Abstract
Hemogenic endothelium (HE) with hematopoietic stem cell (HSC)-forming potential emerge from specialized arterial endothelial cells (AECs) undergoing the endothelial-to-hematopoietic transition (EHT) in the aorta-gonad-mesonephros (AGM) region. Characterization of this AECs subpopulation and whether this phenomenon is conserved across species remains unclear. Here we introduce HomologySeeker, a cross-species method that leverages refined mouse information to explore under-studied human EHT. Utilizing single-cell transcriptomic ensembles of EHT, HomologySeeker reveals a parallel developmental relationship between these two species, with minimal pre-HSC signals observed in human cells. The pre-HE stage contains a conserved bifurcation point between the two species, where cells progress towards HE or late AECs. By harnessing human spatial transcriptomics, we identify ligand modules that contribute to the bifurcation choice and validate CXCL12 in promoting hemogenic choice using a human in vitro differentiation system. Our findings advance human arterial-to-hemogenic transition understanding and offer valuable insights for manipulating HSC generation using in vitro models.
Similar content being viewed by others
Introduction
Hematopoietic stem cells (HSCs) can develop into all blood cell lineages and are vital to individual survival1,2. The first embryonic HSC emerges in the AGM through EHT3, whereby individual HE becomes round form HCs that aggregate into intra-aortic clusters (IACs)4,5,6,7,8,9,10,11,12,13. Within those HCs, CD41+CD45- T1 pre-HSCs further differentiate into CD41+CD45+ T2 pre-HSCs before maturing into definitive HSCs14,15.
During mouse EHT within the AGM, not all AECs differentiate into HSC-forming HEs; some may develop into mature arterial cells16. Moreover, studies indicate that immature HEs need to go through an arterialization process before differentiating into definitive lymphoid–myeloid progenitors9,17. These findings indicate an intimate relationship between arterial specification and HSC formation7. Furthermore, research suggests that the transition from AEC to HE passes through a relatively unexplored intermediate pre-HE stage18, where cells initiate hematopoietic programs while retaining arterial features. Within this AEC to pre-HE to HE transition, a developmental bottleneck exists between pre-HE and HE18. Runx1, a master regulator for EHT, has been shown to assist pre-HEs in overcoming this bottleneck and allowing cells to further develop into HEs18. These findings suggest that pre-HE may serve as a critical stage for hemogenic fate determination, and AECs require driving forces to achieve hemogenic fate. Nevertheless, the mechanisms that specifically facilitate the hemogenic choice of mouse AEC remain poorly understood.
Human HSC-forming HEs may also originate from AECs9,17,19,20, and the existence of a human pre-HE stage has been suggested20. Nonetheless, it remains unclear whether the role of the pre-HE stage during the AEC-to-HE transition is similar between human and mouse. However, for ethical reasons, details regarding the human AEC-to-HE transition are less well characterized compared to that of the mouse. Furthermore, the mouse has long served as a model organism to study various human biological processes, including EHT11,21,22. This highlights the need for cross-species comparative studies that identify cellular differences and explore developmental relationships between species23,24,25,26,27,28,29,30,31, thereby bypassing the limitations constraining human EHT research.
For cross-species comparative studies, homologous genes that share similar DNA sequences and functions across species provide an entry point. Various cross-species analysis tools have been developed utilizing homologous genes24,25,32,33,34. For instance, La Manno et al.24 used a Bayesian generalized linear model (GLM) to identify significantly expressed genes in each cell type and compared analogous cell types across tested species using these genes. However, these approaches are restricted in cases where the annotation of corresponding cell types is unavailable. This requirement is frequently obstructed by subjective assumptions and insufficient markers for cell type identification, especially in non-model species, which constrains cross-species analysis. Consequently, a tool that requires no prior knowledge could circumvent these limitations.
To further elucidate the AEC-to-HE transition, here we introduce HomologySeeker, a cross-species analysis pipeline that detects homologous genes exhibiting highly variable expression in an unbiased manner. Without prior cell type annotation in reference or query species, HomologySeeker accurately captures well-established EHT-related homologous genes between mouse and human EHT ensembles that are constructed from publicly available single-cell transcriptome profiles. We present evidence to show that mouse and human EHT exhibit analogous cell type correspondences, with minimal T1/2 pre-HSC signals observed in human cells. Furthermore, mouse and human exhibit similar developmental trajectories from arterial to hematopoietic groups and display comparable transcriptional expression patterns along the trajectories. Additionally, the pre-HE stage serves as a bifurcation point where cells face hemogenic or arterial choices, and this bifurcation point is conserved between both species. We further examine publicly available human spatial transcriptomics data to identify the ligand modules responsible for the distinct developmental choices of cells in the pre-HE stage between hemogenic and arterial fates. Using a human in vitro hematopoietic differentiation system, we validate the role of CXCL12 cytokine, identified from the module that facilitates further development into the hemogenic fate, in promoting the hemogenic choice of hemogenic precursors. Furthermore, we observed an increased production of HPCs with enhanced multilineage differentiation capability in the CXCL12 group compared to the control group. Our results contribute to a deeper understanding of human AECs and their selection of the hemogenic fate in vivo. Moreover, HomologySeeker provides a valuable tool for comparative transcriptomic studies across various contexts.
Results
HomologySeeker identifies EHT-related highly variable homologous genes
To bypass the requirement of prior annotation, we developed an analysis pipeline called HomologySeeker (“Methods”). HomologySeeker utilizes highly variable expressed homologous genes (herein termed Homologous-HVGs), assuming that genes with high expression variability are more likely to represent genuine biological variation35. Briefly, HomologySeeker identifies homologous genes among tested species, ranks them based on expression variance, and then sets a cutoff using the mean value of all variances to retrieve genuine Homologous-HVGs (Fig. 1a; “Methods”). The calculation of Homologous-HVGs proceeds in an unsupervised manner, requiring no additional information, and is applicable for downstream analysis, thus offering the potential for flexible cross-species analysis application.
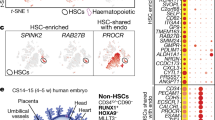
a HomologySeeker workflow. b Pearson correlation heatmap showing the relationship between mouse and human cell types, with colors representing Pearson correlation coefficients. Endo, endothelium section; Hemo, arterial/hemogenic section; Hema, hematopoietic section. c Top-left: Schematic of query projection. Bottom: Heatmap displaying projection scores of human cell types assigned new identities based on mouse reference. Columns represent human cells; rows represent mouse cell types; color intensity represents prediction scores. Unified: combined human cell type annotation; Origin: original human dataset annotation; Stage: cell timepoints. d UMAP visualization of human sub-clusters (C1–C10) post-unsupervised clustering. e Schematic of SingleCellNet model from (Tan et al.47). f SingleCellNet prediction scores of human cells using mouse cell types as a reference; higher scores indicate greater confidence. g Expression levels of marker genes across mouse cell types (upper panel) and human sub-clusters (lower panel). V, venous; A/Endo, arterial or endothelial; Hema, hematopoietic.
To evaluate the performance of HomologySeeker, we re-analyzed La Manno et al.24 scRNA-seq datasets with pre-assigned cell identities removed (Supplementary Fig. 1a; “Methods”; Supplementary Data). We first identified overlapping Homologous-HVGs between human and mouse and then calculated the transcriptome correlation among cell clusters (Supplementary Fig. 1b, c). We then assigned identities to these cell clusters based on the expression level of the marker genes used in La Manno et al.24. Although we observed nearly 50% overlap between our Homologous-HVGs and the homologous genes identified by La Manno et al. (Supplementary Fig. 1d), the transcriptome correlation analysis using Homologous-HVGs reproduced the same paired cell types described in La Manno et al. (Supplementary Fig. 1e, left heatmap). This demonstrated the feasibility of Homologous-HVGs for comparative analysis across species.
We then applied HomologySeeker to identify Homologous-HVGs in mouse and human EHT (“Methods”). To encompass cells at various EHT stages before screening Homologous-HVGs, we constructed human and mouse EHT ensembles using published single-cell RNA-seq datasets generated from surface markers-enriched endothelial cells (ECs), hemogenic ECs (HECs), IACs, hematopoietic stem/progenitor cells (HSPCs), and fetal liver HSCs (FL-HSCs) (“Methods”; Supplementary Data). Directly merging datasets caused cells to cluster based on the dataset rather than cell type (Supplementary Fig. 2a). To mitigate batch effects among various datasets, we employed the “anchor”-based integration method36 for merging datasets. Using the cell identities defined by the original studies (hereafter called pre-defined)18,19,20,37,38, similar cell types tended to cluster together, prompting us to unify corresponding cell types (Supplementary Fig. 2b). We observed a continuous landscape in both mouse and human EHT ensembles, as shown in the two-dimension UMAP (Supplementary Fig. 2c, d). Notably, the mouse EHT ensemble captured the accumulated “bulge” (Supplementary Fig. 2c), identified as the pre-HE stage in Zhu et al.18. These results indicate that our data merging preserved the biological relationships among cells without distorting the original datasets.
