and DNMTs in shaping the DNA methylation dynamics to influence gene expression. These will be important for understanding epigenetic mechanisms of lineage commitment and development with implications for cancers where TETs and DNMTs are mutated or deregulated.
We note that our TET3 occupancy data is limited to only ~1200 peaks, which is fewer compared to TET1 and TET2 peaks in ESCs and in other cell types12,14. The lower number of peaks identified could be due to various biological or technical reasons including: (1) TET3 is expressed at lower levels or is unstable in NE cells, (2) TET3 binds with low affinity to chromatin, and/or (3) TET3 antibodies are not sensitive enough or methods such as CUT&Tag or ChIP-seq are not robust enough for mapping occupancies of large proteins like TET3. All these factors could have contributed to the reduced number of TET3 peaks detected in NE cells, and therefore may have prevented the identification of some key direct target genes of TET3. This may also explain the scarcity of TET3 occupancy data in the literature. Future optimization of antibodies and methodologies may resolve these issues. We also note that TET3 catalytic mutant mRNA levels are significantly reduced in Tet3m/m NE cells compared to TET3 wildtype mRNA levels in Tet+/+ NE cells. It is possible that the TET3 mutant transcript is unstable and that TET3 mutant protein is reduced as well. Using the same conserved mutations in the catalytic pocket of TET1 and TET2 has not resulted in any significant reduction of mutant TET1 or TET2 transcripts12,14. Thus, this reduction in TET3 mutant transcript levels is unique to TET3. An alternative possibility that can explain the reduced Tet3 transcript levels in Tet3m/m NE cells is an auto-regulatory mechanism, where TET3 catalytic activity is needed for proper expression of Tet3. While our CUT&Tag data does not show enrichment of TET3 at Tet3 promoter and gene body, it is possible that TET3 transiently binds to these regions, or that TET3 is enriched at regions distal to the Tet3 gene that are not yet implicated in Tet3 regulation. Regardless, the reduced Tet3 transcript levels in Tet3m/m NE cells should be taken into consideration in data interpretation.
Materials and Methods
Generation of Tet3 m/m and Tet3 –/– mouse ESCs
TET3 catalytic deficient (Tet3m/m) and knockout (Tet3–/–) mouse ESCs were generated following our previous CRISPR/Cas9 gene targeting protocols42. To generate Tet3m/m mouse ESCs: A donor oligo containing two point mutations (H950Y and D952A) in the TET3 iron binding site (in exon 9) and a silent mutation introducing a HaeIII restriction enzyme site was synthesized. A gRNA targeting exon 9 was designed and cloned into a px330-GFP vector. 1.5 μg of gRNA and 3.5 μg of donor oligo were transfected into wild-type mouse ESCs (v6.5 mixed 129/B6 background, male). Correctly targeted clones were validated by RLFP using HaeIII restriction enzyme and sanger sequencing. To generate Tet3–/– mouse ESCs: two gRNA flanking exon 4 were generated and cloned into px330-GFP and -Cherry vectors. 2.5 μg of each gRNA was transfected into wild-type ESCs, and double positive cells were sorted by FACs. Correctly targeted clones were validated by PCR and loss of Tet3 mRNA expression was confirmed by RT-qPCR. Sequences of gRNA, oligos, and primers used for genotyping are in Supplementary Table 1. The Tet3m/m and Tet3–/– mouse ESCs generated in this study are available upon request from the corresponding author.
Mouse ESC culture and differentiation to NE
All mouse ESCs (v6.5 mixed129/B6 background, male) were cultured on feeders in ESC media containing serum/LIF as previously described12. For gene targeting validation, ESCs were trypsinized, pre-plated for 1 h to remove feeder cells, and seeded on gelatin-coated plates for 24 h before harvesting for DNA and RNA extraction. ESCs were differentiated to NE as previously described39. Briefly, pre-plated ESCs were cultured on gelatin-coated plates for 24 h and differentiated for 72 h in the absence of LIF (DMEM supplemented with 10% FBS, 2 mM glutamine, 1x non-essential amino acids, 100 U/mL penicillin, 100 g/mL streptomycin, 1 μM retinoic acid (RA), and 50 mM β-mercaptoethanol). Cells were harvested at 72 h of differentiation for transcriptomic and epigenomic analyzes.
