Abstract
Multiple myeloma (MM) is accompanied by alterations to the normal plasma cell (PC) proteome, leading to changes to the tumor microenvironment and disease progression. There is a great need for understanding the consequences that lead to MM progression and for the discovery of new biomarkers that can aid clinical diagnostics and serve as targets for therapeutics. This study demonstrates the applicability of utilizing the single-cell high-definition liquid biopsy assay (HDSCA) and imaging mass cytometry to characterize the proteomic profile of myeloma. In our study, we analyzed ~87,000 cells from seven patient samples (bone marrow and peripheral blood) across the myeloma disease spectrum and utilized our multiplexed panel to characterize the expression of clinical markers for PC classification, additional potential therapeutic targets, and the tumor microenvironment cells. Our analysis showed BCMA, ICAM3 (CD50), CD221, and CS1 (SLAMF7) as the most abundantly expressed markers on PCs across all myeloma stages, with BCMA, ICAM3, and CD221 having significantly higher expression levels on disease versus precursor PCs. Additionally, we identify significantly elevated levels of expression for CD74, MUM1, CD229, CD44, IGLL5, Cyclin D1, UBA52, and CD317 on PCs from overt disease conditions compared to those from precursor states.
Similar content being viewed by others
Introduction
Multiple myeloma (MM) is the second leading hematologic malignancy accounting for 34,920 new cases and 12,410 deaths annually in the United States alone1. Myeloma initiates in the bone marrow (BM) as a result of the clonal expansion of resident plasma cells (PCs) and is preceded by two precursor states, monoclonal gammopathy of undetermined significance (MGUS) and smoldering MM (SMM). These lead to high tumor burden and organ damage and include MM, relapsed/refractory MM (RRMM), and either primary or secondary plasma cell leukemia (PCL)2,3,4,5. During the past three decades, the global incidence rate for MM has increased by 126%, with mortality increasing by 94%6. Therefore, there is an unmet need to understand the pathway to malignant transformation in myeloma and discover novel biomarkers that can aid clinical diagnostics and serve as targets for therapeutics.
Clonal proliferation of PCs in the BM, the root cause of MM, is accompanied by alterations to the genetic and proteomic profile of PCs, marking a shift from normal to abnormal phenotypes7,8,9,10. Primary genetic events in myelomagenesis include the dysregulation of cyclin D and chromosomal hyperdiploidy11, with secondary events such as multiple chromosomal losses and chromosome 1q amplification occurring as the disease progresses12,13,14,15. Beyond genomics, abnormal PCs express an altered proteomic profile compared to that of normal PCs7,16,17. However, there is currently no single protein marker that can diagnose MM, requiring the clinical workup to rely on multiparameter flow cytometry with varying biomarker combinations for immunophenotyping18,19,20. Moreover, the changes to the proteomic landscape of PCs during MM development, progression, treatment response, and disease relapse have not been fully explored. Given the importance of proteins to serve as targetable markers for diagnosis and treatment, there remains an unmet need for the application of technologies that can provide a more comprehensive proteomic profile of PCs in myeloma settings.
Currently, conventional 4-10 color flow cytometry methods are being used to stratify PCs and monitor disease in the BM of MM patients, mainly during diagnosis, disease monitoring or post-therapy20. There are several methods for MM flow cytometry, of which EuroFlow is the most notable21, utilizing an eight-color assay on two divided tubes from the sample to ensure a sensitivity of 10-5. Studies utilizing flow cytometry for monitoring minimal residual disease (MRD) in MM have shown higher applicability compared to their counterpart genomics-based methods (allele-specific oligonucleotide quantitative PCR (ASOqPCR)/next generation sequencing (NGS)) in patients, demonstrating the relevance of proteomic-based approaches in the clinical assessments of MM22,23. However, despite the advances in the implementation of these techniques, flow cytometry-based methods pose major limitations in standardization due to varying biomarker panels and manual gating strategies22. Additionally, while flow cytometry has the potential for multiplexing markers, the extent of this capability is limited.
Beyond diagnostics, treatment options for MM have expanded rapidly during the past decade, with progress being made in the use of proteasome inhibitors and immunomodulatory drugs24,25. However, despite these advances, myeloma remains mostly a chronic disease with most patients experiencing serial relapse1,24. Recent discoveries in targeted immunotherapy have shown promise in improving clinical outcomes for some patients, but their utility is limited due to adverse side effects from off-target toxicity, increased therapy resistance, and tumor escape26,27,28,29,30,31,32,33,34,35,36,37. Increased immunosuppression of the tumor microenvironment can also negatively impact tumor progression and targeted therapy response38,39,40,41,42,43. Furthermore, a prominent mechanism that allows for MM cells to escape immunotherapy is through tight interactions of PCs with the bone marrow microenvironment cells39,40,41,42. To provide enhanced therapy options for myeloma, we therefore need technologies that can identify additional targetable biomarkers on heterogenous myeloma cells and profile the tumor microenvironment landscape.