Using HomologySeeker, we identified 2456 and 3248 Homologous-HVGs in mouse and human ensembles, respectively (Supplementary Fig. 3a; Supplementary Data). As expected, EHT-associated genes, including SOX1739, RUNX18,40,41, and MYB42,43,44, appeared as Homologous-HVGs (Supplementary Fig. 3a). Among these Homologous-HVGs, 1628 genes are common between mouse and human (hereafter as “common EHT-associated Homologous-HVGs in Supplementary Fig. 3a). In this gene set, we found that 76 out of the top 100 biological pathways enriched in mouse and human were identical (Supplementary Fig. 3c; Supplementary Data), suggesting that a large proportion of these common EHT-associated Homologous-HVGs may participate in similar biological processes. Nonetheless, species-specific terms such as “Coagulation” in human and “Wound healing” in mouse that are both related to the tissue healing process were also observed. These differences could reflect underlying biological differences or variations in nomenclature between species.
Mouse and human EHT display analogous developmental relationship
To evaluate the similarities between mouse and human EHT, we first examined cellular correspondence. We calculated the transcriptome correlation between the pre-defined cell types of mouse and human EHT ensembles using the common EHT-associated Homologous-HVGs (“Methods”, Supplementary Fig. 3b). Based on the relative levels of Pearson correlation coefficients, we grouped corresponding mouse and human pre-defined cell types into three sections on the resulting heatmap: Endo (venous group), Hemo (arterial/hemogenic group), and Hema (Hematopoietic group) (green, orange, and red rectangles, respectively in Fig. 1b). We also conducted “anchor”-based query projection to assign potential human cell identities, using the mouse as a reference (Fig. 1c). Consistent with the transcriptome correlation analysis, almost all human VECs were anchored to mouse Wnt EC, human AEC/HEC to mouse AEC/pre-HE/HEC, and HSPC/HC to mouse IAC/FL-HSC (Fig. 1c).
Interestingly, using pre-defined annotation, we found that the majority of human HSPC1 (~69%, GJA5+ HSPC) and HSPC3 (~79%, GFI1B+ HSPC) exhibited higher mouse FL-HSC scores (Fig. 1c, Supplementary Data). The other human cell types within the Hema group exhibited either higher IAC or FL-HSC scores, indicating diverse hematopoietic potentials within these cells. Notably, a negligible number of human cells were anchored to mouse T1/T2 pre-HSC (T1/T2). Since the “anchor”-based projection relies on the shared nearest neighbors (SNN) of reference cells36, we speculated whether the limited “human T1/T2” signals in “anchor”-based query projection resulted from the large number of mouse IACs co-occupying the T1/T2 in UMAP (Supplementary Fig. 4a, upper panel). However, even with a modified mouse EHT ensemble excluding IACs, we noted minimal T1/T2 assignment of human cells (Supplementary Fig. 4b, lower panel). Considering our EHT ensembles include diverse datasets with various marker combinations for isolating specific cell populations, the weak T1/2 signal observed in human cells may be due to the variability in marker usage and the limited presence of certain cell types (Supplementary Data). Therefore, the existence of pre-HSCs in human EHT remains uncertain, necessitating further exploration.
The observation that each human pre-defined cell type correlates with several mouse pre-defined cell types indicates heterogeneity within human cell types. To delineate potential cell sub-clusters within those heterogeneous pre-defined human cell types, we re-segmented the human EHT ensemble using the Louvain algorithm, a graph-based unsupervised clustering method45 (“Methods”). We employed a “cluster tree”46 to objectively choose a stable clustering resolution, resulting in 10 sub-clusters (Supplementary Fig. 5a; Fig. 1d; C1–10). The correlation between mouse cell types and human sub-clusters reveals additional details (“Methods”, Supplementary Fig. 5b). For example, human C4 exhibits the strongest correlation to mouse AEC while human C6 has the highest correlation to mouse pre-HE (Supplementary Fig. 5b). We then tried to assign cell identities to human sub-clusters, using the mouse as a reference (“Methods”). We applied a machine learning algorithm47 to obtain cell type signatures by using the expression levels of common EHT-associated Homologous-HVGs from mouse to train prediction scores for human sub-clusters (Fig. 1e). We observed high AEC, pre-HE, and HE scores, but low T1, T2, IAC, and FL-HSC scores in human C4–C7 sub-clusters (Fig. 1f). Notably, human C6 was assigned the highest pre-HE score compared to all other human sub-clusters. Conversely, C8–10 exhibited low scores in AEC, pre-HE, T1, and T2 but high IAC and FL-HSC scores (Fig. 1f, Supplementary Fig. 4c). Furthermore, mouse pre-defined cell types and human re-defined sub-clusters displayed comparable EHT marker gene expression patterns (Fig. 1g).
Other than cellular correspondence, we then constructed developmental trajectories for mouse and human EHT ensembles using Monocle348. Given the arterial origin of the definitive HSCs from the AGM region49,50, we assigned mouse AECs and human C4 as the trajectory roots (Fig. 2a, b). Mouse and human EHT ensembles both displayed a continuous trajectory from AEC/C4 to FL-HSC/C10, respectively. This continuous trajectory aligns with the previous findings18,19,20,37,38. We noted that the human developmental trajectory diverged toward C9 and C10 (Fig. 2b). Although both C9 and C10 displayed high IAC scores, C9 had a higher FL-HSC score (Fig. 1f). To investigate the hematopoietic potential of C9 and C10, we utilized marker gene sets identified from the DEGs inference method on publically available hematopoietic progenitor cell (HPC) transcriptome profiles51 (“Methods”; Supplementary Fig. 5b). C9 consistently displayed higher hematopoietic stem cell/multipotent progenitor (HSC/MPP) scores, as well as higher scores for LMPP1/2 that associated with monocyte/dendritic progenitors (MD) and granulocyte-monocyte progenitors (GMP) (Supplementary Fig. 5c, lower panel). Conversely, C10 showed higher lymphoid-primed MPPs (LMPP3) scores, indicating its potential for differentiation toward the lymphoid lineage (Supplementary Fig. 5c, red circle). These results suggest that C9 and C10 likely arise independently from a common precursor (C8) and possess distinct hematopoietic potentials. This finding is consistent with recent research in mouse that HSCs and hematopoietic progenitors may be generated independently of the heterogeneous pre-HSPC population52,53.
a, b Developmental trajectory of the mouse (a) and human (b) EHT ensembles from AEC/C4 to hematopoietic groups, with black lines inferring developmental paths. c Identification of TF modules from Homologous-HVGs set. Venn plot shows the overlap of mouse and human TFs derived from Homologous-HVGs. Lower Pannal: Hierarchical clustering of mouse and human TFs (66 + 160, 114 + 160, respectively), with the blue section representing TF module 1 and the red section representing TF module 2. d TF expression patterns along developmental trajectories. Upper panel: Pseudo-time changes in mouse and human developmental trajectories. Middle panel: Module scores of mouse and human TF modules along the pseudo-time axis. Lower panel: Expression patterns of representative TFs from two modules.
As transcription factors (TFs) play a vital role in EHT21, we further investigated the behavior of TFs along this trajectory. We first selected TFs from Homologous-HVGs (226 and 274 TFs from mouse and human, respectively) (“Methods”; Fig. 2c). More than 50% of these TFs are common to both species. Numerous well-known EHT regulators, including endothelial/arterial TFs SOX17/18 and hematopoietic TFs RUNX1/MYB, are present among these shared TFs (Fig. 2c, upper panel). The combination of both endothelial and hematopoietic TFs highlights the simultaneous regulation of endothelial and hematopoietic programs in EHT16,19,20,37,38. To identify potential TF regulatory modules, we applied hierarchical clustering to analyze the expression changes of these TFs along the EHT trajectory and used DynamicTreeCut54 to discern TF modules (Fig. 2c, lower panel). We found that both species displayed two distinct TF modules. To assess the behavior of these two TF modules behave along the trajectory, we assigned module scores to each cell type (“Methods”; Fig. 2d). For both species, TF module 1 displayed a downward trend along the trajectory, indicating the downregulation of this module as EHT progresses. This TF module 1 contains Nr2f2, Hey2, Sox17, and Sox18, with the majority being endothelial marker genes. TF module 2 comprises Runx1, Myb, Spi1, and Hif, all recognized as positive hematopoietic regulators. TF module 2 showed a consistent pattern along the mouse EHT trajectory while exhibiting an increasing trend in the human EHT trajectory. Together, these results indicate that mouse and human EHT share similarities in corresponding cell types, developmental trajectories, and transcriptional expression patterns.