Transfection of mouse ESCs during differentiation
Hemagglutinin (HA)–tagged wild-type Tet3 catalytic domain (Tet3-CD) was cloned into FUW-HA-2A-tdTomato vector. Myc-tagged Dnmt1 (pCAG-myc-Dnmt1-IRES-blast) and empty vector (pCAG-myc-IRES-blast) were obtained from Dr. Taiping Chen at University of Texas MD Anderson Cancer Center. ESCs were cultured and differentiated as stated above. All transfections were carried out using XtremeGene DNA transfection reagent following manufacturer guidelines. For Tet3-CD overexpression: Tet3+/+, Tet3m/m, and Tet3–/– cells were transfected with either empty vector (FUW-HA-2A-tdTomato) or Tet3-CD (FUW-HA-Tet3-CD-2A-tdTomato) on day 2 of -LIF + RA differentiation. Cells were collected for RNA extraction 48 h later when Tet3-CD expression and tdTomato+ cells were the highest. For Dnmt1 overexpression: Tet3+/+, Tet3m/m, and Tet3–/– ESCs were transfected with either empty vector or myc-Dnmt1 and media was changed to -LIF + RA at day 0. 48 h after transfection, cells were treated with 10 ug blasticidin for 24 h followed by collection for RNA extraction. RNA was extracted using Qiagen RNeasy kit and subjected to RNA-seq at Novogene.
Proliferation and cell cycle assay
For the proliferation assays, 25,000 ESCs of each genotype were seeded onto one well of a gelatin-coated 12-well plate. After 24 h, media was changed to differentiation media (–LIF + RA) and viable cells were counted every 24 h for 3 days. For cell cycle analysis, day 3 differentiated cells (–LIF + RA) were treated with 20uM EdU and 0.1 mg/mL RNase A and 50 µg/mL propidium iodide (PI, P4864, SigmaAldrich, Inc., St. Louis, MO, USA) using the Click-It 647 EdU kit (Thermo-Fisher C10424) following manufacturer’s instructions. Cell cycle analysis was performed using a BD LSR II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and FlowJo software (v.10.8.0).
Teratoma Assay
Pre-plated wild-type, Tet3m/m, and Tet3–/– mouse ESCs were grown on gelatin-coated plates for 24 h. 1 million cells were harvested and injected subcutaneously into SCID mice (Taconic) as previously described12. After 7 weeks, tumors were harvested, fixed in formalin for 2 days, paraffin embedded, sectioned, and stained with Hematoxylin and Eosin at the Einstein Histopathology Core. Slide were imaged using an upright light microscope.
RT-qPCR and ChIP-qPCR
RT-qPCR was performed as previously described12. RNA was extracted from 3 clones of each genotype at day 0 (ESCs cultured on gelatin-coated plates for 24 h) and day 3 of –LIF + RA differentiation using the Qiagen RNeasy mini kit (74104) and subjected to cDNA synthesis using Superscript IV kit (Invitrogen, 18091050), according to manufacturer’s protocols. RT-qPCR was performed using SYBR green and primers in Supplementary Table 2 in a BD Applied Biosystems StepOneTM Real-Time PCR System. Data were normalized to Gapdh and wild-type controls. ChIP-qPCR for TET3 was performed in day 3 -LIF + RA NE cells using anti-TET3 antibody (Millipore ABE290) and primers targeting Dnmt1 promoter as described before12. Dnmt1 primers were designed according to TET3 bound coordinates identified from CUT&Tag analysis. Data was calculated as fold enrichment over IgG and plotted as fold change.
RNA-Seq and data analysis
RNA-seq was performed as previously described12. Day 3 -LIF + RA neuroectoderm cells were collected and total RNA was extracted as described above. Library preparation and mRNA sequencing were done by Novogene using their Illumina Novoseq 6000 platform. Trim galore (v0.6.7) was used to remove adapters and trimmed reads were mapped to the mouse genome (mm39) using STAR (v2.7.9a)43 with default parameters. Gene counts were extracted from mapped reads using featureCounts with --largestOverlap parameter. Raw counts were used to identify differentially expressed genes (DEGs) with DESeq244 (false discovery rate-adjusted p-value < 0.05 and fold-change >1.5), following the package documentation. Gene ontology (GO) enrichment analyzes of DEGs was performed using DAVID45 (https://david.ncifcrf.gov/). All plots were made in R using custom scripts.
