Abstract
Phosphorus (P) is a crucial macronutrient for plant growth and development. Basic metabolic processes regulate growth; however, the molecular detail of these pathways under low phosphorous (LP) in wheat is still unclear. This study aims to elucidate the varied regulatory pathways responses to LP stress in wheat genotypes. Phenotypic, physiological, and transcriptome analyses were conducted on Fielder (P efficient) and Ardito (P inefficient) wheat genotypes after four days of normal phosphorous (NP) and LP stress. In response to LP, Fielder outperformed Ardito, displaying higher chlorophyll content-SPAD values (13%), plant height (45%), stem diameter (12%), shoot dry weight (42%), and root biomass (75%). Root structure analysis revealed that Fielder had greater total root length (50%), surface area (56%), volume (15%), and diameter (4%) than Ardito under LP. These findings highlight Fielder’s superior performance and adaptation to LP stress. Transcriptome analysis of wheat genotype roots identified 3029 differentially expressed genes (DEGs) in Fielder and 1430 in Ardito, highlighting LP-induced changes. Key DEGs include acid phosphatases (PAPs), phosphate transporters (PHT1 and PHO1), SPX, and transcription factors (MYB, bHLH, and WRKY). KEGG enrichment analysis revealed key pathways like plant hormones signal transduction, biosynthesis of secondary metabolites, and carbohydrate biosynthesis metabolism. This study unveils crucial genes and the intricate regulatory process in wheat’s response to LP stress, offering genetic insights for enhancing plant P utilization efficiency.
Similar content being viewed by others
Introduction
Phosphorous (P) is a primary macronutrient central to plant growth and development1. Although soil frequently harbors abundant total phosphorus, its binding with aluminum and iron in acidic soils renders it unavailable to crops2. Utilizing phosphorus fertilizer is a key strategy to enhance crop yield. Despite implementing efficient phosphorus fertilization practices, plants can only absorb about 30% of the applied phosphorus, while the remainder is lost due to fixation and microbial processes. This situation has led to excessive fertilizer application, contributing to environmental problems such as water source eutrophication. Moreover, this issue is exacerbated by the limited availability of rock phosphorus reserves3. Hence, the study of phosphorus nutrition has become of utmost importance. The main focus involves developing cultivars with high phosphorus efficiency and comprehending the morpho-physiological and molecular mechanisms that enable their adaptation to low phosphorus conditions. This panorama is fundamental for sustainable agriculture.
Plants respond to phosphorus deficiency through morphological, physiological, molecular, and metabolic pathways, which enhance both phosphorus uptake from the soil and internal phosphorus utilization. The response to low phosphorus (LP) includes alterations in root system architecture (RSA) and symbiotic associations with arbuscular mycorrhizal fungi, collectively aiding in adaptation to LP stress4. Additionally, plants under LP exhibit increased synthesis and secretion of organic acids (OAs), purple acid phosphatases (PAPs), and phytases3. Phosphate deficiency activates phosphate sensing mechanisms and molecular responses, which involve the coordinated expression of phosphate starvation-response (PSR) genes and microRNAs (miRNAs) in plant tissues5. The PHT1 family of high-affinity phosphate transporters is at the core of phosphate absorption; 9 and 13 PHT1s have been found in Arabidopsis and rice. Functional investigations have been conducted utilizing mutant and transgenic plants of PHT1 genes6,7. A universally conserved master regulator known as the transcription factor PHOSPHATE RESPONSE 1 (PHR1) coordinates the transcriptional activation of a significant portion of phosphate starvation-response (PSR) genes8. The nuclear proteins SYG/PHO81/XPR1 (SPX) serve as Pi-dependent competitive inhibitors of PHR1 activity in both Arabidopsis and rice9,10. Moreover, other Pi-responsive transcription factors like WRKY, MYB, and bHLH families have been identified in several plant species11. Furthermore, miR399 decreases the transcript level of PHO2, which encodes a ubiquitin-conjugating E2 enzyme involved in the ubiquitination and degradation of PHT proteins12.
