Abstract
Ancient DNA is a valuable tool for investigating genetic and evolutionary history that can also provide detailed profiles of the lives of ancient individuals. In this study, we develop a generalised computational approach to detect aneuploidies (atypical autosomal and sex chromosome karyotypes) in the ancient genetic record and distinguish such karyotypes from contamination. We confirm that aneuploidies can be detected even in low-coverage genomes ( ~ 0.0001-fold), common in ancient DNA. We apply this method to ancient skeletal remains from Britain to document the first instance of mosaic Turner syndrome (45,X0/46,XX) in the ancient genetic record in an Iron Age individual sequenced to average 9-fold coverage, the earliest known incidence of an individual with a 47,XYY karyotype from the Early Medieval period, as well as individuals with Klinefelter (47,XXY) and Down syndrome (47,XY, + 21). Overall, our approach provides an accessible and automated framework allowing for the detection of individuals with aneuploidies, which extends previous binary approaches. This tool can facilitate the interpretation of burial context and living conditions, as well as elucidate past perceptions of biological sex and people with diverse biological traits.
Similar content being viewed by others
Introduction
The study of ancient genomes has revolutionised our ability to examine human biology over thousands of years, providing insight into phenotypic variation, social stratification and their impact on health throughout history1,2. In particular, ancient DNA can contribute to our understanding of chromosomal sex in the past, its relationship with gender, and allow for the identification of non-binary chromosomal sex via investigation of the number of sex chromosomes detected in an individual’s karyotype. Deploying genomic data for sex identification can surpass limitations in the sex estimation methods that rely on osteological features, as those can be less accurate or inapplicable when human skeletons are only partially complete3,4,5. Moreover, the subtlety of prepubescent sexual dimorphism means that established osteological sex estimates are rarely applicable to nonadult remains6,7. Several methods have been used to identify chromosomal sex from low-coverage genomes by comparing the proportion of sequences aligning to chromosome Y with sequences aligning to chromosome X or to autosomes8,9,10. However, publicly available tools do not aim to detect other, less frequent, combinations of sex chromosomes or aneuploidies.
Embryonic aneuploidies vary in their severity from pregnancy loss to milder conditions, like Klinefelter syndrome11. Klinefelter syndrome is characterised by infertility and a slightly increased risk for disorders like type 2 diabetes. The majority of individuals with Klinefelter syndrome carry a 47,XXY karyotype, accounting for 1 in 500–650 newborn males, which makes it the most common chromosomal disorder in males12. Although other karyotypes with supernumerary X chromosomes have been described (e.g. 48,XXXY or 49,XXXXY), these occur at much lower frequencies13. Additional copies of chromosome Y (47,XYY), observed in about 1 in 1000 male births, are associated with taller than average stature and an increased risk for asthma, seizures and learning disabilities14,15. On the other hand, individuals with Turner syndrome, a condition that affects 1 in 2500–3000 live female births16, carry only a single chromosome X with karyotype 45,X0 or in mosaic form (e.g. 45,X0/46,XX or 45X0/46,XY). Somatic presentation of Turner syndrome depends on the presence or degree of mosaicism and is associated with shorter stature and a higher incidence of infertility, cardiovascular, renal and endocrine disease17. Autosomal aneuploidies that survive to birth are trisomies 13, 18 and 21 and they are often associated with developmental and heart issues, with trisomy 21 (Down syndrome) being the most common one by far18.
Klinefelter is the only aneuploidy observed multiple times in the ancient genomic record to date, with reports of individuals with a 47,XXY karyotype in Copper Age Iberia19, Early Medieval Finland20, Viking Age Orkney21 and Medieval Portugal22 and Iceland23. Other aneuploidies identified with ancient DNA include a neonate with a 47,XXX karyotype (trisomy X, or triple X syndrome) from Iberia from the second millennium BCE19 and a neonate from Neolithic Ireland with trisomy 2124. The ability to detect these atypical karyotypes in the ancient genomic record has the dual advantage of providing a more detailed insight into the life conditions and societal surroundings of those individuals, while also adding a historical perspective for medical practitioners and patients today.
In this paper, we develop and validate a method that distinguishes between all sex chromosome karyotypes, as well as detecting autosomal aneuploidies such as trisomy 21. We apply this method to new data from ancient Britain and present the oldest known instance of mosaic Turner syndrome dating to the Early Iron Age, three individuals with Klinefelter syndrome, spanning from the Iron Age to the Post-Medieval Period from England, an individual with 47,XYY syndrome in Early Medieval England and an Iron Age infant with Down syndrome.
Results
A computational method to identify aneuploidies using ancient DNA
Our computational approach is based on independently quantifying the number of observed sequences aligning to any chromosome compared to an ‘autosomal baseline’, Na, which we defined as the sum of sequences aligning to chromosomes 1 through 22, excluding chromosomes 13, 18 and 21 which are the only autosomal aneuploidies that survive to birth in appreciable numbers25. We then defined the following metrics. The Rx estimate was calculated by dividing the number of sequences aligning to chromosome X with Na, and the Ry estimate was calculated by dividing the number of sequences aligning to chromosome Y with Na. We estimated the theoretical values for Na, Rx, and Ry expected for chromosome sizes in human genome build 37 and applied our method to published shotgun-sequenced genomes to investigate their overlap with the theoretical values. Leveraging the observed distribution of the empirical data, we optimised the assignment thresholds used to estimate the number of copies of chromosomes X and Y in the karyotype. Additionally, we defined an alternative set of assignment thresholds, optimised for use with target enriched libraries on the widely used “1240k” SNP positions26,27 (see Supplementary Note 4 and Supplementary Figure 3). We obtained standard errors for the assignments using binomial approximations, and used these to test if individuals are significantly deviating from theoretical expectations.