WGBS and data analysis
Whole-genome bisulfite sequencing (WGBS) was performed as described previously12. DNA from day 3 -LIF + RA NE cells was extracted by Quick-DNA miniprep kit (Zymo, D3024) following the manufacturer’s instructions. Bisulfite conversion and sequencing were performed at BGI Genomics (https://en.genomics.cn/). Lamda DNA spike-in confirmed a >99.0% bisulfite conversion efficiency. The libraries were subjected to 100 bp pair-end sequencing on a HiSeq 4000 Illumina platform. Raw reads were filtered by SOAPnuke (https://github.com/BGI-flexlab/SOAPnuke) with the parameters -n 0.001 -l 20 -q 0.4 –adaMR 0.25 –ada_trim –polyX 50 –minReadLen 100 to remove adapters and filter out low-quality reads. Clean reads were mapped to mouse genome mm10 using Bismark (v0.23.1)46 with default parameters. Duplicated reads were removed using deduplicate_bismark and methylation status of each cytosine was determined with bismark_methylation_extractor. The deduplicated BAM files were balanced to the sample with the lowest reads using samtools. The resulting BAM files were then converted to be compatible with methpipe47 (v5.0.1) using methpipe format_reads -f and sorted using samtools. Bisulfite rate conversion was determined by bsrate, single-site methylation levels were identified by methcounts, and symmetric CpGs were merged with symmetric-cpgs. Methylation was compared between samples using methdiff and differentially methylated regions (DMRs, regions >5 CpGs, methylation difference >20%, and FDR < 0.05) between Tet3m/m vs. Tet3+/+ and Tet3–/– vs. Tet3+/+ were identified using hmr and then dmr. DMRs were annotated to genomic features with R package ChIPseeker (v1.36.0). Methylation line plots at DMRs were generated using plotProfile function of deepTools (v3.5.1)48. For visualization on Integrative Genome Browser (IGV), bedGraph files were converted to bigwig files using bedGraphToBigWig from bedtools (v2.30.0)49. To determine the overlapping Tet3m/m and Tet3–/– DMRs, BED files were intersected using bedtools intersect with default parameters.
hmeDIP and data analysis
Tet3+/+, Tet3m/m and Tet3–/– mouse ESCs were differentiated to NE (-LIF + RA for 3 days). DNA was extracted and subjected to hMeDIP (Active Motif hMeDIP Kit #55010) following manufacturer guidelines, and amplified using NEBNext HiFi 2x PCR Master mix. Libraries were cleaned with AMPure XP beads (#A63880) and subjected to 75 bp paired-end sequencing using Illumina NextSeq 500 platform at the Einstein Epigenomics Core. Adapters were trimmed using TrimGalore and sequencing reads were mapped to the mouse genome (mm10) using bowtie2 (v2.4.5)50 with the following parameters: --local --sensitive --very-sensitive-local --no-unal --no-mixed --no-discordant –phred33 -I 10 -X 700. The BAM files were balanced to the sample with the lowest reads using samtools and were used to call peaks using MACS2 (v2.2.7.1)51 with the following parameters: -p 0.0001 -f BAMPE --keep-dup all. BED files with genomic coordinates were annotated using ChIPseeker (v1.36.0). IgG bigwig was subtracted from Tet3+/+, Tet3m/m and Tet3–/– bigwigs using function bigwigCompare --operation subtract from deepTools. The output bigwig files were used to make line plots using function plotProfile from deepTools.