In this study, we demonstrate the applicability of utilizing the high-definition single-cell assay (HDSCA) and imaging mass cytometry (IMC) to identify PCs and further characterize their proteomic expression profiles in a liquid biopsy (Fig. 1). We characterized bone marrow cells in 7 patient samples across the myeloma disease spectrum (2 MGUS, 1 SMM, 2 newly diagnosed MM (NDMM), 1 RRMM, and 1 PCL) and utilized our multiplexed panel to characterize the expression of clinical markers for PC classification, potential therapeutic targets, and the tumor microenvironment cells. Our results from targeted profiling of ~87,000 cells show BCMA, ICAM3, CD221, and CS1 (SLAMF7) as the most abundantly expressed markers on PCs across all myeloma stages, with BCMA, ICAM3, and CD221 having significantly higher expression levels on overt disease conditions, while also being expressed in precursor states. Additionally, we identify significantly elevated levels of expression for CD74, MUM1, CD229, CD44, IGLL5, Cyclin D1, UBA52, and CD317, specifically on PCs from overt disease conditions compared to those from precursor states. Beyond PCs, we were able to further profile the landscape composition of patients’ tumor microenvironment cells.
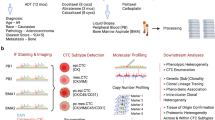
a BMA and PB samples are received, undergo processing by red blood cell lysis, are plated onto custom glass slides at approximately 3 million cells per slide, and are then stained with our 3-color immunofluorescence assay. b Stained slides are scanned using high-throughput fluorescence microscopy at 100X magnification, downstream image processing is done via OCULAR, and a final report of cells is generated for HDSCA. c Slide is stained with a panel of metal-labeled antibodies, laser ablated using the Hyperion IMC system, and multiplexed images are generated. Created with BioRender.com.
Results
Patient demographics and clinical baseline
In total, this study includes analyses of 7 patients’ samples (6 bone marrow aspirates (BMAs) and 1 peripheral blood (PB)) and 1 normal donor’s PB sample. Patient participants enrolled during 04/2019-03/2020 and were between the ages of 38–72 years at the time of enrollment. Patients’ demographics are provided in Table 1. The sample set included a total of eight slides containing an average of (mean ± standard deviation = 2,339,353.7 ± 209,860.6) nucleated cells for the BMA and an average of (2,443,256 ± 213,971.9) nucleated cells for the PB slides.
Identification and morphometric analysis of plasma and non-plasma cells
We identified and categorized the candidate cells using an automated rare cell detection workflow followed by manual classification based on three-color immunofluorescence staining, corresponding to DAPI, CD138, and CD45 (Fig. 2a). Additionally, we considered cellular morphology, consisting of cell size and eccentricity, when classifying cells of interest. PCs were identified as DAPI+|CD138+|CD45− and DAPI+|CD138+|CD45+ events and were included in the liquid biopsy profile for all samples. Additional rare events of interest were detected as morphologically distinct DAPI+|CD138−|CD45+ and DAPI+|CD138−|CD45− cells. Enumeration of the events revealed higher total rare cell count in the overt-disease settings of NDMM1, RRMM, PCL, and NDMM2 (mean ± standard deviation = 1,393.25 ± 634.1) compared to precursor states SMM and MGUS1 (mean ± standard deviation = 709.0 ± 370.0) (Fig. 2b).
Our counts identified lower levels of DAPI+|CD138+|CD45− cells in precursor conditions (mean ± standard deviation = 573 ± 346.0), compared to disease settings (mean ± standard deviation =1324 ± 555.4). However, MGUS2 showed elevated levels of DAPI+|CD138+ cells (1022), compared to SMM (517) and MGUS1 (180), providing a profile more similar to patients diagnosed with overt disease. For the DAPI+|CD138+|CD45+ group, the precursor conditions had similar levels of (mean ± standard deviation = 64.67 ± 42.1) cells to the disease settings (mean ± standard deviation = 53.50 ± 71.9) (Fig. 2b).
For the CD138− groups, precursor settings had an incidence of rare DAPI+|CD138−|CD45+ cells (mean ± standard deviation = 15.67 ± 13.8), whereas overt-disease conditions had none. An overall higher number of cells was also seen in precursor conditions for the DAPI+|CD138−|CD45− group (mean ± standard deviation = 55.67 ± 42.5) compared to overt-disease settings (mean ± standard deviation = 15.75 ± 17.7) (Fig. 2b).
The normal control report identified 2 DAPI+|CD138+|CD45− cells and 1 rare DAPI+|CD138−|CD45+ cell, making it the sample with the least number of rare events. Since some of our disease groups studied only had 1 patient sample, we did not perform statistical tests between the groups.
Multiplexed proteomic profiling of PCs
Selected candidate PCs (DAPI+|CD138+|CD45+ and DAPI+|CD138+|CD45− cells) were included in each region of interest (ROI) alongside ~300 surrounding white blood cells (WBCs) and were subjected to downstream proteomics. Background WBCs (CD45+CD138-CD38- cells) from slides were used as controls, and the expression levels of markers were normalized on a scale of 0–1. PCs were defined as CD138+CD38+ cells to match with the current clinical definition and flow cytometry gating strategy. We then assessed the normalized expression levels of available clinical biomarkers CD81, CD117, CD56, CD27, and CD28 (Fig. 3).
A t-SNE scatter plot of PCs from samples, visualized using only clinical markers (CD45, CD56, CD27, CD81, CD117, CD28), based on 0–1 normalized values. On the left, t-sne is colored by patient ID and on the right, colored by expression levels of biomarkers. B Heatmap illustration of clinical markers on patient samples, based on 0–1 normalized values.