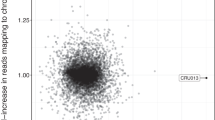
The method described here was applied to 570 published ancient shotgun sequenced genomes from Viking Age Northern Europe21 and ancient Rome28, representing two independent studies from different laboratories to corroborate the absence of batch effects. Individuals with a 46,XX karyotype clustered close to the mean expected Rx value of 0.056 for two copies of chromosome X and their Ry values were close to 0. Meanwhile, individuals with a 46,XY karyotype had Rx values within the typical range expected for a single copy of chromosome X (0.028), while having Ry values close to the mean value of 0.0026, expected of individuals carrying one chromosome Y (Fig. 1a).
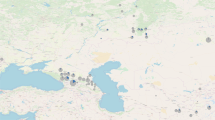
a Rx and Ry estimates, representing the number of sequences aligning to chromosomes X and Y, respectively, as a proportion of the sum of sequences aligned to autosomes, for 134 genomes from Antonio et al.28, 436 genomes from Margaryan et al21. and 5 newly published individuals with sex chromosomal aneuploidies (n = 575) (Supplementary Data 2). b Map of Great Britain with the location of the five sites of origin of individuals presented in this study. c Timeline with archaeological periods of British history and prehistory, spanning from the Iron Age to the present, including dates and karyotypes of newly published individuals with sex chromosomal aneuploidies.
Detection of individuals with 47,XXY and 47,XYY karyotypes
Three individuals, published in this study, clustered in the top right corner in Fig. 1a, having an Rx value within the typical range observed in XX individuals and an Ry value within the typical range for XY individuals. This indicated the presence of two copies of chromosome X and one copy of chromosome Y and suggested that these three individuals (C10427, C11119, C11569) had Klinefelter syndrome (47,XXY). Details about their age and site of origin can be found in Table 1. Contamination estimates based on heterozygosity on chromosome X, commonly used to measure contamination in males, were high (>20%) for C11119 (Sk4205 from the medieval cemetery under Longwall Quad, Magdalen College) and C11569 (Sk30828 from Trinity Burial Ground), consistent with the presence of more than one X chromosome29 (Supplementary Data 1). However, C10427 (WS224 from Wetwang Slack) had a contamination estimate in the typical range of XY individuals (i.e., carrying only one X chromosome). Lack of heterozygosity is a strong indication that the supernumerary chromosome resulted from a copy of the maternal X occurring due to a mitotic error at an early postzygotic stage30. Individual C13582 had the highest observed Ry value out of all studied individuals (Ry = 0.0048) suggesting the presence of an additional copy of Y chromosome in their karyotype, which was also evident using the coverage per chromosome results (Fig. 2).
Proportion of sequences aligned on each chromosome, normalised between chromosomes and across individuals to account for differences in sequencing effort per library (Supplementary Data 3). Individuals C11119 (Sk4205 from Longwall Quad, Magdalen College), C10427 (WS224 from Wetwang Slack), C11569 (Sk30828 from Trinity Burial Ground) with a 47,XXY karyotype carried two copies of chromosome X as well as one copy of chromosome Y. Individual C10462 (WS267 from Wetwang Slack) carried a typical number of sex chromosomes (one X and one Y) and an extra copy of chromosome 21 resulting in a 47,XY, + 21 karyotype. Individual C13582 (Skeleton 16586 from Lincoln Eastern Bypass) had a 47,XYY karyotype, as indicated by the presence of only one copy of chromosome X and two copies of chromosome Y. Individual C10090 (CH163 from Charterhouse Warren) carried no copies of chromosome Y and one copy of chromosome X, with mosaicism (i.e. a minority of cells carried a 46,XX karyotype as evidenced by heterozygosity in chromosome X).
Detection of an Iron Age individual with mosaic Turner syndrome (45,X0/46,XX)
Individual C10090 (CH163 from Charterhouse Warren) had an Rx value (0.033), indicating the presence of one copy of chromosome X and an Ry value (2.32 × 10−5) suggesting the absence of any copies of chromosome Y, thus revealing a 45,X0 karyotype, characteristic of Turner syndrome. Deeper sequencing (final average coverage 9.8x), confirmed the 45,X0 karyotype (Fig. 2).
However, upon closer investigation of the genome of C10090, two observations were suggestive of mosaicism. First, the estimated Rx value was higher than the Rx values of 375 tested individuals identified as XY (5.1 SD from the mean Rx for XY) (Fig. 1a), suggesting the presence of additional chromosome X sequences. Second, heterozygosity estimates29 on polymorphic sites of chromosome X were closer to those of individuals with two copies of chromosome X (16.37%), even when restricting the analysis to authentic ancient DNA sequences to exclude contamination31 (Supplementary Data 1). This does not seem related to structural variation, like the presence of an isochromosome, duplications or deletions of whole regions, that could have explained the observed additional X chromosome sequences, since there was even coverage and consistent levels of heterozygosity across the short and long arms of the chromosome (Supplementary Note 2 and Supplementary Fig. 1).