Mouse and human EHT harbor a bifurcation point during the AEC-to-HE transition
During mouse EHT, subsets of AECs undergo cell fate choices towards either HE or mature arterial fate (late AEC, lAEC)16 (Fig. 3a). Within the mouse ensemble, we observed a bifurcation in the EHT trajectory where pre-HE diverged towards HE or E11.5 EC (hereafter EC) (Fig. 2a; zoomed in Fig. 3b, left). This EC exhibited high expression of Ltbp4, a mature arterial feature gene55 (Fig. 3b, right), leading us to hypothesize that pre-HE may be a bifurcation point for cell fate decisions. To investigate this hypothesis, we projected publicly available single-cell transcriptome profiles, which functionally validate the cell fate choices of early AEC (eAEC) toward HE or lAEC16, onto our mouse EHT ensemble (“Methods”; Fig. 3a). The published eAEC-to-HE trajectory aligned with our AEC-to-HE trajectory (Fig. 3c), and areas where eAEC makes choices between HE or lAEC fates corresponded to pre-HE in our data, indicating that pre-HE possesses a transcriptome comparable to eAEC. In addition, lAEC was projected to the tip of the “bulge” where the EC is located. EC showed the highest module scores measuring the expression level of genes involved in EC development, arterial EC differentiation, and blood vessel EC differentiation (Fig. 3d, left panel), supporting the arterial fate choice of late AEC. Moreover, we observed parallel patterns in the enriched biological pathways during the transition from pre-HE to HE/EC and from early AEC to HE/late AEC (Supplementary Fig. 8a, b). These results indicate that the eAEC identified in previous work16 may correspond to the pre-HE in our mouse EHT ensemble; consequently, pre-HE in our mouse EHT ensemble may possess the ability to choose different cell fates.
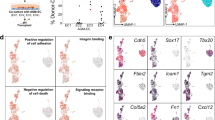
a Developmental trajectory starts from early AEC (eAEC) and bifurcates toward late AEC (lAEC) or HEC. b Left: Close-up of mouse trajectory concerning AEC-Pre-HE-EC/HE transition from Fig. 2a. Right: Expression levels of marker genes for mouse cell types-Nr2f2 (venous marker), Pecam1 (CD31-coding gene) and Cdh5 (endothelial markers), Ltbp4 (mature arterial marker), Ptprc (CD45-coding gene, hematopoietic marker). c Query projection of mouse scRNA-seq data from (Hou et al.16) into mouse ensemble (gray). eAEC (blue), early AEC; lAEC (red), late AEC; HEC (orange). d Module scores for mouse and human cell types/sub-clusters based on gene sets from GO terms and TF module 1. e Query projection of human GJA5+ AEC from Crosse et al.27 into human ensemble (gray). f Human developmental trajectory starting from C4 and branching at C6 toward GJA5+ AEC and C7. g Top five biological pathways enriched by DEGs between mouse pre-HE and HE, and analogous terms enriched by DEGs between pre-HE and EC. Red/blue GO terms indicate enrichment by up/down-regulated DEGs. h Top 5 biological pathways enriched by DEGs between human C6 and C7, and matching terms enriched by DEGs between C6 and GJA5+ AEC. Red/blue GO terms signify enrichment by up/downregulated DEGs. i, j Local regulatory networks among the top ten upstream regulators of upregulated DEGs between mouse pre-HE and HE (i), and between human C6 and C7 (j). Network edges represent co-regulatory relationships with edges involving MYC, highlighted in red.
We also observed a bulge between human C6 and C7 sub-clusters (Fig. 1d). Given the similarity between these two species (Figs. 1 and 2), human EHT might encounter similar cell fate choices during the AEC-to-HE transition. Notably, human C6 showed the highest pre-HE score among all other sub-clusters (Fig. 1f) and exhibited pre-HE signatures in terms of dynamic trajectory, marker genes, and cell composition20 (Supplementary Fig. 6a–c). These findings indicated that C6 likely represents human pre-HE and faces bifurcation choice akin to mouse pre-HE. To explore this, we projected published GJA5+ AECs from CS16 dorsal aorta27 (hereafter called GJA5+ AEC) onto the human EHT ensemble using a similar approach to Fig. 2b (“Methods”). Most GJA5+ AECs clustered at the tip of the C6/C7 bulge, with some scattered at C5/C6 (Fig. 3e), resembling mouse lAECs (Fig. 3c). We then assessed whether C6 exhibits diverging trajectories toward C7 or GJA5+ AEC. After merging human C4-C7 and GJA5+ AEC into a localized cohort, we performed Monocle3 trajectory analysis using a similar strategy to Fig. 2a, b (“Methods”). Comparable to the bifurcation choices encountered by mouse pre-HE, we observed a trajectory starting from C4, reaching C6, and then diverging into C7 or GJA5+ AEC (Fig. 3f; Supplementary Fig. 7a, b). The transition from C6 to C7 revealed the emergence of hemogenic markers (RUNX18,40,41 and KCNK1720) (Supplementary Fig. 7d), whereas the C6 to GJA5+ AEC transition retained pre-HE markers20 ALDH1A1 and IL33 but lacks expression of hematopoiesis-associated genes (HOXA956 and MLLT357). This indicates that the transition from C6 to C7 is involved EHT, while the C6 to GJA5+ AEC transition follows arterial processes. Similar to mouse ECs, human GJA5+ AECs exhibited comparatively high scores in all tested modules (Fig. 3d, right panel). Notably, human C5 appeared as an outlier, suggesting that C5 might not participate in EHT either (Fig. 3f).
We further examined if similar transcriptional networks govern the fate choices in mouse pre-HE and human C6 sub-cluster. We integrated GJA5+ AECs into our human EHT ensemble (Supplementary Fig. 7c) and calculated the differentially expressed genes (DEGs) between these two choices (Supplementary Fig. 8c, d; Supplementary Data, pre-HE vs. HE/EC in mouse, C6 vs. V7/GJA5+ AEC), followed by GO term analysis (Fig. 3g, h; Supplementary Data). Upregulated DEGs in mouse pre-HE-to-EC and human C6-to-GJA5+ AEC transition were enriched for vasculature/angiogenesis development and endothelial development pathways, indicating a vascular fate toward EC/GJA5+ AEC (Fig. 3g, h). These pathways were down-regulated in the pre-HE-to-HE transition in mice and the C6-to-C7 transition in humans (Fig. 3g, h), suggesting an alternative choice toward HE/C7 direction. Upregulated DEGs of the pre-HE-to-HE transition in mouse were mainly enriched for ribosome-related pathways, while in human, they predominantly focused on protein modification-related pathways. (Fig. 3g, h, bar plot in red). This is consistent with prior research that highlighted the role of enhanced ribosomal activity and protein translational processes in the development of HSC-primed HE across both species16,19. We then employed ChEA358 analysis to identify potential upstream regulators of these upregulated DEGs in both the pre-HE-to-HE in mouse and the C6-to-C7 in human. We observed that the top 10 regulators converge on the core factor MYC59 in both species (Fig. 3i, j; Supplementary Data), aligning with a prior study that demonstrated diminished HECs in the aorta upon Myc deletion58. These results indicated that parallel transcriptional networks govern the fate transitions in both mouse and human.
Identification of external signals that facilitate bifurcation choices during the AEC-to-HE transition
Cell fate transition during EHT is guided by the surrounding cellular environment60. To gain a better understanding of how external factors impact transcription networks during bifurcation choice, we analyzed publicly available human spatial transcriptomics data, which provided transcription profiles for the nearby niche of AGM20. We treated each spot on the spatial transcriptomics slide 7 from the CS15 human embryo as a single pseudo cell and used applied unsupervised clustering45 to categorize these pseudo cells into 11 major cell populations, with S1 and S8 derived from AGM (Supplementary Fig. 9a; “Methods”). We then applied NicheNet61, which predicts ligand-target connections through an integrated model encompassing the signaling path from ligands to target genes, to identify potential ligands using DEGs from C6-to-C7 and C6-to-CS16 GJA5+ AEC as downstream targets (“Methods”; Fig. 4a; Supplementary Data). We considered potential ligands for EHT as true only if they were expressed by the S1 or S8 cell populations (Supplementary Fig. 9b, c; Supplementary Data). Notably, various ligands that facilitate C6 to choose distinct fates could affect the same downstream targets (Supplementary Fig. 9c, black rectangle).
a Schematic of signaling pathways from spatial ligands to target genes (DEGs). b Signaling pathway mediated by CXCL12. The signal travels from CXCL12 (ligand, orange) to receptors (pink) through signaling mediators (light blue) and regulators (blue), ending at target genes (purple) (DEGs between C6 and C7 regulated by CXCL12). c Schematic of human in vitro hematopoietic differentiation system from (Shen et al.79). d Representative flow cytometric analysis of the in vitro-defined HEs (CD34+CD144+CD184−CD73−) from day 4 differentiation. e Cell number of in vitro-defined HEs (CD34+CD144+CD184−CD73−) sorted from day 4 (left panel) differentiation with or without CXCL12 treatment (n = 3, *P < 0.05). f Expression of PECAM1, CDH5, TEK, GATA2, and RUNX1 in cells from day 4 with or without CXCL12 treatment. The expression level was normalized to that of β-actin. ns, not significant. (n = 3, *P < 0.05; **P < 0.01). g Representative flow cytometric analysis of the in vitro-defined HPCs (CD34+CD43+) from day 7 differentiation. h Cell number of in vitro-defined HPCs (CD34+CD43+) sorted from day 7 differentiation with or without CXCL12 treatment (n = 3, *P < 0.05). i Colony-forming unit (CFU) assay of HPCs generated with or without CXCL12 treatment. CFUs per 4 × 103 cells plated (n = 3, ***P < 0.001).