CD56 was most abundantly expressed on all PCs throughout the disease spectrum, with the highest level of expression in RRMM and PCL samples. CD45 was highest in MGUS settings, with MGUS1 having the most CD45 enriched cells compared to all patients. MGUS2 had lower CD45 levels compared to MGUS1, in line with the clinical observation of its classification as being CD45 Low/Negative (Fig. 3). Our observations of the clinical marker signals show concordance with the clinical classification, although the precise degree of concordance could not be determined as the exact expression levels from clinical flow cytometry were not available (Supplementary Table 2). Additionally, for the IMC and the clinical flow cytometry, two distinct samples collected at the same time were used for assessment, and the antibody clones may differ between the assays.
To further characterize the proteomic profile of PCs, we investigated the expression of additional biomarkers with the potential to act as MM targets (Fig. 4). Our results demonstrate that BCMA, ICAM3, CD221, and CS1 (SLAMF7) have elevated levels throughout the disease spectrum (Fig. 4a).
a Heatmap representation of CD138+CD38+ PC expression levels for the 14 selected biomarkers (refer to Table 2). Expression levels are depicted using values normalized to a 0–1 scale, which standardizes the different biomarkers for comparison by transforming their expression levels to a common scale. b Correlation matrix of CD138+CD38+ PCs from all samples for the selected 14 proteomic markers. Pearson’s correlation was used, and hierarchical cluster analysis was conducted. c Bar plot comparing the selected proteomic markers between precursor PCs (from MGUS samples) and disease PCs (from NDMM/RRMM/PCL samples). The Kruskal–Wallis H test (one-way ANOVA) was performed, and p-values below *0.05 were considered statistically significant.
To assess the relationships between the selected additional markers, we performed Pearson’s correlation on the expression data on PCs from all samples, followed by hierarchical clustering (Fig. 4b). The results provide 3 distinct cluster sets, with the highest level of overall correlations between CD317/UBA52 (Pearson’s coefficient = 0.78), IGLL5/Blimp1 (Pearson’s coefficient = 0.68), and CD317/MUM1 (Pearson’s coefficient = 0.673). No significant correlation was found between the markers, highlighting the overall heterogeneity of the PCs.
In order to evaluate the potential of the selected markers to serve as MM targets, we set out to compare their expression between normal and abnormal PCs. We pooled PCs from precursor states where no abnormal PCs were detected (<1%) (MGUS1 and MGUS2) and compared their profiles for the selected markers with PCs from patients with NDMM/RRMM/PCL where >95% aberrant PCs were detected by the clinical flow cytometry (Table 1). We performed the Kruskal–Wallis H test (one-way ANOVA), and all p-values equal to or below *0.05 were considered statistically significant. Our results indicate that from the most prevalent markers on PCs across the spectrum of disease, BCMA, ICAM3, and CD221 are significantly expressed at higher levels on PCs from NDMM/RRMM/PCL, compared to those from precursor states (p = 0.05, p < 0.001, p < 0.001, respectively) (Fig. 4c).
We also observed significantly higher levels of expression for CD74 (p = 0.014), MUM1 (p < 0.001), CD229 (p < 0.001), CD44 (p < 0.001), IGLL5 (p < 0.001), Cyclin D1 (p < 0.001), UBA52 (p < 0.001), and CD317 (p < 0.001) on PCs from overt disease conditions compared to those from precursor states (Fig. 4c). Blimp1 and pS6 were also observed to have higher levels of expression on PCs from overt disease versus precursor conditions, however the differences did not reach statistical significance (p = 0.671 and p = 0.590, respectively) (Fig. 4c).
Characterizing the bone marrow microenvironment
The BMA samples from patients diagnosed with MGUS, SMM, NDMM, and RRMM were further analyzed to profile the tumor microenvironment landscape. Representative cells from each BMA sample were profiled based on their marker expression and cell types were determined using multiplexed quantitative proteomic signals (Supplementary Table 3; Fig. 5).
Representative cells from the BM samples are shown and profiled based on enriched biomarker expression. A Global profile of all samples combined, with clusters colored by assigned cell type. B Highlights cells pertaining to each patient sample, with colors assigned to each patient sample, as indicated in the image.
The bone marrow microenvironment analysis showed an overall significantly lower percentage of T cells detected in the BMA of overt disease conditions (NDMM1 = 7.24%/NDMM2 = 10.60%/RRMM = 7.78%; Overt disease = 8.57 ± 1.45), compared to precursor states (MGUS1 = 16.87%/MGUS2 = 14.62%/SMM = 17.0%; Precursor = 16.16 ± 1.09, p = 0.003; Fig. 5).
Discussion
Here, we demonstrate the applicability of utilizing the single-cell high-definition liquid biopsy assay (HDSCA) and imaging mass cytometry (IMC) to identify PCs and further characterize their proteomic expression profiles. By utilizing a panel of antibodies that correspond to current clinical markers used for myeloma cell immunophenotyping, additional biomarkers for targeted therapy, and immune markers, we are able to characterize the proteomic profile of disease for clinical classification, evaluate biomarkers for targeted therapy, and provide an overview of the immune landscape all in one assay.