Hence, an interpretation accommodating a 45,X0 karyotype with a small but substantial excess of chromosome X sequences was mosaicism (45,X0/46,XX) i.e., a smaller proportion of the cells of C10090 carrying a typical 46,XX and the majority of cells carrying a 45,X0 karyotype. Indeed, the majority of adults with Turner syndrome exhibit some degree of mosaicism32,33. Mosaic presentations have also been detected in large-scale genomic datasets like the UK Biobank, where the 45,X0/46,XX karyotype was 6 times more prevalent than the non-mosaic 45,X0 and it was associated with more average stature and fewer reproductive and cardiovascular complications34,35.
Statistical power to detect aneuploidy at low genomic sequencing coverages
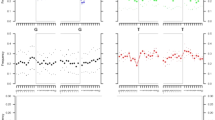
In order to confirm that the detected monosomy could not have been an artefact of poor DNA preservation, the high-coverage genome was downsampled to six different coverage levels 100 times to test the consistency and thresholds of the karyotypic assignment. As shown in Fig. 3b and c, the assignment remained consistent with a 45,X0 karyotype across coverage levels, even with as low as 0.0001x genomic coverage. The Rx and Ry values exhibited higher variability in lower coverage levels, as would be expected by the increasingly limited number of reads, but they remained within the established borders for one copy of X (Fig. 3b) and no copies of Y (Fig. 3c) chromosomes respectively.
a The cranium of the identified individual C10090 (CH163) from Charterhouse Warren (45,X0/46,XX) with mosaic Turner syndrome, exhibiting features consistent with a female morphological characterisation (full description in Supplementary Note 1). Scale bar indicates 5 cm. Image credits: Rick Schulting, Ian Cartwright. b Rx values for C10090 (45,X0/46,XX) across six different coverage levels (n = 507), dashed lines representing Rx boundaries for one copy of chromosome X. Mean is plotted as a circle, median as a horizontal line, lower and upper hinges correspond to the first and third quartiles, error bars represent ± 1 SD (Supplementary Data 4). c Ry values for C10090 (45,X0/46,XX) across six different coverage levels (n = 507), dashed lines representing Ry boundaries for no copies of chromosome Y. Mean is plotted as a circle, median as a horizontal line, lower and upper hinges correspond to the first and third quartiles, error bars represent ± 1 SD (Supplementary Data 4).
Detection of an Iron Age individual with Trisomy 21 (47,XY, + 21)
In addition to estimating the number of sex chromosomes in a karyotype, our approach can be applied to quantify the coverage of any chromosome by estimating the proportion of sequences aligning to that chromosome over the ‘autosomal baseline’ or Na and thus also identifying autosomal aneuploidies. Indeed, a neonate with trisomy 21 was identified with this method (C10462, WS267 from Wetwang Slack) from Iron Age Yorkshire with a 47,XY, + 21 karyotype (Fig. 2, Supplementary Note 3 and Supplementary Fig. 2).
Discussion
The identification of individuals with autosomal and sex chromosomal aneuploidies in the ancient genomic record contributes to a more detailed reconstruction of past societies and highlights the limitations of approaches focusing on binary classification of karyotypic sex. Examining the genomic information from these individuals in conjunction with osteological and archaeological data is crucial in providing a comprehensive perspective of aneuploidy across time.
Certain aneuploidies affecting the sex chromosomes can lead to disorders of sex development (DSDs) which result in mixed or ambiguous sex-related physical characteristics36. Many people with mild disorders experience few effects and are often unaware that they have a DSD. However, severe cases can produce stronger physical effects, including factors, which may be relevant to their own and society’s perception of their sex or gender20. There is evidence that people with DSDs exhibit elevated rates of gender dysphoria37, highlighting the potentially complex relationship between the physical manifestations of DSDs and a person’s sex and social gender as understood by themselves and their wider community38,39,40. It is difficult to know an ancient individual’s conception of their own gender identity and gender norms in the past may not align with those of the present day. However, while most people with sex chromosomal aneuploidies in the past may have lived their lives within conventional gender norms, it is possible that an elevated proportion would have been seen to transgress gender boundaries. Therefore, consideration of past people with sex chromosomal aneuploidies alongside accompanying gendered grave goods may provide a way of exploring past gender variability and how this may or may not relate to the physical effects of DSDs20. This idea was explored by Moilanen and colleagues20 in relation to an individual with possible Klinefelter Syndrome from early medieval Finland who was buried with both typically masculine- and feminine-gendered grave goods.
