Abstract
ZNF462 haploinsufficiency is linked to Weiss–Kruszka syndrome, a genetic disorder characterized by neurodevelopmental defects, including autism. Though conserved in vertebrates and essential for embryonic development, the molecular functions of ZNF462 remain unclear. We identified its murine homologue ZFP462 in a screen for mediators of epigenetic gene silencing. Here we show that ZFP462 safeguards neural lineage specification of mouse embryonic stem cells (ESCs) by targeting the H3K9-specific histone methyltransferase complex G9A/GLP to silence meso-endodermal genes. ZFP462 binds to transposable elements that are potential enhancers harbouring pluripotency and meso-endoderm transcription factor binding sites. Recruiting G9A/GLP, ZFP462 seeds heterochromatin, restricting transcription factor binding. Loss of ZFP462 in ESCs results in increased chromatin accessibility at target sites and ectopic expression of meso-endodermal genes. Taken together, ZFP462 confers lineage and locus specificity to the broadly expressed epigenetic regulator G9A/GLP. Our results suggest that aberrant activation of lineage non-specific genes in the neuronal lineage underlies ZNF462-associated neurodevelopmental pathology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data Availability
High-throughput sequencing data produced in this study are deposited at the Gene Expression Omnibus (GEO) database under super series accession number GSE175369 (RNA-seq: GSE176321, QuantSeq: GSE176319, ATAC-seq: GSE176322 and ChIP–seq: GSE177058). G9a/GLP DKO ESC ATAC-seq and RNA-seq data are obtained from GSE138102 (ref. 52). REST and ADNP ChIP–seq data are obtained from GSE27148 (ref. 95) and GSE97945 (ref. 51), respectively. Meso-endoderm cell ATAC-seq data are obtained from GSE116262 (ref. 66). Processed single-cell RNA-seq data of mouse embryo stage E4.5 to E7.0 are obtained from ftp://ftp.ebi.ac.uk/pub/databases/scnmt_gastrulation43. Mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD037238. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Eckersley-Maslin, M. A., Alda-Catalinas, C. & Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018).
Burton, A. & Torres-Padilla, M.-E. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 15, 723–735 (2014).
Gökbuget, D. & Blelloch, R. Epigenetic control of transcriptional regulation in pluripotency and early differentiation. Development 146, dev164772 (2019).
Iwafuchi-Doi, M. & Zaret, K. S. Cell fate control by pioneer transcription factors. Development 143, 1833–1837 (2016).
Padeken, J., Methot, S. P. & Gasser, S. M. Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/s41580-022-00483-w (2022).
Nishibuchi, G. & Nakayama, J. Biochemical and structural properties of heterochromatin protein 1: understanding its role in chromatin assembly. J. Biochem. 156, 11–20 (2014).
Larson, A. G. & Narlikar, G. J. The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry 57, 2540–2548 (2018).
Kumar, A. & Kono, H. Heterochromatin protein 1 (HP1): interactions with itself and chromatin components. Biophys. Rev. 12, 387–400 (2020).
Nicetto, D. & Zaret, K. S. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 55, 1–10 (2019).
Chen, J. et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34–42 (2013).
Ragunathan, K., Jih, G. & Moazed, D. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348, 1258699 (2015).
Audergon, P. N. C. B. et al. Restricted epigenetic inheritance of H3K9 methylation. Science 348, 132 LP–132135 (2015).
Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012).
Shinkai, Y. & Tachibana, M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 25, 781–788 (2011).
Tachibana, M. et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 (2002).
Lienert, F. et al. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet. 7, e1002090 (2011).
Wen, B., Wu, H., Shinkai, Y., Irizarry, R. A. & Feinberg, A. P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250 (2009).
Liu, N. et al. Recognition of H3K9 methylation by GLP is required for efficient establishment of H3K9 methylation, rapid target gene repression, and mouse viability. Genes Dev. 29, 379–393 (2015).
Zylicz, J. J. et al. G9a regulates temporal preimplantation developmental program and lineage segregation in blastocyst. eLife 7, e33361 (2018).
Feldman, N. et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8, 188–194 (2006).