Our analysis showed upregulated expression of BCMA, ICAM3, CD221, and CS1 (SLAMF7) in patients across all disease states. BCMA, a member of the TNF17 receptor family, has been found to be expressed on late-stage B-cell lymphocytes (memory B cells mainly), normal as well as abnormal plasma cells44,45,46. Targeted BCMA therapies have revolutionized the myeloma treatment landscape with unparalleled efficacy leading to improved clinical outcomes for late relapse patients32,33,34,37. However, there are currently no approved diagnostic tools that specifically measure cellular BCMA levels, limiting our ability to evaluate its expression throughout the course of the disease and routinely monitor the effects of anti-BCMA therapies. Utilizing targeted panels that measure BCMA could improve anti-BCMA therapy response monitoring in such patients. ICAM3, a type I transmembrane glycoprotein, has been previously identified to be upregulated on the surfaceome of MM cell lines and may play a role in immune evasion of natural killer cell-initiated lysis17,47. Our results identified ICAM3 as an upregulated biomarker in patients across MM stages, with significantly higher expression in overt disease compared to precursor states supporting its potential significance. Insulin-like growth factor-1 receptor (CD221) has been previously shown to be upregulated on human myeloma cells, with Bataille et al. demonstrating CD221 expression level as a negative prognostic marker for MM patients48. CS1 (SLAMF7) has demonstrated uniformly high expression on MM cells, regardless of genomic heterogeneity and disease state49,50. Importantly, Elotuzumab, a humanized anti-CS1 (SLMAF7), is approved for the treatment of relapsed myeloma with new target products in development51. Our analysis confirms both CD221 and CS1 (SLAMF7) to be highly expressed in myeloma cells, in agreement with previous studies.
Furthermore, we were also able to identify additional biomarkers that were more specifically expressed in overt disease conditions compared to precursor states. Markers such as CD74, MUM1, CD229, CD44, IGLL5, Cyclin D1, UBA52, and CD317 have the potential to act as emerging biomarkers for targeted therapies, pending future studies that can demonstrate their utility on a larger scale.
The core limitation of our study was the low number of patients we were able to include and not being able to provide a longitudinal study of each patient. Additionally, for the PCL patient, we were not able to receive a BMA sample, as the clinical workup was limited to a blood draw. In the future, we hope to recruit a higher number of patients and obtain matched PB and BMA samples across the MM spectrum to better characterize the disease. Validation in a larger cohort of MM patients can further help to understand the potential utility of our identified proteins as early diagnostic and prognostic markers.
Our goal was to demonstrate the utility of the HDSCA and IMC to provide a comprehensive profile of MM disease. MM is primarily diagnosed in the advanced stages, and currently, the medical interventions for treatment are limited. Early diagnosis and identification of additional therapeutic targets of MM are crucial as they provide chances for better disease management and can improve patient care and survival. We believe that a multiplexed proteomic panel could assist in future MM disease management, providing alternative therapeutic targets that have the potential to improve the survival expectancy of patients. Identifying such protein-based biosignature of MM from the liquid biopsy has the potential to serve as an early predictor of disease development and progression.
Methods
Patient enrollment
A total of 7 patients and one normal blood donor (ND) are included in this study. All patients, except the ND, were recruited at The University of Texas MD Anderson Cancer Center (Houston, TX) (IRB: UP-19-0033) between 2019 and 2020 (Table 1). For diagnosis purposes, all participating patients received a bone marrow biopsy and serological testing. A corresponding sample from each patient underwent standard-of-care flow cytometry analysis by MD Anderson as part of the MM diagnostic workup. At the time of sample collection, 2 patients were diagnosed with MGUS, 1 with SMM, 2 with NDMM, 1 with RRMM, and 1 with PCL (Table 1). The NBD sample was acquired from the Scripps Clinic Normal Blood Donor Service from an individual with no known pathology. Bone marrow aspirates (BMA) were collected from MGUS, SMM, NDMM, and RRMM patients, while peripheral blood (PB) was collected for PCL and ND. Recruitment took place according to institutional review board-approved protocols at MD Anderson Cancer Center, and all study participants provided written informed consent.
Sample collection and processing
All PB and BMA samples (8 mL) were collected in Streck tubes (Cell-free DNA Blood Collection Tube, Streck, La Vista, NE, USA) at MD Anderson and shipped to the Convergent Science Institute in Cancer at University of Southern California within a 48-h time period, as previously described52,53. Immediately upon receipt, all samples underwent red blood cell lysis in isotonic ammonium chloride solution, and the remaining nucleated cell population was plated in a monolayer on custom-made cell adhesive glass slides (Marienfeld, Lauda, Germany). The WBC count of the sample was used to approximate plating 3 million cells per slide. The prepped slides were subsequently incubated in 7% BSA, dried, and stored at −80 °C52,53,54.