The three skeletons with Klinefelter syndrome from this study were buried in different contexts and time periods, but with certain similarities, the most important one being that their burials did not reveal any differences in how they were perceived by their contemporaries. The individual with Klinefelter syndrome from Wetwang Slack41 (C10427, Skeleton ID: WS224) was identified osteologically as male, around 18–19 years at death without associated grave goods. The individual found under Longwall Quad, Magdalen College (C11119, Skeleton ID: Sk4205) possessed sexually dimorphic features, which were consistently male; nearly all of them strongly male and two, probably male42. With an estimated stature of 176 cm (+/−3.27 cm), the individual was tall compared with the rest of the burial assemblage. The individual from Trinity Burial Ground (C11569, Skeleton ID: Sk30828) was an adolescent, found without additional artefacts and occupying a plain wooden coffin lacking metal fittings, in a part of the burial ground where many of the burials may have represented poorer members of society. By employing observations of epiphyseal fusion43 and dental development and eruption44, C11569 was estimated to have been between 16 and 19 years of age. Generally, many of the epiphyses were unfused and this suggested delayed skeletal development, or a prolonged growth period. Had the individual lived to attain skeletal maturity, this may have resulted in a taller-than-average stature. Similarly, the individual from Lincoln Eastern Bypass (C13582, Skeleton ID: 16586) with a 47,XYY karyotype had a stature of 184.2 cm, taller than average but within the male height range for the cemetery and was osteologically characterised as male42 (more details on the archaeological contexts and osteological assessment for all individuals in Supplementary Note 1).
Regarding the individual with mosaic Turner syndrome from Charterhouse Warren (C10090, Skeleton ID: CH163), it was possible to confidently match the cranium and mandible (the latter yielding the tooth analysed in this study), but no postcranial elements could be associated. Relatively few formal burials are known for the Early Iron Age of southern Britain, so it is difficult to assess how unusual Charterhouse Warren was for its period. Indeed, considerable variability seems to be the norm45 and disarticulated Iron Age human remains have been found in other caves in southwest England46,47. The main skeletal manifestation of Turner syndrome is short stature48, which could not be assessed given this lack of association. The clinical literature also records some changes in skull morphology49; while these are difficult to address given the skull’s incomplete and reconstructed state (see Supplementary Note 1), the cranium does appear to have a high-arched palate which is one of the skeletal changes seen in Turner syndrome (Fig. 3a). Morphological characteristics of the cranium including slight brow ridges, sharp orbital margins, rounded orbits, high frontal, and smooth nuchal area support a female biological sex estimation42 (Fig. 3a). The Charterhouse Warren individual died as a young adult, ca. 18–22 years old (see Supplementary Note 1). Interestingly, there is a reported relationship between spheno-occipital synchondrosis closure and the onset of puberty50, and in the Charterhouse Warren individual the spheno-occipital synchondrosis is unfused, implying that they would not have experienced menarche, despite their estimated age. A delay in, or absence of, menarche is a feature of Turner syndrome, caused by the absence or low levels of oestrogen16,17.
Identification of aneuploidies such as Down syndrome, which can result in neurodevelopmental problems51 can provide insights into care within ancient societies, as well as how people with these conditions, which have characteristic physical manifestations, were perceived by their peers. The individual with Down syndrome from Wetwang Slack41 (C10462, Skeleton ID: WS267) was a male neonate, buried in the ditch of a square barrow containing a primary adult female burial. However, the burial of neonates within the ditches of pre-existing barrows appears to have been relatively common at Wetwang Slack and is not likely to relate directly to the genetic condition of this individual.
Overall, individuals with aneuploidy from this study were largely buried in accordance with the customs that prevailed in their lifetimes, with the exception of CH163 from Charterhouse Warren (C10090), suggesting that they were most likely considered ordinary members of their community. Inferences regarding their view of themselves and their gender are limited, not least due to the lack of associated gendered grave goods or records. Of course, our understanding of attitudes on identity and gender in Medieval England, surpasses that of the Iron Age, primarily due to the abundance of historical records available for the former. Thus, while patriarchal stereotypes and strictly defined gender roles prevailed in the Medieval period52, female individuals from the Iron Age have intriguingly often been found in elite burials with “objects of power” typically associated with male occupants53,54. An additional aspect to consider, beyond gender identity, is the fact that people with sex chromosomal aneuploidies are substantially more likely to experience later onset of puberty and childlessness55 and the influence these would exert on their daily lives. Indeed, there were osteological indications of delayed growth in one of the three individuals with Klinefelter (C11569), as well as in the individual with mosaic Turner syndrome (C10090) and none of the six individuals in this study were buried in proximity to progeny, although this is not a rare occurrence. Lastly, we would advise caution when interpreting the frequency of aneuploidies in the past, as certain aspects of ancient DNA studies could introduce bias and influence our observations; namely the preferential targeting of crania (often treated differently to postcranial elements in mortuary contexts) due to better DNA preservation56, the scarcity of skeletal material covering certain periods and regions (for example due to widespread cremation) or variable infant representation in skeletal assemblages57.
The method presented here provides a straightforward approach that can identify chromosomal sex as well as autosomal and sex chromosomal aneuploidies (XXY, X0, XYY, XXX and +21) on genomic data of varying quality, while also flagging up likely contamination. In principle, our method can be expected to detect around half of contamination scenarios, i.e. those cases when the contamination originates from an individual with a different chromosomal sex to the individual under study. One individual from a published study (VK12521, who was excluded from downstream analysis in the original paper due to low coverage) had intermediate values both for Rx and Ry (Fig. 1a), indicating contamination with modern-day DNA from someone with different chromosomal sex to the individual under study, which could be relatively common in low-yield ancient DNA samples that have been handled extensively58,59,60. The presence of different-sex contamination tends to inflate the observed Rx values while decreasing Ry values for XY individuals and inflate the Ry values while decreasing Rx values for XX individuals, due to the presence of additional sets of autosomes in the denominator. Importantly, these values are different from those observed in any type of sex chromosomal aneuploidy and thus they can be distinguished with this method, unlike previously8. We provide the classification of putatively contaminated libraries in our implementation of the methods in this paper. A potential limitation of the method is its lack of sensitivity to structural variation, such as micro-deletions in the Y chromosome61. Rare cases of aneuploidy that can be attributed to chromosomal rearrangements18 can also not be excluded, as they cannot be identified by this method currently.