CUT&Tag and data analysis
To map the genomic occupancy of TET3, CUT&Tag was applied to day 3 -LIF + RA differentiated wild-type cells as described previously12. 500,000 cells (per antibody) were collected at 72 h of differentiation and crosslinked with 0.5% formaldehyde for 5 mins. Fixed cells were bound to Concavalin A-coated beads, permeabilized, and incubated with primary antibodies (anti-Tet3 Millipore ABE290 and rabbit IgG isotype control CST 3900) overnight at 4 °C. Samples were then incubated with secondary antibody (guinea pig anti-rabbit Antibodies Online ABIN101961) at room temperature followed by incubation with pre-loaded pA-Tn5. Transposase was activated at 37 °C with magnesium to fragment bound DNA. DNA was isolated by phenol-chloroform isoamyl alcohol extraction and amplified using NEBNext HiFi 2x PCR Master mix. Libraries were cleaned with AMPure XP beads (#A63880) and subjected to 75 bp paired-end sequencing using Illumina NextSeq 500 platform at the Einstein Epigenomics Core. Sequencing reads were mapped to the mouse genome (mm10) using bowtie2 (v2.4.5) with the following parameters: --local --sensitive --very-sensitive-local --no-unal --no-mixed --no-discordant –phred33 -I 10 -X 700. Duplicates were removed using Picard tools MarkDuplicates function. The BAM files were balanced to the sample with the lowest reads using samtools and were used to call peaks using MACS2 (v2.2.7.1) with the following parameters: -p 0.00001 -f BAMPE --keep-dup all. BED files with genomic coordinates were annotated using ChIPseeker (v1.36.0). Line plots and heatmaps were generated using deepTools.
Statistics and reproducibility
All experiments were performed with three biological replicates unless otherwise described in the methods. GraphPad Prism 8 was used to perform statistical analyzes. One way ANOVA test or unpaired t-test was applied to calculate statistical significance. Differences were considered statistically significant when p < 0.05. Statistical methods used for bioinformatics analyzes are explained in detail under the respective methods sub-sections.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq, CUT&Tag and WGBS data have been deposited in the Gene Expression Omnibus (GEO) database. Accession number (GSE237800). The uncropped images of gels can be found under Supplementary Information as Supplementary Fig. 5. The source data for all figures are provided in Supplementary Data 1. All other relevant data and the ESC lines used to generate the data in this study are available upon request from the corresponding author.
References
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Maiti, A. & Drohat, A. C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338 (2011).
Pastor, W. A., Aravind, L. & Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013).
Bachman, M. et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 6, 1049–1055 (2014).
Hashimoto, H. et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 40, 4841–4849 (2012).
Wu, H. & Zhang, Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68 (2014).
Ross, S. E. & Bogdanovic, O. TET enzymes, DNA demethylation and pluripotency. Biochem. Soc. Trans. 47, 875–885 (2019).
Dawlaty, M. M. et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 29, 102–111 (2014).
Li, X. et al. Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling. Proc. Natl. Acad. Sci. 113, E8267–E8276 (2016).
Chrysanthou, S. et al. The DNA dioxygenase Tet1 regulates H3K27 modification and embryonic stem cell biology independent of its catalytic activity. Nucleic Acids Res. 50, 3169–3189 (2022).
Dawlaty, M. M. et al. Tet1 is dispensable for maintaining pluripotency and Its loss Is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 (2011).
Flores, J. C., Sidoli, S. & Dawlaty, M. M. Tet2 regulates Sin3a recruitment at active enhancers in embryonic stem cells. iScience 26, 107170 (2023).
Dawlaty, M. M. et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24, 310–323 (2013).
Gu, T.-P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011).
Kang, J. et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc. Natl. Acad. Sci. USA 112, E4236–E4245 (2015).
Yan, R. et al. Dynamics of DNA hydroxymethylation and methylation during mouse embryonic and germline development. Nat. Genet. 55, 130–143 (2023).
Iqbal, K., Jin, S. G., Pfeifer, G. P. & Szabó, P. E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA 108, 3642–3647 (2011).
Kang, J., Kalantry, S. & Rao, A. PGC7, H3K9me2 and Tet3: regulators of DNA methylation in zygotes. Cell Res. 23, 6–9 (2013).
Peat, J. R. et al. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep. 9, 1990–2000 (2014).
Tsukada, Y., Akiyama, T. & Nakayama, K. I. Maternal TET3 is dispensable for embryonic development but is required for neonatal growth. Sci. Rep. 5, 15876 (2015).
Tanaka, S. et al. Tet2 and Tet3 in B cells are required to repress CD86 and prevent autoimmunity. Nat Immunol. 21, 950–961 (2020).
Xue, S. et al. TET3 inhibits Type I IFN Production Independent of DNA Demethylation. Cell Rep. 16, 1096–1105 (2016).
Santiago, M. et al. Tet3 regulates cellular identity and DNA methylation in neural progenitor cells. Cell Mol. Life Sci. 77, 2871–2883 (2020).
Montalbán-Loro, R. et al. TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn. Nat. Commun. 10, 1726 (2019).