Among the true potential ligands that facilitate the C6-to-CS16 GJA5+ AEC transition, TGFB1 is the top candidate (Supplementary Fig. 9b, arterial module). However, TGFB1 also contributes to the C6-to-C7 transition, implying its divergent roles in cell fate selection, as previous work has shown that the interplay between TGFβ and Notch signaling directs AECs to adopt a hemogenic identity62. SPP1 has been shown to target CD4419,27, a receptor that marks the HSPC-forming AECs63. Our analysis further suggests that SPP1 promotes the arterial fate of those HSPC-forming arterial ECs.
All of the top five true potential ligands that facilitate the C6-to-C7 transition (Supplementary Fig. 9b, Hemogenic module) are pivotal during EHT62,64,65. BMP signals (BMP4, BMP5, and BMP7), especially BMP4, are required for HSC emergence and maturation within AGM66,67,68. VEGFA is required for NOTCH signaling, which activates the hematopoietic program65,69,70,71. Besides maintaining the quiescent HSC pool72,73, the CXCL12-CXCR4 axis has been found to either suppress the EC program of mouse HE74 or facilitate the generation of engrafting HSCs from E9-to-E10 hemogenic precursors75, highlighting a vital role for CXCL12 in EHT. PCDH7, one of the potential ligands that facilitate the C6-to-CS16 GJA5+ AEC transition (Supplementary Fig. 9b), also interacts with CXCR4 (Supplementary Fig. 9c), implying a distinct function for CXCR4 in cell fate choice under different conditions.
In our ligand-target analysis, CXCL12 activates several key regulators (Fig. 4b). Among these key regulators, MEIS1 has been shown to promote the hemogenic specification of APLNR+ mesoderm progenitors in human76. Consistent with this, MEIS1 was also identified as a TF that specifically regulates the DEGs between human C6 and C7 (Supplementary Fig. 9c, C7 vs. C6 specific). Moreover, GATA2, which is vital in HSC generation77,78, participated in the CXCL12 signaling pathway. One of the CXCL12 target regulators is MYC, which we identified as the core upstream regulator of upregulated DEGs in both species (Fig. 3h, i). Considering its prominent role among the true potential ligands facilitating the C6-to-C7 transition, we hypothesize that CXCL12 may selectively promote the hemogenic choice in human embryonic hematopoietic development.
CXCL12 promotes hemogenic fate
Given the potential role of CXCL12 during hemogenic fate determination of pre-HE, we wondered if CXCL12 treatment can truly promote HE formation from the hemogenic precursor. To this end, we took advantage of a human pluripotent stem cell (hPSC) in vitro system79 (Fig. 4c) that mimics hematopoietic differentiation, allowing us to bypass the ethical restrictions on human embryos. In this monolayer-based in vitro system that underwent chemically defined culture, hPSCs (Day 0, referred to as D0) progress through mesoderm (D2) and endothelial (D4) specification before developing into hematopoietic progenitors (HPCs) (D7) with multilineage differentiation capability. CD144+CD34+CD73−CD184− cells at D4 are considered as HEs80 (hereafter called in vitro-defined HEs), and CD34+CD43+ cells at D7 as HPCs (hereafter called in vitro-defined HPCs). As in vitro-defined HEs are mainly enriched at D4, hence we anticipate that the hemogenic precursor-to-HE transition happens between D2 and D4 in this system. Therefore, we added CXCL12 (Peprotech, 300-28A) into the culture medium on day 2 to determine whether it promoted the hemogenic precursor-to-HE transition. After two days of differentiation until D4, we quantified the abundance of in vitro-defined HEs (CD144+CD34+CD73−CD184−) using flow cytometric analysis (FACS) (“Methods”; Fig. 4d; Supplementary Fig. 10a). We observed that CXCL12 treatment (referred to as the CXCL12 group) significantly increased the amount of in vitro-defined HEs as compared to the control group (no CXCL12 treatment) (Fig. 4e, left panel; “Methods”, *P value < 0.05), supporting the promoting role of CXCL12 in hemogenic fate determination.
Quantitative real-time polymerase chain reaction (qRT-PCR) analyses of the D4 cells from the CXCL12 group showed significantly increased RNA expression of hematopoietic markers, such as GATA2 and RUNX1 (Fig. 4f; “Methods”, *P value < 0.05). Meanwhile, the RNA expression of endothelial markers, such as PECAM1 (CD31-coding gene) and TEK (TIE2-coding gene), showed no significant change (Fig. 4f). Notably, KDR, which encodes a VEGFR2 receptor that marked the endothelial subset with hematopoietic potential81,82, exhibits significantly increased RNA expression under CXCL12 treatment. Additionally, the increased MYC expression agrees with our analysis that CXCL12 might positively regulate MYC during the C6-to-C7 transition (Fig. 3h, i; Fig. 4f). These results indicated that CXCL12 facilitates hemogenic fate by promoting the hematopoietic program instead of repressing the endothelial program. We then evaluated the hematopoietic potential for these HEs. Given that in both our FACS results (Fig. 4d) and the original paper80,83, at D4, CD34+ cells encompassed all CD34+CD144+CD73−CD184− HE cells. Therefore, for subsequent hematopoietic potential analysis, we used Magnetic-Activated Cell Sorting (MACS) to isolate CD34+ cells at D4 as a representation of the CD34+CD144+CD73−CD184− HEs. These CD34+ cells were cultured in STEMdiff APEL 2 medium until day 7 (D7) and followed by assessing the formation of HPCs. Using FACS, we observed a significantly higher amount of in vitro-defined HPCs (CD34+CD43+) from the CXC12 group (Fig. 4g, h; Supplementary Fig. 10b; “Methods”, *P value < 0.05), supporting the enhanced hematopoietic potential of those in vitro-defined HEs from the CXCL12 group.
Given the increased production of in vitro-defined HEs and HPCs, we hypothesized that CXCL12 treatment could promote the multilineage potential of those in vitro-defined HPCs. To test this hypothesis, we used MACS to sort an equal number of in vitro-defined HPCs (CD34+CD43+) at D7 from both the CXCL12 and control groups and followed by measuring their colony-forming potential using a colony-forming unit (CFU) assay (“Methods”). In line with our hypothesis, we observed significantly higher numbers of hematopoietic colonies of myeloid and erythroid lineages from the CXCL12 treatment group (Fig. 4i; “Methods“, ***P value < 0.001), indicating an enhanced multilineage differentiation capability of HPCs from the CXCL12 group. In summary, our results not only support the role of CXCL12 in facilitating the hemogenic fate of HE precursors but also highlight its role in promoting the hematopoietic potential of HEs (Fig. 5).
During mouse and human EHT, a specific subset of AECs differentiates into pre-HEs. In response to interactions between specific cell fate determinants (like CXCL12) and their corresponding receptors, pre-HEs may advance toward either a hemogenic or arterial fate. Those hemogenic precursors that acquire the CXCL12 signal can further develop into a hematopoietic population with multilineage differentiation capability aggregated in clusters. AEC arterial endothelial cell, pre-HE pre-hemogenic endothelium, HE hemogenic endothelium, IAC intra-aortic cluster.
Discussion
Here we introduce a cross-species analysis method (HomologySeeker) based on homologous genes exhibiting high levels of expression variability (Fig. 1a). Compared to state-of-the-art methods that require prior cell type annotation, including a recent model (CAME)34 that advances the utilization of non-one-to-one homologous gene mapping, HomologySeeker avoids prior cell type annotation for cross-species comparisons. We utilize HomologySeeker to study EHT transcriptome ensembles that we constructed from publicly available single-cell RNA-seq datasets (Supplementary Fig. 2). These ensembles could serve as an expandable repository for the scientific community. We showed that human and mouse EHT display analogous cell type correspondences, similar developmental trajectories, and comparable transcription expression patterns (Figs. 1 and 2), substantiating the conserved nature between these two species. However, due to the diversity of datasets and potential omissions in the data integrated into our human ensemble, the minimal T1/2 signals we observed in human cells (Fig. 1c–f; Supplementary Fig. 4) could suggest a scarce presence of pre-HSC cells if any. Moreover, we do not observe mouse cells equivalent to the human C5 population (Fig. 1f, g), warranting further exploration. We observe that pre-HEs have the potential to differentiate into either HEs or lAECs, a phenomenon conserved between mouse and human (Fig. 3), which refines our understanding of the human arterial-to-hemogenic transition. We further identify ligand modules that contribute to pre-HE choices and demonstrate that CXCL12 significantly enhances the HSPC-forming potential of HE precursors using an in vitro differentiation system (Fig. 4).