As indicated from our results, the ever-increasing number of ancient genomes will only multiply the opportunities to observe and contextualise genomic diversity and more importantly, methods like the one presented here will provide another layer of information that can contribute to a more detailed reconstruction of the human past.
Methods
Ancient DNA extraction and sequencing
For the genomic data generated in this study, DNA was extracted from auditory ossicles, dentine or the petrosal part of the temporal bone62 and double-indexed single-stranded libraries were prepared and amplified using Agilent Bravo Workstations63,64 before sequencing approximately 2.5–5 million read-pairs per library on the Illumina NextSeq 500 platform (with the exception of C13582 that was sequenced directly on the Illumina NovaSeq 6000 to approximately 70 million reads). The same libraries, except C13582, were then sequenced for a second round on the Illumina NovaSeq 6000 to increase the sequencing depth per library, obtaining a final average coverage of ~0.85x, except in the case of C10090 which was sequenced to 9.8x coverage.
Bioinformatic processing
Sequenced libraries were preprocessed and analysed using the nf-core/eager pipeline65 version 2.3.3 and the version-matched Singularity image. The hs37d5 genome and version 50.0 of AADR (v50.0_1240K)66 were used as references. Read trimming and adapter removal was applied within the pipeline with the following options specified:
“--complexity_filter_poly_g”,
“--clip_forward_adaptor AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC”,
“--clip_reverse_adaptor GGAAGAGCGTCGTGTAGGGAAAGAGTGT”,
“--preserve5p” and “--clip_readlength 35”.
Paired reads were collapsed (“--mergedonly”) and aligned using bwa aln67 with maximum edit distance 0.01 (“--bwaalnn 0.01”) and subsequently filtered (“--run_bam_filtering”). PCR duplicates were identified and removed with DeDup68 (“--dedupper dedup”). Contamination estimates based on heterozygosity on the X chromosome were calculated using ANGSD29 and contamination estimates based on mitochondrial DNA were calculated using schmutzi version 1.5.669.
Y-chromosome haplogroups were inferred with Yleaf version 3.170, using only reads with mapping quality over 30. To determine mitochondrial haplogroups, the deduplicated BAM files output by nf-core/eager were first filtered for mitochondrial reads and realigned using bwa aln (“-n 0.01”, “-l 1024”, and ”-k 2”) to rCRS (rCRS_NC_012920). Variants in mitochondrial reads were identified with BCFtools version 1.10.271,72: BAM files were filtered with “bcftools view” for minimum depth 1 (“--include ‘FMT/DP > = 1”) and variants called with “bcftools call” specifying “--multiallelic-caller”. Mitochondrial haplogroups were then assessed using “haplogrep classify” version 2.2.873 with the best 3 hits calculated (“--hits 3”).
Published data
Published data21,28 for 570 individuals were downloaded from the European Nucleotide Archive.
Sex identification
Sex identification was done using the method reported in this study. The karyotype assignment thresholds were defined separately for shotgun sequenced and for target-enriched libraries and details on their determination can be found in Supplementary Note 4. BAM files for both published and newly generated genomes were filtered for mapping quality (-q30) using SAMtools version 1.1374 and the sex identification script (karyo_RxRy_script.py, from https://github.com/kyriaki-anast/karyo_RxRy) was applied using this command:
“samtools view -q30 bam_file.bam | python2 karyo_RxRy_script.py > output_file”.
Chromosomal coverage
Chromosomal coverage for all newly published individuals (Fig. 2) was calculated by counting the number of sequences mapping to the human reference genome hs37d5, filtering only for regions included in the 1000 Genomes Strict Mask75. The counts of sequences mapping to each chromosome were then normalised between chromosomes and across individuals to account for differences in sequencing effort per library.
Downsampling
The high coverage genome (9.8x) of individual C10090 was randomly downsampled to six different coverage levels, in 100 independent repetitions for each level. Final genomic coverage levels were expressed as proportions of initial coverage. The sex identification method was applied to all downsampled BAM files as described above (applying a mapping quality filter (-q) of 30). Raw output is in Supplementary Data 4 and data is plotted in Fig. 3b and c.
Statistics and reproducibility
Sample sizes were not determined in advance. Methods were applied to published data generated from different groups to control for batch effects. Raw data, analysis output, newly developed scripts and software versions have been made available.