Immunostaining and image acquisition
Slides were thawed prior to immunofluorescent staining. All steps were performed at room temperature using an IntelliPATH FLX™ autostainer (Biocare Medical LLC, Irvine, CA, USA). Slides received 2% neutral buffered paraformaldehyde solution (VWR, San Dimas, CA) for 20 min for cell fixation and were subsequently incubated with 10% goat serum (Millipore, Billerica, MA) for 20 min to block non-specific binding sites. The slides were then stained with a primary antibody cocktail containing mouse anti-human CD138 (B-A38, MCA2459GA, Bio-Rad, Hercules, CA) and mouse anti-human CD45 Alexa Fluor® 647 monoclonal antibody (F10-89-4, MCA87A647, AbD Serotec, Raleigh, NC), for 1 h. Antibodies had previously been validated as a part of the assay development study for MM55. The slides were washed with TBS after primary staining. Thereafter, slides were incubated with goat anti-mouse Alexa Fluor® 555 (A21127, Invitrogen, Carlsbad, CA) and counterstained with 4, 4-diamidino-2-phenylindole (DAPI; D1306, Thermo Fisher, Waltham, MA) for 40 min. Finally, all the slides were mounted with a glycerol-based media, cover-slipped and sealed for subsequent imaging52,54,55.
The immunofluorescent-stained slides were then imaged using automated high-throughput fluorescence scanning microscopy at 100X magnification, which was used to obtain 2304 frame images per fluorescence channel per side, as previously described54. Background noise levels on all slides, the gain and exposure times for all channels, DAPI (DNA), Alexa647 (CD45), and AlexaFluor®555 (CD138) were standardized for background normalization by the scanner software. After images were captured, 2304 frames per slide, cells were segmented and had their features extracted via customized EBImage56 and R software (R version 4.1.2, R core team, 2021)57.
Cell classification and region of interest identification
We utilized a customized algorithm known as OCULAR (Outlier Clustering Unsupervised Learning Automated Report) to identify and categorize rare cells in our study. This innovative image-processing pipeline combines dimensionality reduction, segmentation, and unsupervised clustering methodologies, using principal component analysis and unsupervised learning 55,57,58.
Initially, OCULAR employs the ‘EBImage’ R package to segregate DAPI-positive cells and DAPI-negative events. It then extracts features for each cell, generating a comprehensive array of 761 cellular and nuclear parameters. Utilizing principal component analysis (PCA) on these parameters and conducting hierarchical clustering based on the top 350 components, OCULAR can identify both common and rare cells from all DAPI+ events. This process is deterministic, ensuring the repeatability and robustness of the results.
OCULAR further enhances its categorization by performing a K-nearest neighbor analysis on the cellular morphological features, which include marker signal intensity and cellular and nuclear shape and size 55,57,58. It subsequently categorizes cells into three main groups: (1) Plasma cells, identified using CD138 as the marker (DAPI+|CD138+|CD45− and DAPI+|CD138+|CD45+), (2) DAPI+ only cells (DAPI+|CD138−|CD45−), and (3) non-Plasma Cell hematopoietic cells (DAPI+|CD138−|CD45+). In addition to these, OCULAR identifies ‘morphologically distinct’ cells that possess unique size, shape, and eccentricity compared to surrounding White Blood Cells 55,57,58.
For further characterization, an average of 34 regions of interest (ROI) on the slide were selected in the BMA and PB samples to undergo downstream targeted proteomics analysis (Supplementary Table 1). ROIs were selected as regions on the slide that had at least 1 DAPI+|CD138+ cell.
Targeted proteomics using imaging mass cytometry
For the downstream multiplexed proteomics, we utilized the Hyperion imaging mass cytometry (IMC) system, as previously described59,60. Metal-labeled antibodies were validated in the HDSCA workflow to ensure the specificity, selectivity, and reproducibility of antibodies through testing on biomarker-specific cell lines spiked into normal blood donor samples and spread on slides59.
The slides were stained with an MM-specific panel that utilized 35 metal-labeled antibodies and 2 DNA intercalators to characterize normal and abnormal PCs and BM microenvironment cells and to further characterize MM (Table 2). All antibodies were prepared at a standard dilution of 1:100. For BCMA, two antibody clones were tagged to the same metal (Nd150). Prior to staining with the metal-labeled antibody cocktail for IMC analysis, the slides were stored at 4 °C for 15-78 days (mean 50.4 ± 25.1). Metal-labeled antibodies were retrieved as either direct conjugates from Fluidigm (now Standard BioTools; San Francisco, CA) or underwent in-house conjugation, as per Maxpar’s antibody labeling protocol instructions (Table 2). The cocktail of metal-labeled antibodies was prepared in 1% BSA and 0.1% Tween in PBS59. The slides were removed from the 4 °C refrigerator, had their coverslips taken off, and were dipped in fresh PBS twice to wash off the glycerol-based mounting media prior to staining. Slides were then blocked with a buffer containing 1% BSA and IgG mouse Fc fragments (31205, Thermo Scientific, Waltham, MA) for 1 h and then incubated with the antibody cocktail for 1.5 hr on an orbital shaker at room temperature. Afterward, the slides were washed with fresh PBS and subsequently incubated with DNA intercalators (Ir191/Ir193, 201192A, Fluidigm, San Francisco, CA) for 30 min. Finally, the slides were washed with PBS and ddH2O, dried, and stored at room temperature until ablation (approximately for 1–4 days).
Laser ablation was conducted on previously identified ROIs across the specimen slides. The protein expression images had a resolution of 1 μm2 across the region of interest (400 × 400 μm), allowing for limited characterization of sub-cellular localization. Ion mass data were collected and used for reconstruction of the 1 μm2 ROI spatial resolution, 36-dimensional images of the ROI. The number of ROIs run per slide is listed in the supplementary information (Supplementary Table 1).