Our finding that mouse pre-HEs can further differentiate into HEs or E11.5 ECs (Fig. 3) harmonizes with parallel studies16,18. These studies show that the cell fate choice of eAECs toward either HEs or lAECs16 co-occurs with the pre-HE stage, an intermediate stage between AEC and HE18. Zhu et al.18 knocked down Runx1, and we observed increased pre-HE numbers but a reduced amount of HE. This pre-HE-to-HE transition is similar to that of the eAEC-to-HE transition, as reported by Hou et al.16. Additionally, the transcriptome profile of lAECs in Hou et al.16 overlaps with E11.5 ECs that reside at the end of the trajectory, away from the path toward HEs (Fig. 3c). This pre-HE-to-E11.5 EC transition resembles that of the eAEC-to-lAEC transition in Hou et al.16. Furthermore, cells from this human C6 sub-cluster exhibit similar fate choices as mouse pre-HE towards the C7 sub-cluster, which contains HE signatures, or GJA5+ AECs (Fig. 3f). Taken together, the pre-HE stage may serve as a bifurcation point for cell fate decisions, and this phenomenon is conserved across species.
Our study shows that CXCL12 not only facilitates hemogenic fate but also promotes hematopoietic potential, as evidenced by the increased number of in vitro-defined HPCs, indicating the formation of genuine HSPC-forming HEs under CXCL12 treatment (Fig. 4). Furthermore, hematopoietic progenitors treated with CXCL12 exhibited enhanced multilineage differentiation capability (Fig. 4i), consistent with a previous study that CXCL12-CXCR4 signaling enables the generation of long-term engrafting HSCs from mouse E9-to-E10 AGM derived hemogenic precursors75. Additionally, our study identifies CXCL12 as a shared upstream effector for hemogenic regulators, like MEIS175 and MYC59 (Fig. 4b), both involved in mouse hemogenic fate choice decisions. These findings suggest that CXCL12 plays a conserved role during the pre-HE-to-HE transition between mouse and human, potentially serving as a critical checkpoint for hematopoiesis manipulation. Nevertheless, further exploration is needed to elucidate the precise mechanism of how CXCL12 promotes hemogenic fate. Studies72,73 have shown that CXCL12 functions through the CXCR4 receptor, which is also involved in EHT9,19,74,75. Interestingly, only a subset of the CXCR4+ population shows hemogenic potential, while others exhibit arterial features80. Our ligand network analysis indicates that CXCR4 interacts only with either CXCL12 or PCDH7 (Supplementary Fig. 9c), the latter belongs to ligand modules that facilitate arterial choices in pre-HEs (Supplementary Fig. 9b, c). Consequently, PCDH7 may compete with CXCL12 for CXCR4, resulting in a mutually antagonistic relationship between CXCL12 and PCDH7 that co-regulates pre-HE fate selection.
Our work presents HomologySeeker as a new approach for investigating cell fate transitions across species. By applying this method to the study of EHT, we have advanced our understanding of this critical developmental stage, particularly during the human arterial to hemogenic transition. Our findings provide valuable insight into the regulation of hematopoiesis and the enhancement of hematopoietic efficiency in human in vitro differentiation.
Methods
Maintenance and hematopoietic differentiation of hPSCs
The H1 hPSC line was obtained from the WiCell Research Institute (Madison, WI, http://www.wicell.org). Cultured cells were maintained on Matrigel-coated 6-well plates (Corning) containing E8 medium (Gibco)84,85, and the medium was replaced daily. The hPSCs were sub-cultured every 3–4 days with a treatment of 0.5 mM ethylenediaminetetraacetic acid (EDTA; Life Technologies) for passaging when cells reached 60–70% confluence. For hematopoietic differentiation, single hPSCs were obtained for sequential EC–HC induction. Briefly, single-cell suspensions of hPSCs were obtained by treating the hPSC cultures at 70–80% confluency with TrypLE (Thermo Fisher Scientific). Single cells were then plated at an optimized density of 6 × 103 cells/well onto 12-well plates (Corning) coated with vitronectin (Peprotech) in STEMdiff APEL 2 Medium (STEMCELL Technologies) supplemented with 3 μM GSK3 inhibitor, CHIR99021 (ABM Inc), 4 ng/ml ActivinA (Peprotech), 10 ng/ml BMP4 (Peprotech), and 10 μM Rho kinase inhibitor, Y-27632 (STEMCELL Technologies) on day 0. After 48 h (day 2), the medium was changed to STEMdiff APEL 2 Medium supplemented with 40 ng/ml VEGF (Peprotech). For the following 24 h (day 3), recombinant Human FGF2 (ABM Inc.) was added to a final concentration of 40 ng/ml until day 4. CD34+ cells were isolated from differentiated cells on day 4 by magnetic-activated cell sorting (MACS, Miltenyi Biotec.). We re-seeded the isolated CD34+ cells on vitronectin (Peprotech)-coated 12-well plates (Corning) at a density of 1.25 × 105 cells/well in STEMdiff APEL 2 Medium (STEMCELL Technologies) supplemented with 40 ng/ml VEGF (PeproTech) and 40 ng/ml FGF2 (ABM Inc) until day 7. The entire differentiation process was incubated at 37 °C in 5% CO2 with 100% humidity.
Flow cytometry analysis
Cells were dissociated to form a single-cell suspension by TrypLE treatment and washed with FACS buffer PBE (2% FBS and 0.5 mM EDTA in PBS). The dissociated cells were then resuspended in PBE and labeled with fluorochrome-conjugated anti-human CD73-PE-Cy7 (BioLegend, clone: AD2), CD184-APC (Invitrogen, clone: 12G5), CD144-PE (Invitrogen, clone: 16B1), CD34-APC-Cy7 (BioLegend, clone: 561), CD34-PE (Invitrogen, clone: 4H11), and CD43-PE (BioLegend, clone: 10G7). Dead cells were excluded according to DAPI (BD Biosciences) staining. Isotype-matched control antibodies were used to determine the background. Flow cytometry was performed using Canto II analyzer (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star, Inc.).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR) assay
Total RNA was extracted using a TRizol reagent (Roche). cDNA was synthesized from 2 μg of total RNA using the GoScript™ Reverse Transcriptase Kit (Promega) and stored at -80°C until use. Real-time PCR was performed using a ChamQ SYBR Color qPCR Master Mix (Low ROX Premixed) (Vazyme) on a QuantStudio™ 3 (Applied Biosystems). Amplification of β-actin was conducted in parallel to control for the quantity of loaded cDNA in each reaction. Primer sequences are listed in Supplementary Data.
Hematopoietic colony-forming unit (CFU) assays
4000 CD34+CD43+ HPC single cells in 0.1 ml IMDM (Life Technologies) with 2% FBS were mixed with MethoCult H4034 Optimum (STEMCELL Technologies). The mixture was then transferred to ultra-low attachment 12-well plates (Corning). The cells were incubated at 37 °C in 5% CO2 with 100% humidity for 14 days before counting colonies. Each type of colony was classified according to morphology. Each assay was performed in triplicate.
scRNA-seq data collection and pre-processing
For mouse and human midbrain scRNA-seq datasets from La Manno et al.24, gene expression matrices deposited in the NCBI Gene Expression Omnibus (GEO) were downloaded under the accession number GSE76381 (STRT-seq).
For mouse EHT scRNA-seq datasets, gene expression matrices under accession numbers: GSE112642 (Baron et al.38, Cel-seq) and GSE137117 (Zhu et al.18, 10× Genomics droplet-based scRNA-seq) were downloaded (Supplementary Data). For the scRNA-seq data of Zhou et al.37 (GSE135202, STRT-seq) and Hou et al.16 (GSE139389, STRT-seq), raw reads were split by barcode sequence attached in Read 2. The TSO sequence and adapter contaminants were trimmed using trim_galore (v0.6.7)86 for Read 1. Trimmed Reads 1 were aligned against mm10 mouse genome using STAR (v2.6.0c)87 (Parameters: outFilterMatchNminOverLread = 0.3, outFilterScoreMinOverLread = 0.3). Uniquely mapped reads were counted using HTSeq (v0.13.5)88 and grouped by the cell-specific barcodes. For each barcode, the copy number of transcripts of a given gene was taken as the number of distinct UMIs of that gene.
Human EHT scRNA-seq data were collected under accession numbers: GSE135202 (Zeng et al.19, STRT-seq and 10X Genomics droplet-based scRNA-seq), GSE162950 (Calvanese et al.20, 10× Genomics droplet-based scRNA-seq), and GSE151877 (Crosse et al.27, 10× Genomics droplet-based scRNA-seq) (Supplementary Data). Briefly, sequencing data from 10× genomics was processed using CellRanger (v2.1.1) with default mapping arguments. The sequencing data of STRT-seq were processed as mouse STRT-seq datasets, but using the GRCh38/hg38 human genome for reads mapping. To keep the consistency of gene annotation, all gene names from mouse and human datasets were converted to official gene symbols using the alias2Symbol function from limma (v3.18.10)89. Only CDH5+GJA5+HEY2+APLNR-NR2F2-PDGFRA-PDGFRB-GYPA-EPCAM- cells from the single-cell dataset (Crosse et al.) were selected as GJA5+ AECs.