Ethics declaration – sample provenance
Permissions to study the archaeological samples presented in this study were obtained as follows. CH163 - Permissions acquired from David Walker, archaeological curator at Wells Museum. WS 224 (Gen Lab 199) and WS 267 (Gen Lab 162) - Permissions acquired from Jo Buckberry, Reader in Biological Anthropology at the University of Bradford. Sk 4205 - Permissions acquired from Louise Loe, head human osteologist at Oxford Archaeology South and David Radford, archaeologist at Oxford City Council. Sk 30828 - Permissions acquired from Louise Loe, head human osteologist at Oxford Archaeology South and Stephen Rowland, project manager at Oxford Archaeology. Sk 16586 – Permissions acquired from Malin Holst, Managing Director at York Osteoarchaeology. Minimally destructive sampling for aDNA analysis followed guidelines issued by the Department for Culture, Media and Sport (DCMS) and the Advisory Panel on the Archaeology of Burials in England (APABE) (apabe.archaeologyuk.org).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Sequencing data (BAM files) are available on the European Nucleotide Archive (accession code: PRJEB65239). Public datasets21,28 were accessed through the European Nucleotide Archive (accession codes: PRJEB37976 and PRJEB32566). Source data used to plot Fig. 1a are in Supplementary Data 2. Source data used to plot Fig. 2b, c are in Supplementary Data 4. Source data used to plot Fig. 3 are in Supplementary Data 3.
Code availability
The sex identification method76 described and used in this paper can be accessed at https://github.com/kyriaki-anast/karyo_RxRy. This includes an option for target-enriched libraries and autosomal aneuploidy detection.
References
Skoglund, P. & Mathieson, I. Ancient Genomics of Modern Humans: The First Decade. Annu. Rev. Geno. Hum. Genet. 19, 381–404 (2018).
Racimo, F., Sikora, M., Vander Linden, M., Schroeder, H. & Lalueza-Fox, C. Beyond broad strokes: sociocultural insights from the study of ancient genomes. Nat. Rev. Genet. 21, 355–366 (2020).
Kjellström, A. Evaluations of sex assessment using weighted traits on incomplete skeletal remains. Int. J. Osteoarchaeol. 14, 360–373 (2004).
Meindl, R. S., Lovejoy, C. O., Mensforth, R. P. & Don Carlos, L. Accuracy and direction of error in the sexing of the skeleton: implications for paleodemography. Am. J. Phys. Anthropol. 68, 79–85 (1985).
Armit, I. et al. Kinship practices in Early Iron Age South-east Europe: genetic and isotopic analysis of burials from the Dolge njive barrow cemetery, Dolenjska, Slovenia. Antiquity. 97, 403–418 (2023).
Ubelaker, D. H. Estimating age at death from immature human skeletons: an overview. J. Forensic Sci. 32, 1254–1263 (1987).
Stull, K. E., Cirillo, L. E., Cole, S. J. & Hulse, C. N. Subadult sex estimation and KidStats. in Sex Estimation of the Human Skeleton 219–242 (Elsevier, 2020).
Skoglund, P., Storå, J., Götherström, A. & Jakobsson, M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 40, 4477–4482 (2013).
Mittnik, A., Wang, C.-C., Svoboda, J. & Krause, J. A Molecular Approach to the Sexing of the Triple Burial at the Upper Paleolithic Site of Dolní Věstonice. PLoS One 11, e0163019 (2016).
Lamnidis, T. C. et al. Ancient Fennoscandian genomes reveal origin and spread of Siberian ancestry in Europe. Nat. Commun. 9, 5018 (2018).
Gu, C. et al. Chromosomal Aneuploidy Associated With Clinical Characteristics of Pregnancy Loss. Front. Genet. 12, 667697 (2021).
Kanakis, G. A. & Nieschlag, E. Klinefelter syndrome: more than hypogonadism. Metabolism 86, 135–144 (2018).
Visootsak, J. & Graham, J. M. Jr. Klinefelter syndrome and other sex chromosomal aneuploidies. Orphanet. J. Rare Dis. 1, 42 (2006).
Bardsley, M. Z. et al. 47,XYY syndrome: clinical phenotype and timing of ascertainment. J. Pediatr. 163, 1085–1094 (2013).
Ross, J. L., Zeger, M. P. D., Kushner, H., Zinn, A. R. & Roeltgen, D. P. An extra X or Y chromosome: contrasting the cognitive and motor phenotypes in childhood in boys with 47,XYY syndrome or 47,XXY Klinefelter syndrome. Dev. Disabil. Res. Rev. 15, 309–317 (2009).
Ranke, M. B. & Saenger, P. Turner’s syndrome. Lancet 358, 309–314 (2001).
Gravholt, C. H., Viuff, M. H., Brun, S., Stochholm, K. & Andersen, N. H. Turner syndrome: mechanisms and management. Nat. Rev. Endocrinol. 15, 601–614 (2019).
Cuckle, H. & Benn, P. Review of epidemiological factors (other than maternal age) that determine the prevalence of common autosomal trisomies. Prenat. Diagn. 41, 536–544 (2021).
Villalba-Mouco, V. et al. Genomic transformation and social organization during the Copper Age-Bronze Age transition in southern Iberia. Sci. Adv. 7, eabi7038 (2021).
Moilanen, U. et al. A Woman with a Sword? – Weapon Grave at Suontaka Vesitorninmäki, Finland. Eur. J. Archaeol. 25, 42–60 (2022).
Margaryan, A. et al. Population genomics of the Viking world. Nature 585, 390–396 (2020).