Li, T. et al. Critical role of Tet3 in neural progenitor cell maintenance and terminal differentiation. Molecular Neurobiol. 51, 142–154 (2015).
Fang, S. et al. Tet inactivation disrupts YY1 binding and long-range chromatin interactions during embryonic heart development. Nat. Commun. 10, 4297 (2019).
Cao, T., Pan, W., Sun, X. & Shen, H. Increased expression of TET3 predicts unfavorable prognosis in patients with ovarian cancer-a bioinformatics integrative analysis. J. Ovarian Res. 12, 101 (2019).
Carella, A. et al. Epigenetic downregulation of TET3 reduces genome-wide 5hmC levels and promotes glioblastoma tumorigenesis. Int. J. Cancer 146, 373–387 (2020).
Beck, D. B. et al. Delineation of a human Mendelian disorder of the DNA demethylation machinery: TET3 deficiency. Am. J. Hum. Genet. 106, 234–245 (2020).
Sager, S. G. et al. A novel de novo TET3 loss-of-function variant in a Turkish boy presenting with neurodevelopmental delay and electrical status epilepticus during slow-wave sleep. Brain Dev. 45, 140–145 (2023).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Wu, H. et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 (2011).
Ito, K. et al. Non-catalytic roles of Tet2 are essential to regulate hematopoietic stem and progenitor cell homeostasis. Cell Rep. 28, 2480–2490.e2484 (2019).
Chrysanthou, S., Flores, J. C. & Dawlaty, M. M. Tet1 Suppresses p21 to Ensure Proper Cell Cycle Progression in Embryonic Stem Cells. Cells 11, 1366 (2022).
van der Veer, B. K. et al. Dual functions of TET1 in germ layer lineage bifurcation distinguished by genomic context and dependence on 5-methylcytosine oxidation. Nucleic Acids Res 51, 5469–5498 (2023).
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
Semrau, S. et al. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat. Commun. 8, 1096 (2017).
Lopez-Moyado, I. F. et al. Paradoxical association of TET loss of function with genome-wide DNA hypomethylation. Proc. Natl. Acad. Sci. USA 116, 16933–16942 (2019).
Ma, L. et al. Tet-mediated DNA demethylation regulates specification of hematopoietic stem and progenitor cells during mammalian embryogenesis. Sci. Adv. 8, eabm3470 (2022).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinform. 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Jiao, X. et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinform. 28, 1805–1806 (2012).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinform. 27, 1571–1572 (2011).
Song, Q. et al. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One 8, e81148 (2013).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinform. 26, 841–842 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Acknowledgements
We thank the Einstein Epigenomics Cores for help with next-generation sequencing and the Skoultchi lab at Einstein for providing reagents for CUT&Tag. We thank Dr. Taiping Chen at the University of Texas MD Anderson Cancer Center for providing Dnmt1 plasmids. We are grateful to Hui Su and Joun Lee for help with tissue culture and cell line generation and Ian MacArthur, Blake Ebert and Halley Pierce for critical reading of the manuscript. This work was supported by funds to M.M.D from NIH R01GM122839 (ESC work and gene regulation mechanisms) and NYSDOH/NYSTEM Contract C34877GG (DNA methylation analysis). M.M.D is also supported by NIH R01HL148852, Hirschle Trust Funds and funds from Albert Einstein College of Medicine Stem Cell Institute and Genetics Department. H.C.K. was supported by an NIH T32 training grant in cellular and molecular biology and genetics (T32 GM007491) and Paul S. Frenette Scholar Awards Program of the Ruth L. and David S. Gottesman Institute for Stem Cell Research and Regenerative Medicine at Albert Einstein College of Medicine.
Author information
Authors and Affiliations
Contributions
H.C.K generated the cell lines, performed the experiments, analyzed data, and prepared the figures. M.S. assisted with next generation sequencing analyzes. M.M.D. and H.C.K. designed the study and wrote the manuscript with input from M.S. M.M.D. supervised the study and secured funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Wee Wei Tee and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ketchum, H.C., Suzuki, M. & Dawlaty, M.M. Catalytic-dependent and -independent roles of TET3 in the regulation of specific genetic programs during neuroectoderm specification. Commun Biol 7, 415 (2024). https://doi.org/10.1038/s42003-024-06120-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06120-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.