Cell boundary segmentations and pixel values were determined by a customized pipeline that utilizes CellProfiler61, ilastik pixel classifier62, and histoCAT63. CellProfiler (version 3.18) was used to remove strong outlier pixel signals, “hot pixels”, scale the images at 2x, and prepare image crops for ilastik pixel classifier training. Classification of nuclei, membrane, and background segmentations was done using ilastik (version 1.3.3) and exported as probabilities masks. After visual confirmation of masks, the classifier was applied to the entire dataset using batch processing. After the generation of segmentation masks, files were generated for histoCAT with masks for all ROIs. Single-cell measurements were extracted from all channels using the scikit-image package64 in Python 3 programming language (Python Software Foundation, Scotts Valley, CA). We calculated the signal output as the mean ion counts detected over the cell area, with the background signal—determined from the negative mask space—being subtracted from the cell values. We included all cells that demonstrated DNA signals in the final analysis. We then assessed the distribution of the signal for each marker in relation to negative control cell subsets, using background WBCs (defined as CD45+CD138−CD38− cells) from slides as controls. To facilitate comparison across different biomarkers, we normalized the proteomic expression levels for each biomarker to a scale ranging from 0 to 1.
Data analysis and visualization
Data analysis and visualization were conducted using R (Version 4.1.1., Boston, MA), the Python programming language (Version 3.0, Python Software Foundation, Scotts Valley, CA), and the Orange 3.0 data-mining toolbox in Python65. Groups were compared using the Kruskal–Wallis test (a one-way ANOVA on ranks) to detect non-parametric rank-based dependence between multiple groups. This test was used to determine whether the distributions have a median shift greater than the null hypothesis. p-values below 0.05 were considered statistically significant. No correction was performed, as the comparisons were planned comparisons. Finally, Pearson’s correlation was used to evaluate the relationship between parameters.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data discussed in this manuscript are included in the main manuscript text and the Supplementary Materials. The images of the single cells are available through the BloodPAC Data Commons Accession ID “BPDC000132” and the permalink https://data.bloodpac.org/discovery/BPDC000132. Additionally, the immunofluorescence image data is available in figshare at and the image mass cytometry data is available in figshare at https://doi.org/10.6084/m9.figshare.23929635.
Code availability
The code used to analyze the single data uses standard third-party open-source libraries and packages in R and Python. The code used to identify CTCs in the HDSCA workflow uses custom code and is proprietary and licensed to Epic Sciences for commercial use.
References
American Cancer Society. Cancer Statistics Center, 2021. http://cancerstatisticscenter.cancer.org (2021).
International Myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br. J. Haematol. 121, 749–757 (2003).
Landgren, O. et al. Prevalence of monoclonal gammopathy of undetermined significance among men in Ghana. Mayo Clin. Proc. 82, 1468–1473 (2007).
Landgren, O. & Waxman, A. J. Multiple myeloma precursor disease. JAMA 304, 2397–2404 (2010).
Kyle, R. A. et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N. Engl. J. Med. 356, 2582–2590 (2007).
Cowan, A. J. et al. Global burden of multiple myeloma: a systematic analysis for the Global Burden of Disease Study 2016. JAMA Oncol. 4, 1221–1227 (2018).
Chanukuppa, V. et al. Proteomic alterations in multiple myeloma: a comprehensive study using bone marrow interstitial fluid and serum samples. Front. Oncol. 10, 566804 (2021).
Liang, Y. et al. Malignant clonal evolution drives multiple myeloma cellular ecological diversity and microenvironment reprogramming. Mol. Cancer 21, 182 (2022).
Laganà, A. et al. Integrative network analysis identifies novel drivers of pathogenesis and progression in newly diagnosed multiple myeloma. Leukemia 32, 120–130 (2018).
Jiménez, C. et al. A next-generation sequencing strategy for evaluating the most common genetic abnormalities in multiple myeloma. J. Mol. Diagn. 19, 99–106 (2017).
Bergsagel, P. L. et al. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 106, 296–303 (2005).
Burroughs Garcìa, J. et al. Role of 1q21 in multiple myeloma: from pathogenesis to possible therapeutic targets. Cells 10, 1360 (2021).
Hanamura, I. et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood 108, 1724–1732 (2006).
Shaughnessy, J. Amplification and overexpression of CKS1B at chromosome band 1q21 is associated with reduced levels of p27Kip1 and an aggressive clinical course in multiple myeloma. Hematology 10, 117–126 (2005).
Nemec, P. et al. Gain of 1q21 is an unfavorable genetic prognostic factor for multiple myeloma patients treated with high-dose chemotherapy. Biol. Blood Marrow Transplant. 16, 548–554 (2010).
Dytfeld, D. et al. Proteomic profiling of multiple myeloma plasma cells and normal plasma cells reveals differential expression of Clu1 and Basp1 proteins. Blood 116, 4034 (2010).
Ferguson, I. D. et al. The surfaceome of multiple myeloma cells suggests potential immunotherapeutic strategies and protein markers of drug resistance. Nat. Commun. 13, 4121 (2022).