Roca-Rada, X. et al. A 1000-year-old case of Klinefelter’s syndrome diagnosed by integrating morphology, osteology, and genetics. Lancet 400, 691–692 (2022).
Ebenesersdóttir, S. S. et al. Ancient genomes from Iceland reveal the making of a human population. Science 360, 1028–1032 (2018).
Cassidy, L. M. et al. A dynastic elite in monumental Neolithic society. Nature 582, 384–388 (2020).
Hassold, T. & Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (2001).
Fu, Q. et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl Acad. Sci. USA 110, 2223–2227 (2013).
Rohland, N. et al. Three assays for in-solution enrichment of ancient human DNA at more than a million SNPs. Genome Res. 32, 2068–2078 (2022).
Antonio, M. L. et al. Ancient Rome: A genetic crossroads of Europe and the Mediterranean. Science 366, 708–714 (2019).
Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinforma. 15, 356 (2014).
Tüttelmann, F. & Gromoll, J. Novel genetic aspects of Klinefelter’s syndrome. Mol. Hum. Reprod. 16, 386–395 (2010).
Skoglund, P. et al. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl Acad. Sci. USA 111, 2229–2234 (2014).
Hook, E. B. & Warburton, D. The distribution of chromosomal genotypes associated with Turner’s syndrome: livebirth prevalence rates and evidence for diminished fetal mortality and severity in genotypes associated with structural X abnormalities or mosaicism. Hum. Genet. 64, 24–27 (1983).
Zhong, Q. & Layman, L. C. Genetic considerations in the patient with Turner syndrome-45,X with or without mosaicism. Fertil. Steril. 98, 775–779 (2012).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Tuke, M. A. et al. Mosaic Turner syndrome shows reduced penetrance in an adult population study. Genet. Med. 21, 877–886 (2019).
Hughes, I. A. Disorders of sex development: a new definition and classification. Best. Pract. Res. Clin. Endocrinol. Metab. 22, 119–134 (2008).
Furtado, P. S. et al. Gender dysphoria associated with disorders of sex development. Nat. Rev. Urol. 9, 620–627 (2012).
Fisher, A. D. et al. Gender identity, gender assignment and reassignment in individuals with disorders of sex development: a major of dilemma. J. Endocrinol. Invest. 39, 1207–1224 (2016).
Lee, P. A. et al. Global Disorders of Sex Development Update since 2006: Perceptions, Approach and Care. Horm. Res. Paediatr. 85, 158–180 (2016).
Garofalo, E. M. & Garvin, H. M. The confusion between biological sex and gender and potential implications of misinterpretations. in Sex Estimation of the Human Skeleton 35–52 (Elsevier, 2020).
Armit, I. The Wetwang/Garton Slack Project (WGSP). York: Archaeology Data Service. https://doi.org/10.5284/1030285 (2021).
Ubelaker, D. H. & Buikstra, J. E. Standards for Data Collection from Human Skeletal Remains: Proceedings of a Seminar at the Field Museum of natural history. (Arkansas Archeological Survey, 1994).
Scheuer, L. & Black, S. Developmental Juvenile Osteology. (Elsevier Academic Press, 2000).
AlQahtani, S. J., Hector, M. P. & Liversidge H. M. Brief communication: The London atlas of human tooth development and eruption. Am. J. Phys. Anthropol. 142, 481–490 (2010).
Harding, D. W. Death and burial in Iron Age Britain / D.W. Hardin. (Oxford University Press, 2016).
Cox, M. & Loe, L. The Human skeletal remains from Fishmonger’s Swallet, Alveston, Gloucestershire: Evidence for anthropogenic modification. Proc. Univ. Bristol Spelaeol. Soc. 29, 33–66 (2022).
Langford, F. Third report on Read’s Cavern (Keltic Cavern). Proceedings of the University of Bristol Spelaeological Society 1, 135–143 (1922).
Binder, G. Short stature due to SHOX deficiency: genotype, phenotype, and therapy. Horm. Res. Paediatr. 75, 81–89 (2011).
Doswell, B. H., Visootsak, J., Brady, A. N. & Graham, J. M. Jr. Turner syndrome: an update and review for the primary pediatrician. Clin. Pediatr. 45, 301–313 (2006).
Alhazmi, A. et al. Timing and rate of spheno-occipital synchondrosis closure and its relationship to puberty. PLoS One 12, e0183305 (2017).
Bull, M. J. Down Syndrome. N. Engl. J. Med. 382, 2344–2352 (2020).
Hill, B. Women, Work And Sexual Politics In Eighteenth-Century England. (Routledge, 1993).
Arnold, B. “Honorary Males” or Women of Substance? Gender, Status, and Power in Iron-Age Europe. J. Eur. Archaeol. 3, 153–168 (1995).
Mays, S. et al. Sex identification of a Late Iron Age sword and mirror cist burial from Hillside Farm, Bryher, Isles of Scilly, England. J. Archaeol. Sci. Rep. 52, 104099 (2023).
Zhao, Y. et al. Detection and characterization of male sex chromosome abnormalities in the UK Biobank study. Genet. Med. 24, 1909–1919 (2022).
Charlton, S., Booth, T. & Barnes, I. The problem with petrous? A consideration of the potential biases in the utilization of pars petrosa for ancient DNA analysis. World Archaeol. 51, 574–585 (2019).