Kellner, S. & Kikyo, N. Transcriptional regulation of the Oct4 gene, a master gene for pluripotency. Histol. Histopathol. 25, 405–412 (2010).
Epsztejn-Litman, S. et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 15, 1176–1183 (2008).
Benevento, M., van de Molengraft, M., van Westen, R., van Bokhoven, H. & Nadif Kasri, N. The role of chromatin repressive marks in cognition and disease: a focus on the repressive complex GLP/G9a. Neurobiol. Learn. Mem. 124, 88–96 (2015).
Roopra, A., Qazi, R., Schoenike, B., Daley, T. J. & Morrison, J. F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 14, 727–738 (2004).
Mozzetta, C. et al. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol. Cell 53, 277–289 (2014).
Mulligan, P. et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol. Cell 32, 718–726 (2008).
Schaefer, A. et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64, 678–691 (2009).
Deimling, S. J., Olsen, J. B. & Tropepe, V. The expanding role of the Ehmt2/G9a complex in neurodevelopment. Neurogenesis 4, e1316888 (2017).
Kleefstra, T. et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am. J. Hum. Genet. 79, 370–377 (2006).
Kramer, J. M. Regulation of cell differentiation and function by the euchromatin histone methyltranserfases G9a and GLP. Biochem. Cell Biol. 94, 26–32 (2015).
Scheer, S. & Zaph, C. The lysine methyltransferase G9a in immune cell differentiation and function. Front. Immunol. 8, 429 (2017).
Weiss, K. et al. Haploinsufficiency of ZNF462 is associated with craniofacial anomalies, corpus callosum dysgenesis, ptosis, and developmental delay. Eur. J. Hum. Genet. 25, 946–951 (2017).
Kruszka, P. et al. Phenotype delineation of ZNF462 related syndrome. Am. J. Med. Genet. A 179, 2075–2082 (2019).
González-Tarancón, R. et al. A novel mutation in the ZNF462 gene c.3306dup; p.(Gln1103Thrfs*10) is associated to Weiss–Kruszka syndrome. A case report. Acta Clin. Belg. https://doi.org/10.1080/17843286.2020.1780391 (2020).
Athanasiadou, R. et al. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS ONE 5, e9937 (2010).
Niwa, H., Miyazaki, J. & Smith, A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 (2000).
Michlits, G. et al. CRISPR-UMI: single-cell lineage tracing of pooled CRISPR–Cas9 screens. Nat. Methods 14, 1191–1197 (2017).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Rose, N. R. & Klose, R. J. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta Gene Regul. Mech. 1839, 1362–1372 (2014).
Ren, W. et al. DNMT1 reads heterochromatic H4K20me3 to reinforce LINE-1 DNA methylation. Nat. Commun. 12, 2490 (2021).
Ueda, J., Tachibana, M., Ikura, T. & Shinkai, Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP*. J. Biol. Chem. 281, 20120–20128 (2006).
Olsen, J. B. et al. G9a and ZNF644 physically associate to suppress progenitor gene expression during neurogenesis. Stem Cell Rep. 7, 454–470 (2016).
Argelaguet, R. et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576, 487–491 (2019).
McDonald, A. C. H., Biechele, S., Rossant, J. & Stanford, W. L. Sox17-mediated XEN cell conversion identifies dynamic networks controlling cell-fate decisions in embryo-derived stem cells. Cell Rep. 9, 780–793 (2014).
Wamaitha, S. E. et al. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev. 29, 1239–1255 (2015).
Schrode, N., Saiz, N., Di Talia, S. & Hadjantonakis, A.-K. GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev. Cell 29, 454–467 (2014).
Wang, B. et al. Zfp462 deficiency causes anxiety-like behaviors with excessive self-grooming in mice. Genes Brain Behav. 16, 296–307 (2017).
Bibel, M., Richter, J., Lacroix, E. & Barde, Y.-A. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc. 2, 1034–1043 (2007).
Venere, M. et al. Sox1 marks an activated neural stem/progenitor cell in the hippocampus. Development 139, 3938–3949 (2012).
Ecco, G., Imbeault, M. & Trono, D. KRAB zinc finger proteins. Development 144, 2719–2729 (2017).