Jeong, T. D. et al. Simplified flow cytometric immunophenotyping panel for multiple myeloma, CD56/CD19/CD138(CD38)/CD45, to differentiate neoplastic myeloma cells from reactive plasma cells. Korean J. Hematol. 47, 260–266 (2012).
Chatterjee, G., Gujral, S., Subramanian, P. G. & Tembhare, P. R. Clinical relevance of multicolour flow cytometry in plasma cell disorders. Indian J. Hematol. Blood Transfus. 33, 303–315 (2017).
Rajkumar, S. V. et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 15, e538–e548 (2014).
Turner, R. et al. The utility of EuroFlow MRD assessment in real-world multiple myeloma practice. Front. Oncol. 12, 820605 (2022).
Takamatsu, H. Comparison of minimal residual disease detection by multiparameter flow cytometry, ASO-qPCR, droplet digital PCR, and deep sequencing in patients with multiple myeloma who underwent autologous stem cell transplantation. J. Clin. Med. 6, 91 (2017).
Sánchez, R., Ayala, R. & Martínez-López, J. Minimal residual disease monitoring with next-generation sequencing methodologies in hematological malignancies. Int. J. Mol. Sci. 20, 2832 (2019).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020).
Gandhi, U. H. et al. Outcomes of patients with multiple myeloma refractory to CD38-targeted monoclonal antibody therapy. Leukemia 33, 2266–2275 (2019).
Greil, C., Engelhardt, M., Finke, J. & Wäsch, R. Allogeneic stem cell transplantation in multiple myeloma. Cancers 14, 55 (2021).
Garfall, A. L. et al. Chimeric antigen receptor T cells against CD19 for multiple myeloma. N. Engl. J. Med. 373, 1040–1047 (2015).
Garfall, A. L. et al. Anti-CD19 CAR T cells with high-dose melphalan and autologous stem cell transplantation for refractory multiple myeloma. JCI Insight 3, e120505 (2018).
Peinert, S. et al. Gene-modified T cells as immunotherapy for multiple myeloma and acute myeloid leukemia expressing the Lewis Y antigen. Gene Ther. 17, 678–686 (2010).
Baumeister, S. H. et al. Phase I trial of autologous CAR T cells targeting NKG2D ligands in patients with AML/MDS and multiple myeloma. Cancer Immunol. Res. 7, 100–112 (2019).
Brudno, J. N. et al. T cells genetically modified to express an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of poor-prognosis relapsed multiple myeloma. J. Clin. Oncol. 36, 2267–2280 (2018).
Raje, N. et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N. Engl. J. Med. 380, 1726–1737 (2019).
Berdeja, J. G. et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): a phase 1b/2 open-label study. Lancet 398, 314–324 (2021).
Shah, N., Chari, A., Scott, E., Mezzi, K. & Usmani, S. Z. B-cell maturation antigen (BCMA) in multiple myeloma: rationale for targeting and current therapeutic approaches. Leukemia 34, 985–1005 (2020).
Smith, E. L. et al. GPRC5D is a target for the immunotherapy of multiple myeloma with rationally designed CAR T cells. Sci. Transl. Med. 11, eaau7746 (2019).
Huang, H. et al. Phase I open-label single arm study of GPRC5D CAR T-cells (OriCAR-017) in patients with relapsed/refractory multiple myeloma (POLARIS). J. Clin. Oncol. 40, 8004 (2022).
Raje, N. S. et al. Updated clinical and correlative results from the phase I CRB-402 study of the BCMA-targeted CAR T cell therapy bb21217 in patients with relapsed and refractory multiple myeloma. Blood 138, 548 (2021).
Desantis, V. et al. The leading role of the immune microenvironment in multiple myeloma: a new target with a great prognostic and clinical value. J. Clin. Med. 11, 2513 (2022).
Holthof, L. C. & Mutis, T. Challenges for immunotherapy in multiple myeloma: bone marrow microenvironment-mediated immune suppression and immune resistance. Cancers 12, 988 (2020).
Shah, N. N. & Fry, T. J. Mechanisms of resistance to CAR T cell therapy. Nat. Rev. Clin. Oncol. 16, 372–385 (2019).
Cheng, J. et al. Understanding the mechanisms of resistance to CAR T-cell therapy in malignancies. Front. Oncol. 9, 1237 (2019).
Van de Donk, N., Themeli, M. & Usmani, S. Z. Determinants of response and mechanisms of resistance of CAR T-cell therapy in multiple myeloma. Blood Cancer Discov. 2, 302–318 (2021).
Rodríguez-Lobato, L. G. et al. CAR T-cells in multiple myeloma: state of the art and future directions. Front. Oncol. 10, 1243 (2020).
Avery, D. T. et al. BAFF selectively enhances the survival of plasmablasts generated from human memory B cells. J. Clin. Investig. 112, 286–297 (2003).
Chiu, A. et al. Hodgkin lymphoma cells express TACI and BCMA receptors and generate survival and proliferation signals in response to BAFF and APRIL. Blood 109, 729–739 (2007).
O’Connor, B. P. et al. BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med. 199, 91–98 (2004).
Garg, TarunK. et al. Multiple myeloma cells modulate ICAM-3 to evade natural killer cell-mediated lysis. Blood 122, 3105 (2013).