McFadden, C., Muir, B. & Oxenham, M. F. Determinants of infant mortality and representation in bioarchaeological samples: A review. Am. J. Biol. Anthropol. 177, 196–206 (2022).
Green, R. E. et al. Analysis of one million base pairs of Neanderthal DNA. Nature 444, 330–336 (2006).
Malmström, H., Storå, J., Dalén, L., Holmlund, G. & Götherström, A. Extensive human DNA contamination in extracts from ancient dog bones and teeth. Mol. Biol. Evol. 22, 2040–2047 (2005).
Krause, J. et al. A Complete mtDNA Genome of an Early Modern Human from Kostenki, Russia. Curr. Biol. 20, 231–236 (2010).
Kohn, T. P., Kohn, J. R., Owen, R. C. & Coward, R. M. The Prevalence of Y-chromosome Microdeletions in Oligozoospermic Men: A Systematic Review and Meta-analysis of European and North American Studies. Eur. Urol. 76, 626–636 (2019).
Rohland, N., Glocke, I., Aximu-Petri, A. & Meyer, M. Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc. 13, 2447–2461 (2018).
Gansauge, M.-T., Aximu-Petri, A., Nagel, S. & Meyer, M. Manual and automated preparation of single-stranded DNA libraries for the sequencing of DNA from ancient biological remains and other sources of highly degraded DNA. Nat. Protoc. 15, 2279–2300 (2020).
Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, https://doi.org/10.1101/pdb.prot5448 (2010).
Fellows Yates, J. A. et al. Reproducible, portable, and efficient ancient genome reconstruction with nf-core/eager. PeerJ 9, e10947 (2021).
Mallick, S. et al. The Allen Ancient DNA Resource (AADR): A curated compendium of ancient human genomes. bioRxiv 2023.04.06.535797. https://doi.org/10.1101/2023.04.06.535797 (2023).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Peltzer, A. et al. EAGER: efficient ancient genome reconstruction. Genome Biol. 17, 60 (2016).
Renaud, G., Slon, V., Duggan, A. T. & Kelso, J. Schmutzi: estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 16, 224 (2015).
Ralf, A., Montiel González, D., Zhong, K. & Kayser, M. Yleaf: Software for Human Y-Chromosomal Haplogroup Inference from Next-Generation Sequencing Data. Mol. Biol. Evol. 35, 1291–1294 (2018).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, (2021).
Weissensteiner, H. et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–W63 (2016).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Anastasiadou, K. karyo_RxRy.v1.0.1. https://doi.org/10.5281/zenodo.10186749 (2023).
Acknowledgements
This study was supported by the Wellcome Trust (217223/Z/19/Z) and Francis Crick Institute core funding (FC001595) from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust. P.Sk. is supported by the European Molecular Biology Organization, the Vallee Foundation, and the European Research Council (grant no. 852558). L.S. acknowledges support provided by a Sir Henry Wellcome fellowship [220457/Z/20/Z]. Analysis of the Charterhouse Warren assemblage was supported by the British Academy (SG163375). Analysis of the samples from Wetwang Slack forms part of the COMMIOS project which has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 834087). Access to the Wetwang Slack human remains collections was provided by the Biological Anthropology Research Centre (BARC), University of Bradford. The excavation and analysis of the medieval cemetery under Longwall Quad, Magdalen College was commissioned by Magdalen College and excavation and analysis of the Trinity Burial Ground Hull assemblage was commissioned by Balfour Beatty for National Highways. The excavation and analysis of the Lincoln Eastern Bypass assemblage was commissioned by the Lincolnshire County Council and conducted by Network Archaeology and York Osteoarchaeology. Image credits for Fig. 3a: Rick Schulting, Ian Cartwright. The authors thank the staff of the Advanced Sequencing Facility and the Bioinformatics and Biostatistics STP at The Francis Crick Institute for their contributions to this study. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Funding
Open Access funding provided by The Francis Crick Institute.
Author information
Authors and Affiliations
Contributions
Conceptualization: K.A., P.Sk. Data curation: K.A., C.B. Formal Analysis: K.A. Methodology: K.A., P.Sk. Software: K.A., P.Sk. Validation: K.A. Funding acquisition: L. L., I. A., R. S., P. Sk. Investigation: K.A., M.S., T.B., L.S., M.K., M.W., F.T., J.M., P.Sw., I.G., J.B., M.H., M.M., D.F., P.P., J.R.V.,T.A., B.F., M.G., A.G., L.M., S.R., D.W., H.W., A.W., P.Sk. Supervision: L.L., I.A., R.S., P.Sk. Visualisation: K.A., R.S. Writing – original draft & revisions: K.A., M.S., T.B., C.B., R.S., A.W., L.M., H.W.,K.K., P.P., J.R.V., J.B., M.H., S.R., L.L., I.A., P.Sk.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Zuzana Hofmanova and George Inglis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Anastasiadou, K., Silva, M., Booth, T. et al. Detection of chromosomal aneuploidy in ancient genomes. Commun Biol 7, 14 (2024). https://doi.org/10.1038/s42003-023-05642-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-05642-z
This article is cited by
-
Ancient DNA reveals first known case of sex-development disorder
Nature (2024)
-
Cases of trisomy 21 and trisomy 18 among historic and prehistoric individuals discovered from ancient DNA
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.