Ostapcuk, V. et al. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature 557, 739–743 (2018).
Jiang, Q. et al. G9a plays distinct roles in maintaining DNA methylation, retrotransposon silencing, and chromatin looping. Cell Rep. 33, 108315 (2020).
Kunarso, G. et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 42, 631–634 (2010).
Sundaram, V. & Wysocka, J. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos. Trans. R. Soc. B Biol. Sci. 375, 20190347 (2020).
Sundaram, V. et al. Functional cis-regulatory modules encoded by mouse-specific endogenous retrovirus. Nat. Commun. 8, 14550 (2017).
Todd, C. D., Deniz, Ö., Taylor, D. & Branco, M. R. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. eLife 8, e44344 (2019).
Bourque, G. et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762 (2008).
Niwa, H. How is pluripotency determined and maintained? Development 134, 635–646 (2007).
Thomson, M. et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145, 875–889 (2011).
Wang, Z., Oron, E., Nelson, B., Razis, S. & Ivanova, N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440–454 (2012).
Le Bin, G. C. et al. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development 141, 1001–1010 (2014).
Frum, T. et al. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev. Cell 25, 610–622 (2013).
Pintacuda, G. et al. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 68, 955–969.e10 (2017).
Ernst, J. & Kellis, M. Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 12, 2478–2492 (2017).
Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020).
Cernilogar, F. M. et al. Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 47, 9069–9086 (2019).
Nissim, S. et al. Iterative use of nuclear receptor Nr5a2 regulates multiple stages of liver and pancreas development. Dev. Biol. 418, 108–123 (2016).
Seitz, C. et al. The orphan nuclear receptor LRH-1/NR5a2 critically regulates T cell functions. Sci. Adv. 5, eaav9732 (2022).
Olivieri, D. et al. Cooperation between HDAC3 and DAX1 mediates lineage restriction of embryonic stem cells. EMBO J. 40, e106818 (2021).
Soufi, A. et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 (2015).
Morgunova, E. & Taipale, J. Structural insights into the interaction between transcription factors and the nucleosome. Curr. Opin. Struct. Biol. 71, 171–179 (2021).
Michael, A. K. et al. Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 368, 1460–1465 (2020).
Isbel, L., Grand, R. S. & Schübeler, D. Generating specificity in genome regulation through transcription factor sensitivity to chromatin. Nat. Rev. Genet. https://doi.org/10.1038/s41576-022-00512-6 (2022).
King, H. W. & Klose, R. J. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife 6, e22631 (2017).
Frank, J. A. & Feschotte, C. Co-option of endogenous viral sequences for host cell function. Curr. Opin. Virol. 25, 81–89 (2017).
Bourque, G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr. Opin. Genet. Dev. 19, 607–612 (2009).
Carbon, S. et al. AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 (2009).
Blomfield, I. M. et al. Id4 promotes the elimination of the pro-activation factor Ascl1 to maintain quiescence of adult hippocampal stem cells. Elife 8, e48561 (2019).
Baubec, T., Ivánek, R., Lienert, F. & Schübeler, D. Methylation-dependent and -Independent genomic targeting principles of the MBD protein family. Cell 153, 480–492 (2013).
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
Moussa, H. F. et al. Canonical PRC1 controls sequence-independent propagation of Polycomb-mediated gene silencing. Nat. Commun. 10, 1931 (2019).
Mayer, D. et al. Zfp281 orchestrates interconversion of pluripotent states by engaging Ehmt1 and Zic2. EMBO J. 39, e102591 (2020).
Batki, J. et al. The nascent RNA binding complex SFiNX licenses piRNA-guided heterochromatin formation. Nat. Struct. Mol. Biol. 26, 720–731 (2019).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 (2007).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Fursova, N. A. et al. Synergy between variant PRC1 complexes defines Polycomb-mediated gene repression. Mol. Cell 74, e8 (2019).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Arnold, P. et al. Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res. 23, 60–73 (2013).