For the 10× Genomics droplet-based scRNA-seq dataset from Huo et al.51, the gene expression matrix was downloaded from GEO under the accession number GSE224714. Only cells sampled from healthy controls were retained for further analysis.
HomologySeeker method
As comparative analyses using all homologous genes may include genes that are not expressed across all cells or unrelated to the development system in question, making downstream interpretation challenging. To avoid this, we sought to take advantage of the concept of highly variable genes (HVGs), which is widely used in single-cell RNA-seq analysis to select genuine biological variations. Furthermore, HVGs can be identified in an unsupervised and low-calculation-cost manner that applies to various kinds of development systems. HomologySeeker is designed to identify homologous gene sets with highly variable expression (Homologous-HVGs) for cross-species analysis while keeping species-specific homologous/non-homologous genes for additional purposes.
HomologySeeker consists of two main steps: (i) Homologous gene collection and filtering, (ii) homologous-HVGs identification. Briefly, homologous genes between species are collected from Ensemble databases using “getLDS” function in biomaRt (v2.46.3 was used in this study). HomologySeeker only keeps genes with one-to-one orthology and high orthology confidence introduced by the Ensemble database in “Ortholog_qc_manual” section (https://ensembl.org/info/genome/compara/Ortholog_qc_manual.html). Next, returned gene sets are fed into an HVG selection method (Seurat v4.1.136 was used in this study) to get variation levels (i.e., standardized variance) of all genes for each species. Finally, to objectively select variable genes, HomologySeeker utilizes the mean values of the variation levels of gene sets as the cutoff for selecting “genuine” highly variable genes, which results in species Homologous-HVG sets for further comparative analysis.
EHT ensembles construction
Expression matrices from Zhou et al., Baron et al., and Zhu et al. were used to construct the mouse EHT ensemble (Supplementary Data). Only cells annotated as venous/arterial EC, EC, HE, IAC, T1/2 pre-HSC, and FL-HSC were included. The ensemble was constructed based on the instruction of “Performing integration on datasets normalized with SCTransform”90 (https://satijalab.org/seurat/articles/integration_introduction.html) in Seurat. Briefly, normalization and highly variable genes selection were performed for each dataset using “SCTransform” function (Parameter: method = “glmGamPoi”, min_cells = 1). Integration features and objects were prepared using “SelectIntegrationFeatures” and “PrepSCTIntegration” with default settings, respectively. Then anchors identified using the “FindIntegrationAnchors” function among datasets were used for data integration using “IntegrateData” function with default parameters. The resulting integrated dataset was called the “EHT ensemble”.
For human, expression matrices from Zeng et al. and Calvanese et al. were used to construct the human EHT ensemble. Only cells annotated as venous/arterial EC, HE, and HSPC/HC were included for further analysis. The human EHT ensemble construction was performed as a mouse EHT ensemble.
To maintain the consistency of cell annotation, mouse cell types are unified as Wnt_EC, AEC, EC, Pre-HE, HE, IAC, T1, T2, and FL-HSC according to original annotations, whereas human cell types were unified as VEC, AEC, HEC, HC, and HSPC (Supplementary Fig. 2c).
For merging GJA5+ AECs into the human EHT ensemble, STACAS (v2.0.1)91, a sub-type anchoring correction method for alignment in Seurat, was used to prevent batch effect overcorrection. Briefly, each dataset (Zeng et al. (STRT-seq + 10×), Calvanese et al., and GJA5+ AEC) was normalized using the “NormalizeData” function in Seurat. Then “Run.STACAS” function (Parameters: dims = 1:50) was used to perform the integration analysis of all normalized datasets.
Dimension reduction and unsupervised clustering
Dimension reduction and unsupervised clustering were done by Seurat unless otherwise mentioned.
To visualize single cells in 2D space, the dimension of both EHT ensembles was first reduced based on principal component analysis using the “RunPCA” function with default settings. EHT ensembles were visualized by projecting cells in 2D space using UMAP implemented in “RunUMAP” function (Parameters: dims=1:50).
To cluster the human single cells, the nearest-neighbor graph of the human EHT ensemble was first constructed using “FindNeighbors” function, and sub-clusters were identified by the Louvain algorithm using “FindClusters” function (Parameters: resolution = 0.8, clustree (v0.4.4)46 were used to determine the optimal clustering resolution).
For the 10x Genomics droplet-based scRNA-seq dataset from Huo et al., datasets from each healthy donor were integrated based on the instruction of “Performing integration on datasets normalized with SCTransform”. Then the dimension of the integrated dataset was reduced based on principal component analysis using the “RunPCA” function with default settings. Cells were projected into 2D space using UMAP implemented in “RunUMAP” function (Parameters: dims = 1:50).
GO enrichment analysis
GO term enrichment was performed using clusterProfiler (v4.5.0.992)92 with default parameters.
Pearson correlation analysis
For Pearson correlation analysis between mouse and human midbrain data, Homologous-HVGs sets for mouse and human were calculated based on single cell matrices from La Manno et al. using HomologySeeker. After Homologous-HVG identification, median matrices constructed by La Manno et al. (genes as rows and cell types as columns with the median value of that cell type as the matrix value) were used for Pearson correlation analysis. median matrices (x) were normalized by “log(1 + x)-rowMeans(log(1 + x))” ahead according to La Manno et al. (“rowMeans” equal to the mean value of each row). Overlapped Homologous-HVGs between mouse and human were used to calculate Pearson correlation using the “cor” function implemented in the R base package (v4.0.3).
For Pearson correlation analysis between mouse and human EHT ensembles, Homologous-HVGs sets for mouse and human were selected using HomologySeeker (based on the residual variance of each gene returned by Seurat integration using the “SCT” method (according to “EHT ensemble construction” section)). Median matrices and Pearson correlations were calculated based on corrected single-cell matrices.
Single-cell projection
Single-cell projection analysis was performed following the instruction of “Mapping and annotating query datasets” (https://satijalab.org/seurat/articles/integration_mapping.html).
For intra-species projection, the query single-cell dataset was normalized using the “SCTransform” function. Transfer anchors between query and reference datasets were identified using “FindTransferAnchors” function. Anchors were then used to project the query dataset into reference using the “MapQuery” function. For inter-species projection, the PCA space of EHT ensembles was re-calculated using shared Homologous-HVGs between mouse and human EHT ensembles using the “RunPCA” function. Then anchors between query and reference were identified using the “FindTransferAnchors” function. Anchors were then used to project the query dataset into the reference using “MapQuery” function. Prediction scores were visualized in heatmap using ComplexHeatmap93 (v2.6.2).
Developmental trajectory inference
Developmental trajectory of EHT ensembles was inferred using Monocle3 (v1.2.7) according to “Calculating Trajectories with Monocle 3 and Seurat” (http://htmlpreview.github.io/?https://github.com/satijalab/seurat-wrappers/blob/master/docs/monocle3.html). Briefly, mouse Wnt EC and human C1–3 (venous EC sub-clusters) were excluded from further analysis. The Seurat object was first converted to Monocle3 cell_data_set object. Unsupervised clustering of cells was performed using “cluster_cells” function (Parameter: reduction_method = “UMAP”, cluster_method = “louvain”). The principal graph was learned from UMAP space using “learn_graph” function (Parameter: close_loop=F). Cell order according to pseudo time was inferred using the “order_cells” function.
For the TF expression patterns along the developmental trajectory, we fitted a local regression to the expression level for each cell at their value of pseudo time using ggplot2 (v3.3.6) (“geom_smooth” function with method = “loess”) (https://ggplot2.tidyverse.org).
The developmental trajectory of single-cell RNA-seq data from Hou et al. was inferred using Monocle (v2.9.0)16. Briefly, the normalization factors and variability of scRNA-seq data were calculated using “estimateSizeFactors” and “estimateDispersions” functions, respectively. Only genes that expressed at least 10 cells were retained. Then the highly variable genes of scRNA-seq data calculated by “FindVariableFeatures” function from Seurat were fed into “setOrderingFilter” function to acquire features for further trajectory inference. Genes from the cell cycle GO term (GO:0007049) were filtered out from the highly variable genes to reduce the influence of the cell cycle effect. Then cells were projected into lower dimensional space using “reduceDimension” function. The final trajectory was inferred using “orderCells” function.
TF module identification
Mouse and human TF lists were downloaded from AnimalTFDB3.0 and HumanTFDB3.0 (http://bioinfo.life.hust.edu.cn/)94, respectively. All TFs were selected from mouse and human Homologous-HVG lists. To identify potential TF modules, Pearson distance was calculated according to Pijuan-Sala et al.95. Briefly, the Pearson correlation distance between TFs was calculated as “([1 − x]/2)0.5”, where x is the Pearson correlation among TFs. Then hierarchical clustering was performed using the unweighted pair group method with arithmetic mean (UPGMA), and modules were identified using the “dynamicTreeCut” function in dynamicTreeCut (v1.63-1)54.