Bataille, R., Robillard, N., Avet-Loiseau, H., Harousseau, J. L. & Moreau, P. CD221 (IGF-1R) is aberrantly expressed in multiple myeloma, in relation to disease severity. Haematologica 90, 706–707 (2005).
Malaer, J. D. & Mathew, P. A. CS1 (SLAMF7, CD319) is an effective immunotherapeutic target for multiple myeloma. Am. J. Cancer Res. 7, 1637–1641 (2017).
Chu, J. et al. CS1-specific chimeric antigen receptor (CAR)-engineered natural killer cells enhance in vitro and in vivo antitumor activity against human multiple myeloma. Leukemia 28, 917–927 (2014).
Prommersberger, S. et al. CARAMBA: a first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free Sleeping Beauty gene transfer to treat multiple myeloma. Gene Ther. 28, 560–571 (2021).
Shishido, S. N. et al. Preanalytical variables for the genomic assessment of the cellular and acellular fractions of the liquid biopsy in a cohort of breast cancer patients. J. Mol. Diagn. 22, 319–337 (2020).
Rodríguez-Lee, M. et al. Effect of blood collection tube type and time to processing on the enumeration and high-content characterization of circulating tumor cells using the high-definition single-cell assay. Arch. Pathol. Lab. Med. 142, 198–207 (2018).
Marrinucci, D. et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys. Biol. 9, 016003 (2012).
Ndacayisaba, L. J. et al. Enrichment-free single-cell detection and morphogenomic profiling of myeloma patient samples to delineate circulating rare plasma cell clones. Curr. Oncol. 29, 2954–2972 (2022).
Pau, G., Fuchs, F., Sklyar, O., Boutros, M. & Huber, W. EBImage—an R package for image processing with applications to cellular phenotypes. Bioinformatics 26, 979–981 (2010).
Chai, S. et al. Platelet-coated circulating tumor cells are a predictive biomarker in patients with metastatic castrate-resistant prostate cancer. Mol. Cancer Res. 19, 2036–2045 (2021).
Shishido, S. N. et al. Characterization of cellular and acellular analytes from pre-cystectomy liquid biopsies in patients newly diagnosed with primary bladder cancer. Cancers 14, 758 (2022).
Gerdtsson, E. et al. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg. Sci. Phys. Oncol. 4, 015002 (2018).
Gerdtsson, A. S. et al. Large extracellular vesicle characterization and association with circulating tumor cells in metastatic castrate resistant prostate cancer. Cancers 13, 1056 (2021).
McQuin, C. et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018).
Berg, S., Kutra, D. & Kroeger, T. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017).
van der Walt, S. et al. scikit-image: image processing in Python. PeerJ 2, e453 (2014).
Demsar, J. et al. Orange: data mining toolbox in Python. J. Mach. Learn. Res. 14, 2349–2353 (2013).
Acknowledgements
We thank the patients and their caregivers who consented to this study. We also thank the clinical research staff who contributed to the study. We are grateful to past and current technical staff at CSI Cancer for processing the samples. This work was supported by: the Miriam and Sheldon G. Adelson Medical Research Foundation (R.Z.O., D.S., E.E.M., P.K.); the Anderson Cancer Center Support Grant (P30 CA016672) (E.E.M.); the Leukemia and Lymphoma Society Specialized Center of Research (LLS SCOR-12206-17) (E.E.M., R.Z.O.); the Riney Family Multiple Myeloma Research Fund at MD Anderson from the Paula and Rodger Riney Foundation (E.E.M., R.Z.O.); the University of Texas Anderson Moon Shot Program (E.E.M.); the NCI’s USC Norris Comprehensive Cancer Center (CORE) Support 5P30CA014089-40 (P.K.); the Winnie and James Hart Endowed Fellowship (S.M.S.); the Vassiliadis Research Fund (P.K.); and institutional support from the USC Michelson Center Convergent Science Institute in Cancer (P.K.).
Author information
Authors and Affiliations
Contributions
Conceptualization: S.M.S. and L.N.; Data curation: S.M.S., L.N., K.R., V.H.; Formal analysis: S.M.S.; Funding acquisition: R.Z.O., D.E.S., E.E.M. and P.K.; Investigation: S.M.S., S.N.S., P.K.; Methodology: S.M.S., V.H., L.Y.M., P.L.; Project administration: S.M.S., S.N.S., P.K.; Resources: R.Z.O., D.E.S., E.E.M., P.K.; Supervision: S.M.S., S.N.S., P.K.; Validation: S.M.S., S.N.S., P.K.; Visualization: S.M.S.; Writing—original draft: S.M.S., S.N.S., P.K.; Writing—review and editing: S.M.S., L.N., K.R., V.H., L.Y.M., G.T., P.L., R.Z.O., D.E.S., E.E.M., S.N.S., and P.K. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing non-financial interests but the following competing financial interests: P.K. holds ownership interests in Epic Sciences and is a paid consultant/advisory board member for Epic Sciences. P.K. is a royalty recipient for related technologies licensed to Epic Sciences for development.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Setayesh, S.M., Ndacayisaba, L.J., Rappard, K.E. et al. Targeted single-cell proteomic analysis identifies new liquid biopsy biomarkers associated with multiple myeloma. npj Precis. Onc. 7, 95 (2023). https://doi.org/10.1038/s41698-023-00446-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-023-00446-0