Acknowledgements
We are grateful to all members of the Bell and Brennecke laboratories for support, feedback and discussions. HP1γ and GLP Avi-tagged cell lines are kind gifts from M. Bühler and J. Betschinger labs at FMI, Switzerland. We thank N. Urban for providing mouse NSCs. We thank C. Buecker, F. Mohn and K. Jensen for sharing unpublished date, experimental advice and helpful discussions. We thank F. Berger, L. Cochella and J. Schnabl for feedback on the manuscript. We thank the Vienna Biocenter Core Facility Next Generation Sequencing. The GMI/IMBA/IMP Scientific Service units, especially the BioOptics facility and the Mass Spectrometry unit, provided outstanding support. DNA methylation analysis by LC–MS/MS was performed by the Metabolomics Facility at Vienna BioCenter Core Facilities (VBCF), funded by the City of Vienna through the Vienna Business Agency. We thank Life Science Editors for editorial assistance. We apologize to colleagues whose work could not be cited due to space limitations. Funding: O.B., J.B., U.E. and S.M. were supported by the Austrian Academy of Sciences. O.B. was supported by the New Frontiers Group of the Austrian Academy of Sciences (NFG-05), the Human Frontiers Science Programme Career Development Award (CDA00036/2014-C), the National Institute of Mental Health (R01MH122565) and the Norris Comprehensive Cancer Center of USC and its NCI Award (P30CA014089). R.Y. was supported by EMBO Long-Term Fellowship (ALTF 256-2015). D.S. acknowledges support from the Novartis Research Foundation and the European Research Council under the European Union’s (EU) Horizon 2020 research and innovation program grant agreement (ReadMe-667951). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
R.Y., K.S. and O.B. initiated and designed the study. R.Y and K.S. generated cell lines, performed CRISPR–Cas9 genetic screen and differentiation assays. R.Y. performed all molecular biology experiments and revisions. S.M. and P.H. provided experimental advice and P.H. carried out immunohistochemistry. R.Y. and M.N analysed transcriptome and epigenome data. J.W. and G.M. analysed CRISPR–Cas9 genetic screen data. U.E., G.M and G.V. provided the CRISPR sgRNA library and helped with the screen design. L.M. and C.P. participated in experiments. L.I. and D.S. shared reagents and experimental expertise. J.B. co-supervised part of the project in his laboratory. O.B. supervised all aspects of the project. The manuscript was prepared by R.Y. and O.B. D.S. and J.B. edited the manuscript. All authors discussed results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of WT and mutant CiA Oct4 dual reporter cells.
a) Western blot confirms expression of TetR-FLAG and TetR-FLAG-HP1 proteins in WT and Dnmt1 knockout (KO) CiA Oct4 dual reporter ESCs. LMNB1 is used as protein loading control. b) FACS gating strategy used to analyse the GFP fluorescence in cells. c) Flow cytometry histograms show GFP expression in CiA Oct4 dual reporter cells after TetR-FLAG recruitment and after TetR-FLAG release following Dox addition for four days. d) ChIP-qPCR shows relative enrichment of HP1γ upstream of the TetO site before tethering, in the presence of TetR-FLAG-HP1 and after Dox-dependent release of TetR-FLAG-HP1 for four days. n = 2 independent biological replicates. e) Bar plot shows fraction of cytosine methylation (5mC) in WT and Dnmt1 KO CiA Oct4 dual reporter ESCs measured by LC-MS. n = 3 independent biological replicates. Data are presented as mean values + /− SD. f) Western blot shows expression of TetR-FLAG-HP1 and ZFP462 in WT and two independent Zfp462 KO CiA Oct4 dual reporter ESC lines. LMNB1 is used as loading control.
Extended Data Fig. 2 ZFP462 is conserved across vertebrates and acts as transcriptional repressor.
a) Phylogenetic tree of ZFP462 protein orthologues in vertebrate species. Bootstrap values are shown on branches. b) Western blot shows expression ZFP462 fusions with TetR-FLAG in CiA Oct4 dual reporter ESCs. Bottom image shows long exposure. Hashtag indicates cleaved TetR-FLAG protein from TetR-FLAG-ZFP462-FL (full-length). TetR-FLAG fusions initially express mCherry for selection which is cleaved via P2A signal. Asterisk marks P2A uncleaved protein product. c) Representative flow cytometry histograms show GFP expression in CiA Oct4 dual reporter ESCs expressing TetR-FLAG fusions with full-length or truncated ZFP462 proteins. Each histogram is average profile of 100,000 analysed cells.