Differential expression analysis
To find DEGs, Wilcoxon Rank Sum tests implemented using “FindMarkers” function in Seurat were performed to identify DEGs. DEGs with adjusted P values less than 0.0001 were deemed significant. For the DEGs between human C6 and GJA5+ AECs, only the aggregated part (Fig. 3d, shadow in blue) of GJA5+ AECs was used for differential expression analysis. For the marker genes modules of HSC/MPP and LMPP from Huo et al.51, DEGs between HSC/MPP or LMPP and all other cells are calculated. The top 20 upregulated DEGs that ranked by fold change were used for subsequent module score calculation.
Module score calculation
Module scores of TFs, marker genes, and gene sets from GO terms were estimated by using the “AddModuleScore” function in Seurat. The gene sets encompassed by EC development (GO:0001885), Arterial EC differentiation (GO:0060842), and Blood vessel EC differentiation (GO:0060837) were collected from AmiGO 2 (http://amigo.geneontology.org/amigo).
Identification of potential upstream regulators of DEGs
The upstream regulators of DEGs between pre-HEs and HEs (C7 vs. C6 in human) were predicted by TF enrichment analysis using ChIP-X Enrichment Analysis 3 (ChEA3; https://maayanlab.cloud/chea3/). The TF local network was constructed using the top 10 returned regulators interaction mined from the ENCODE ChIP-seq project.
SingleCellNet analysis
SingleCellNet (v0.1.0)47 was used to assign human cells with potential identities inferred from mice based on differentially expressed homologous genes. Briefly, cell types classifiers were built using the “scn_train” function using mouse cell types as a reference (Parameter: nTopGenes = 100). Human cells were classified using a trained classifier using the “scn_predict” function with default settings.
RNA velocity analysis
Velocyto (v0.17.17)96 was used for RNA velocity analysis of the human EHT ensemble. To annotate spliced, unspliced, and spanning reads in the measured cells, “run_smartseq2” and “run10x” commands were used to generate loom files for human STRT-seq and 10× genomics drop-based single-cell data with GRCh38/hg38 reference genome. The output loom files were combined and analyzed using the “velocyto.R” package (v0.6). RNA velocity was estimated using the “RunVelocity” function with default settings. RNA velocities were visualized on the human EHT ensemble using the shared nearest-neighbor graph calculated in “Dimension reduction and unsupervised clustering” section using the “show.velocity.on.embedding.cor” function (Parameter: n = 100, which equals neighborhood size).
Analysis of spatial transcriptomics data
The spatial transcriptomics matrix of the CS15 human embryo (slide7) was downloaded from GitHub deposited by Calvanese et al., and analyzed by Seurat. Briefly, the “SCTransform” function was used to normalize and find variable genes within the spatial transcriptomics data. Dimension reduction and unsupervised clustering were then performed according to the “Dimension reduction and unsupervised clustering” section with some modifications (Parameters: dims = 1:30 in “FindNeighbors” function, resolution = 1.2 in “FindClusters” function and dims = 1:30 in “RunUMAP” function).
Ligand-target signaling inference
NicheNet (v1.1.0)61 was used to infer potential ligands that share active links with target genes (DEGs between human C6 and C7/late AEC). Briefly, pseudo cells from spatial transcriptomics data located in the AGM region were defined as sender cells. Potential ligands expressed by sender cells were ranked based on how well they interacted with target genes (evaluated by the Pearson correlation coefficient).
The signaling paths from ligands to target genes were inferred based on the instructions for “NicheNet Results: Ligand-Targets interesting paths” introduced by Saez lab that combine NicheNet and OmnipathR (v3.5.21) (https://github.com/saezlab/NicheNet_Omnipath/blob/master/07_LigandTargetPaths.md). The resulting pathways were visualized in Cytoscape (v3.8.2)97.
Statistics and reproducibility
Data obtained from multiple experiments were reported as the mean ± SEM. An unpaired t-test was used to compare the means from two groups, and ANOVA was used to compare the means from three or more groups. Results with a value of P < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-seq data is existing data available in GEO under accession numbers: (Zhou et al.37, GSE135202), (Baron et al.38, GSE112642), (Zhu et al.18, GSE137117), (Hou et al.16, GSE139389), (Zeng et al.19, GSE135202), (Crosse et al.27, GSE151877), (Calvanese et al.20, GSE162950), (La Manno et al.24, GSE76381), (Huo et al.51, GSE224714). Details are listed in Supplementary Data. The highly variable homologous gene sets, differential expressed gene sets between different cell types or subclusters, GO biological pathways enriched by differentially expressed gene sets, upstream regulators of the differentially expressed genes set, NicheNet singling pathways components, primer sequences for real-time polymerase chain reaction (QPCR), and cell metadata for mouse and human ensembles are available as Excel sheets in Supplementary Data. Single-cell analysis code used in this study is available upon reasonable request.
Code availability
HomologySeeker is openly available as an R package. The code, documentation, and examples are accessible at https://github.com/YenLab/HomologySeeker. Interfaces for mouse and human ensembles are also available.
References
Orkin, S. H. & Zon, L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008).
Doulatov, S., Notta, F., Laurenti, E. & Dick, J. E. Hematopoiesis: a human perspective. Cell Stem Cell 10, 120–136 (2012).
Ivanovs, A. et al. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J. Exp. Med. 208, 2417–2427 (2011).
Zhao, S., Feng, S., Tian, Y. & Wen, Z. Hemogenic and aortic endothelium arise from a common hemogenic angioblast precursor and are specified by the Etv2 dosage. Proc. Natl Acad. Sci. USA 119, e2119051119 (2022).
Lange, L., Morgan, M. & Schambach, A. The hemogenic endothelium: a critical source for the generation of PSC-derived hematopoietic stem and progenitor cells. Cell Mol. Life Sci. 78, 4143–4160 (2021).
Fadlullah, M. Z. et al. Murine AGM single-cell profiling identifies a continuum of hemogenic endothelium differentiation marked by ACE. Blood https://doi.org/10.1182/blood.2020007885 (2021).
Slukvin, I. I. & Uenishi, G. I. Arterial identity of hemogenic endothelium: a key to unlock definitive hematopoietic commitment in human pluripotent stem cell cultures. Exp. Hematol. 71, 3–12 (2019).
Yzaguirre, A. D., Howell, E. D., Li, Y., Liu, Z. & Speck, N. A. Runx1 is sufficient for blood cell formation from non-hemogenic endothelial cells in vivo only during early embryogenesis. Development https://doi.org/10.1242/dev.158162 (2018).
Park, M. A. et al. Activation of the arterial program drives development of definitive hemogenic endothelium with lymphoid potential. Cell Rep. 23, 2467–2481 (2018).
Garcia-Alegria, E. et al. Early human hemogenic endothelium generates primitive and definitive hematopoiesis in vitro. Stem Cell Rep. 11, 1061–1074 (2018).
Gritz, E. & Hirschi, K. K. Specification and function of hemogenic endothelium during embryogenesis. Cell Mol. Life Sci. 73, 1547–1567 (2016).
Tavian, M. et al. Aorta-associated CD34+ hematopoietic cells in the early human embryo. Blood 87, 67–72 (1996).
Tavian, M., Hallais, M. F. & Peault, B. Emergence of intraembryonic hematopoietic precursors in the pre-liver human embryo. Development 126, 793–803 (1999).
Rybtsov, S. et al. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J. Exp. Med. 208, 1305–1315 (2011).
Taoudi, S. et al. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell 3, 99–108 (2008).
Hou, S. et al. Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res. 30, 376–392 (2020).
Uenishi, G. I. et al. NOTCH signaling specifies arterial-type definitive hemogenic endothelium from human pluripotent stem cells. Nat. Commun. 9, 1828 (2018).
Zhu, Q. et al. Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood 136, 845–856 (2020).
Zeng, Y. et al. Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res. 29, 881–894 (2019).
Calvanese, V. et al. Mapping human haematopoietic stem cells from haemogenic endothelium to birth. Nature https://doi.org/10.1038/s41586-022-04571-x (2022).
Ottersbach, K. Endothelial-to-haematopoietic transition: an update on the process of making blood. Biochem. Soc. Trans. 47, 591–601 (2019).
Dzierzak, E. & Bigas, A. Blood Development: Hematopoietic Stem Cell Dependence and Independence. Cell Stem Cell 22, 639–651 (2018).
Gerstein, M. B. et al. Comparative analysis of the transcriptome across distant species. Nature 512, 445–448 (2014).
La Manno, G. et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e519 (2016).
Tosches, M. A. et al. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888 (2018).
Pollen, A. A. et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756.e717 (2019).
Crosse, E. I. et al. Multi-layered spatial transcriptomics identify secretory factors promoting human hematopoietic stem cell development. Cell Stem Cell 27, 822–839.e828 (2020).
Bakken, T. E. et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021).
Du, J. et al. Integrative transcriptomic analysis of developing hematopoietic stem cells in human and mouse at single-cell resolution. Biochem. Biophys. Res. Commun. 558, 161–167 (2021).
Popova, G. et al. Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell https://doi.org/10.1016/j.stem.2021.08.015 (2021).
Emont, M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature https://doi.org/10.1038/s41586-022-04518-2 (2022).