Extended Data Fig. 3 Zfp462 expression analysis and impact of Zfp462 deletion on ESC morphology and gene expression.
a) UMAP plots visualize lineage assignments of cells in mouse embryonic developmental stages (embryonic day (E) E4.5 cells, E5.5 cells and E6.5-7 cells) as previously described (Wolf Reik et.al) (top). UMAP plots of Zfp462 RNA expression at corresponding developmental stages (bottom). b) Sanger sequence chromatograms of heterozygous Zfp462 mutants. Heterozygous non-sense mutations are highlighted in grey. c) Alkaline phosphatase staining of Zfp462 +/Y1195* and Zfp462 −/− cl.2 ESCs (scale bar = 100 μm). d) Correlation plot shows principal component analysis (PCA) of replicate RNA-seq experiments from WT, two Zfp462 +/− and two Zfp462 −/− ESC lines. e) Volcano plot shows differential gene expression of Zfp462 −/− cl.2 ESCs compared to WT ESCs. (n = three replicates). f) Bar plots show gene ontology (GO) terms enriched in the four clusters of heatmap in Fig. 3e.
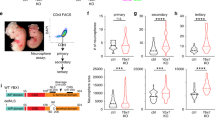
Extended Data Fig. 4 Lineage-specifying genes are deregulated during neuronal differentiation.
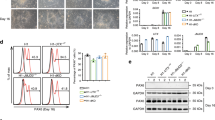
a) Alkaline phosphatase staining of WT and Zfp462 mutant ESCs cultured in 2i/LIF/Serum medium (scale bar = 180 μm). b) Representative bright field images show Zfp462 +/Y1195* and Zfp462 −/− cl.2 at corresponding stages of neural differentiation. Scale bar = 100μm. c) Western blot analysis shows levels of ZFP462, G9A and H3K9me2 in WT and Zfp462 −/− ESCs during neural differentiation. LMNB1 is used as loading control. d) Line plot shows RT-qPCR analysis of Zfp462 RNA expression during neural differentiation (n = two replicates). Expression level is shown relative to ESCs (2i/S/L). e) Heatmaps show differential expression of selected marker genes specific for endodermal (EN), mesodermal (ME) and neural lineages (EC) in heterozygous and homozygous Zfp462 mutant cells during neural differentiation.
Extended Data Fig. 5 ChIP-seq profiles of ZFP462, REST, ADNP, HP1γ and H3K9me2 in WT and Zfp462 KO ESCs.
a) Heatmap shows Spearman correlation between input controls and two independent ZFP462 ChIP-seq experiments. b) Heatmap shows Spearman correlation between input controls and two independent ChIP-seq experiments of GLP and HP1γ in WT and Zfp462 KO ESCs. c) Western blot shows ZFP462 expression in WT and Zfp462 KO Avi-FLAG-tagged GLP and HP1γ ESCs. d) Heatmap shows Spearman correlation between input controls and two independent ChIP-seq experiments of WIZ and H3K9me2 in WT and Zfp462 KO ESCs. e) Heatmaps of ZFP462, REST, ADNP, HP1γ and H3K9me2 ChIP-seq signals at ZFP462, REST and ADNP peaks in WT and Zfp462 KO ESCs. Blue-to-red scaled heatmap represents ChIP-seq enrichment ratios between Zfp462 KO versus WT (KO/WT). Each heatmap represents a 10 kb window centred on peak midpoints, sorted by ZFP462, REST and ADNP signal in their respective clusters. Below are scale bars (n = average distribution of two replicates).
Extended Data Fig. 6 Rescue of Zfp462 KO ESCs with ZFP462 full-length and ZFP462 C-terminal truncation.
a) Heatmap shows Spearman correlation between three independent ATAC-seq experiments in WT and Zfp462 KO ESCs. b) Scatter plot shows pairwise correlation of gene expression changes between Zfp462 KO vs WT and G9a/Glp dKO vs WT. coefficient of determination (R²): black - all genes, red – differentially regulated genes in Zfp462 KO vs WT (padj. ≤ 0.05, LFC ≥ 1). c) Cartoon depicts CRISPR strategy to knock-in coding sequences for ZFP462 full-length and ZFP462 NT+Mid proteins at the Zfp462 gene locus. Insertions were targeted in-frame with exon 3. d) Bar plot shows RT-qPCR analysis of Zfp462 RNA transcript levels in WT, KO and rescue ESCs. n = 2 independent biological replicates. e) Western blot analysis shows ZFP462 protein levels in WT, Zfp462 KO and rescue ESCs. LMNB1 is used as loading control. f) Alkaline phosphatase staining of WT and Zfp462 KO and rescue ESCs (scale bar = 100μm). g) Heatmaps of ATAC-seq signal ratios at ZFP462 and REST peaks of Zfp462 KO vs WT (KO/WT) and Zfp462 rescue ESCs (ZFP462 FL/WT) or (ZFP462 NT+Mid/WT). Each row represents a 10 kb window centred on peak midpoints, sorted by KO/WT enrichment ratio (n = average distribution of two ATAC-seq replicates). h) Line plots show RT-qPCR analysis of lineage marker expression during neural differentiation (n = two replicates) in WT, Zfp462 KO and rescue ESCs. Expression levels are shown relative to ESCs (2i/S/L).
Extended Data Fig. 7 Correlation between genomic distribution of ZFP462 and pluripotency transcription factors.
a) Heatmaps show ATAC-seq and ChIP-seq signal of ZFP462, H3K9me2, OCT4, SOX2, NANOG, ESRRB and NR5A2 at peaks of ZFP462, OSN (shared OCT4-SOX2-NANOG peaks not overlapping with ZFP462 peaks) and REST. Red-to-blue scaled heatmaps represent signal ratios between Zfp462 KO versus WT (KO/WT). Each row represents a 10 kb window centred on peak midpoints, sorted by H3K9me2 KO/WT ChIP signal loss. (n = average distribution of two ChIP-seq replicates / ATAC-seq three replicates). b) Genomic screenshot shows DNA accessibility (ATAC-seq) and ChIP-seq signals of H3K27ac and ZFP462 at the Oct3/4 locus in WT ESCs. ChIP-seq signals of OCT4, SOX2 and NANOG in WT (black line) and Zfp462 KO ESCs (green fill) are superimposed. ATAC-seq and ChIP-seq profiles are normalized to library size. c) Bar plot shows percentage of ZFP462-bound TE families overlapping with ChromHMM-annotated enhancers in ESCs. ZFP462-bound TEs contribute a total of 35.37% of ChromHMM-annotated enhancers in ESCs. d) HOMER analysis of known DNA sequence motifs enriched at ZFP462 peaks overlapping ChromHMM-annotated ME/EN-specific enhancers. Top ranked DNA sequence motifs and respective significance values are shown in the table. e) Box plots shows enrichment of NR5A2 and ESRRB ChIP signal in WT and Zfp462 KO ESCs at ZFP462 peaks overlapping with Mesoderm (ME)/Endoderm (EN)- and Ectoderm (EC)-specific enhancers. n = 553 (EN/ME), n = 314 (EC). Shown are median (horizontal line), 25th to 75th percentiles (boxes), and 90% (whiskers). Outliers are not shown. f) Bar plot shows frequency distribution of significantly up- and down-regulated genes located proximal to ZFP462 peaks annotated as ME/EN-specific enhancers (KO/WT, LFC ≥ 1 and padj. ≤ 0.05). g) Western blot shows ZFP462 protein expression in NSCs isolated from mouse brain and NPCs differentiated from WT ESCs. LMNB1 is used as loading control.
Supplementary information
Supplementary Table
List of genes enriched in genetic screen. Proteins enriched in IP–MS experiments. Information about oligos used in this study.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yelagandula, R., Stecher, K., Novatchkova, M. et al. ZFP462 safeguards neural lineage specification by targeting G9A/GLP-mediated heterochromatin to silence enhancers. Nat Cell Biol 25, 42–55 (2023). https://doi.org/10.1038/s41556-022-01051-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-01051-2