Shekhar, K. et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e1330 (2016).
Pandey, S., Shekhar, K., Regev, A. & Schier, A. F. Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-seq. Curr. Biol. 28, 1052–1065.e1057 (2018).
Liu, X., Shen, Q. & Zhang, S. Cross-species cell-type assignment from single-cell RNA-seq data by a heterogeneous graph neural network. Genome Res. 33, 96–111 (2023).
Yip, S. H., Sham, P. C. & Wang, J. Evaluation of tools for highly variable gene discovery from single-cell RNA-seq data. Brief. Bioinform. 20, 1583–1589 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
Zhou, F. et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature 533, 487–492 (2016).
Baron, C. S. et al. Single-cell transcriptomics reveal the dynamic of haematopoietic stem cell production in the aorta. Nat. Commun. 9, 2517 (2018).
Clarke, R. L. et al. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat. Cell Biol. 15, 502–510 (2013).
North, T. E. et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16, 661–672 (2002).
Chen, M. J., Yokomizo, T., Zeigler, B. M., Dzierzak, E. & Speck, N. A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891 (2009).
Dai, G. et al. Over-expression of c-Myb increases the frequency of hemogenic precursors in the endothelial cell population. Genes Cells 11, 859–870 (2006).
Sakamoto, H. et al. Proper levels of c-Myb are discretely defined at distinct steps of hematopoietic cell development. Blood 108, 896–903 (2006).
Oh, I. H. & Reddy, E. P. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18, 3017–3033 (1999).
Waltman, L. & van Eck, N. J. A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B https://doi.org/10.1140/epjb/e2013-40829-0 (2013).
Zappia, L. & Oshlack, A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience https://doi.org/10.1093/gigascience/giy083 (2018).
Tan, Y. & Cahan, P. SingleCellNet: a computational tool to classify single cell RNA-Seq data across platforms and across species. Cell Syst. 9, 207–213.e202 (2019).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis (monocle3). Nature 566, 496–502 (2019).
Bertrand, J. Y. et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010).
Boisset, J. C. et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010).
Huo, Y. et al. Single-cell dissection of human hematopoietic reconstitution after allogeneic hematopoietic stem cell transplantation. Sci. Immunol. 8, eabn6429 (2023).
Yokomizo, T. et al. Independent origins of fetal liver haematopoietic stem and progenitor cells. Nature 609, 779–784 (2022).
Patel, S. H. et al. Lifelong multilineage contribution by embryonic-born blood progenitors. Nature 606, 747–753 (2022).
Langfelder, P., Zhang, B. & Horvath, S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720 (2008).
Zilberberg, L. et al. Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: role of fibrillins and fibronectin. J. Cell Physiol. 227, 3828–3836 (2012).
Dou, D. R. et al. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nat. Cell Biol. 18, 595–606 (2016).
Calvanese, V. et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature 576, 281–286 (2019).
He, C. et al. c-myc in the hematopoietic lineage is crucial for its angiogenic function in the mouse embryo. Development 135, 2467–2477 (2008).
Dignum, T. et al. Multipotent progenitors and hematopoietic stem cells arise independently from hemogenic endothelium in the mouse embryo. Cell Rep. 36, 109675 (2021).
Mirshekar-Syahkal, B., Fitch, S. R. & Ottersbach, K. Concise review: from greenhouse to garden: the changing soil of the hematopoietic stem cell microenvironment during development. Stem Cells 32, 1691–1700 (2014).
Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162 (2020).
Monteiro, R. et al. Transforming growth factor beta drives hemogenic endothelium programming and the transition to hematopoietic stem cells. Dev. Cell 38, 358–370 (2016).
Oatley, M. et al. Single-cell transcriptomics identifies CD44 as a marker and regulator of endothelial to haematopoietic transition. Nat. Commun. 11, 586 (2020).
Souilhol, C. et al. Inductive interactions mediated by interplay of asymmetric signalling underlie development of adult haematopoietic stem cells. Nat. Commun. 7, 10784 (2016).
Leung, A. et al. Uncoupling VEGFA functions in arteriogenesis and hematopoietic stem cell specification. Dev. Cell 24, 144–158 (2013).
McGarvey, A. C. et al. A molecular roadmap of the AGM region reveals BMPER as a novel regulator of HSC maturation. J. Exp. Med. 214, 3731–3751 (2017).
Durand, C. et al. Embryonic stromal clones reveal developmental regulators of definitive hematopoietic stem cells. Proc. Natl Acad. Sci. USA 104, 20838–20843 (2007).
Kirmizitas, A., Meiklejohn, S., Ciau-Uitz, A., Stephenson, R. & Patient, R. Dissecting BMP signaling input into the gene regulatory networks driving specification of the blood stem cell lineage. Proc. Natl Acad. Sci. USA 114, 5814–5821 (2017).
Lawson, N. D., Vogel, A. M. & Weinstein, B. M. sonic hedgehog and vascular endothelial growth factor Act Upstream of the Notch Pathway during Arterial Endothelial Differentiation. Dev. Cell 3, 127–136 (2002).
Robert-Moreno, A. et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 27, 1886–1895 (2008).
Kumano, K. et al. Notch1 but Not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity 18, 699–711 (2003).
Mendt, M. & Cardier, J. E. Role of SDF-1 (CXCL12) in regulating hematopoietic stem and progenitor cells traffic into the liver during extramedullary hematopoiesis induced by G-CSF, AMD3100 and PHZ. Cytokine 76, 214–221 (2015).
Zheng, Z. et al. Uncovering the emergence of HSCs in the human fetal bone marrow by single-cell RNA-seq analysis. Cell Stem Cell 29, 1562–1579.e1567 (2022).
Ahmed, T., Tsuji-Tamura, K. & Ogawa, M. CXCR4 signaling negatively modulates the bipotential state of hemogenic endothelial cells derived from embryonic stem cells by attenuating the endothelial potential. Stem Cells 34, 2814–2824 (2016).
Hadland, B. et al. Engineering a niche supporting hematopoietic stem cell development using integrated single-cell transcriptomics. Nat. Commun. 13, 1584 (2022).
Wang, H. et al. MEIS1 regulates hemogenic endothelial generation, megakaryopoiesis, and thrombopoiesis in human pluripotent stem cells by targeting TAL1 and FLI1. Stem Cell Rep. 10, 447–460 (2018).
de Pater, E. et al. Gata2 is required for HSC generation and survival. J. Exp. Med. 210, 2843–2850 (2013).
Eich, C. et al. In vivo single cell analysis reveals Gata2 dynamics in cells transitioning to hematopoietic fate. J. Exp. Med. 215, 233–248 (2018).
Shen, J. et al. Single-cell transcriptome of early hematopoiesis guides arterial endothelial-enhanced functional T cell generation from human PSCs. Sci. Adv. 7, eabi9787 (2021).
Ditadi, A. et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat. Cell Biol. 17, 580–591 (2015).
Kabrun, N. et al. Flk-1 expression defines a population of early embryonic hematopoietic precursors. Development 124, 2039–2048 (1997).
Nishikawa, S. I., Nishikawa, S., Hirashima, M., Matsuyoshi, N. & Kodama, H. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development 125, 1747–1757 (1998).
Chen, X. et al. Integrative epigenomic and transcriptomic analysis reveals the requirement of JUNB for hematopoietic fate induction. Nat. Commun. 13, 3131 (2022).
Shen, J. et al. Defining early hematopoietic-fated primitive streak specification of human pluripotent stem cells by the orchestrated balance of Wnt, activin, and BMP signaling. J Cell Physiol. https://doi.org/10.1002/jcp.28272 (2019).
Shen, J. et al. Sequential cellular niches control the generation of enucleated erythrocytes from human pluripotent stem cells. Haematologica 105, e48–e51 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology 20, https://doi.org/10.1186/s13059-019-1874-1 (2019).
Andreatta, M. & Carmona, S. J. STACAS: Sub-Type Anchor Correction for Alignment in Seurat to integrate single-cell RNA-seq data. Bioinformatics 37, 882–884 (2021).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov. (Camb.) 2, 100141 (2021).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Hu, H. et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38 (2019).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFA0800200), the National Natural Science Foundation of China (31872843 and 31701160), and the National Key Research and Development Program of China (2018YFA0801003).
Author information
Authors and Affiliations
Contributions
S.M., J.H., and K.Y. designed the research; S.M. developed the HomologySeeker package. S.M. and J.H. performed the analysis. S.M., K.Q., and Q.L. performed the in vitro verification assays. S.M. and K.Y. analyzed the results, made figures, and wrote the paper. K.Y., J.H., and W.Z. commented on the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Ryohichi Sugimura, Emanuele Azzoni, and Zongcheng Li for their contribution to the peer review of this work. Primary Handling Editors: Eirini Trompouki and Manuel Breuer. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mo, S., Qu, K., Huang, J. et al. Cross-species transcriptomics reveals bifurcation point during the arterial-to-hemogenic transition. Commun Biol 6, 827 (2023). https://doi.org/10.1038/s42003-023-05190-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-05190-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.