Wheat (Triticum aestivum L., BBAADD, 2n = 6x = 42) is the most widely cultivated staple food crop13. In recent years, advancements have uncovered several molecular mechanisms governing Pi signaling and homeostasis in wheat. Genes encoding Al-activated malate and citrate transporters (such as TaALMT1 and TaMATE1B) play a crucial role in facilitating the efflux of organic acids (OAs), thereby improving the efficiency of Pi utilization in agricultural settings14. Phytases are phosphatases that can initiate the stepwise hydrolysis of phytate and thereby provide phosphate. The presence of phytase activity in the wheat can be attributed to three genes (rTaPAPhy_a1, rTaPAPhy_b1, and rTaPhyIIa2)15. In wheat, TaPHT1;2, TaPht1;4, TaPHT1;9-4B, and TaPT2 (all members of the PHT1 family) facilitate Pi uptake under low-Pi conditions16,17,18,19, whereas TaPHT2;1 (from the PHT2 family) and TaPHO2-A1 are crucial for regulating Pi translocation20,21. Multiple transcription factors (TFs) play a role in wheat’s response to phosphate deficiency. For instance, TaPHR-A1, TaNFYA-B1, TaPHR3-A1-A, and TaMYB4-7D have been demonstrated to enhance the expression of several TaPHT1 genes in transgenic wheat plants19,22,23,24. Furthermore, wheat miRNAs such as TaMIR1139 and TaMIR399 target genes across different families, playing pivotal roles in regulating plant tolerance to Pi starvation20,25. Metabolomic analysis presents a valuable tool in plant research for gaining deeper insights into plant responses to stress, particularly Pi starvation. This approach sheds light on the metabolic effects of Pi deficiency in wheat, revealing alterations in pathways related to carbohydrates, amino acids, and secondary metabolism26,27. Additionally, proteomic analysis revealed significant alterations in the abundance of proteins associated with nitrogen and phosphorus, small molecule, and carboxylic acid metabolic processes in response to Pi deficiency in wheat28. Plant hormones, such as auxin, cytokinin (CK), ethylene (ETH), abscisic acid (ABA), gibberellin (GA), jasmonic acid (JA), and salicylic acid (SA), undergo changes that play crucial roles in integrating Pi signaling and regulating root growth29,30,31. However, most studies have only investigated the effects of LP stress on one or two phytohormones in roots, with minimal available data on the comprehensive effects of LP stress on the phytohormone metabolome in wheat roots.
The plant’s response to low phosphorus (LP) stress is a multifaceted process regulated by the interplay of numerous genes. Despite this complexity, the transcriptional and metabolic mechanisms in wheat under LP stress remain relatively understudied. Advances in sequencing technologies, including metabolome and transcriptome analyses, offer promising avenues to accelerate the identification of molecular mechanisms governing plant responses to LP stress. This study delved into the physiological, transcriptomic, and targeted metabolomic responses of Fielder (P-efficient) and Ardito (P-inefficient) to elucidate key differentially expressed genes and metabolic pathways essential for LP response in wheat. These findings establish a fundamental groundwork and serve as a valuable resource for further exploration of gene regulatory mechanisms and a deeper understanding of the molecular processes involved in LP response and adaptation.
Result
Phenotypic changes and phosphorus efficiency under LP stress
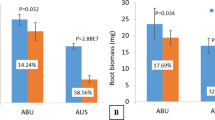
After undergoing low phosphorus (LP) treatment, two wheat genotypes experienced significant (p < 0.05) reductions in plant growth. Parameters such as SPAD, plant height, root dry weight, and shoot dry weight were significantly lower following LP treatment compared to normal phosphorus (NP) treatment in both genotypes (Table 1). Root structure analysis revealed decreased total root length, surface area, volume, and diameter in both genotypes under LP stress (Table 2). Notably, Fielder exhibited increased phenotypic indices under low-P stress compared to Ardito. Root images depicted elongated roots (greater root length) in Fielder compared to Ardito under LP (Figure S1). Overall, Fielder demonstrated superior performance to Ardito under LP stress, as evidenced by higher values and lower degradation rates of phenotypic indicators. Regarding root P contents, Fielder and Ardito experienced reductions of 59% and 70% under LP compared to NP, with Fielder showing a 95% increase in roots compared to Ardito. Additionally, shoots exhibited a 59% reduction in Fielder and a 56% reduction in Ardito under LP compared to NP, while shoot P was 79% higher in Fielder than Ardito under LP. Fielder accumulated more total P in both shoots and roots than Ardito, particularly in the roots (Fig. 1). These findings underscore Fielder's enhanced tolerance to Pi starvation and efficient Pi accumulation, designating it as the P-efficient line, while Ardito exhibited lower phosphorus tolerance.
Wheat root transcriptome profiling in response to LP stress
Transcriptome sequencing analysis was applied to investigate the molecular responses of wheat roots to Pi starvation. Twelve cDNA libraries were constructed, combining two genotypes, two treatments, and three replicates. These libraries yielded an average of 51.88 M clean reads (ranging from 44.29 M to 61.88 M). Additionally, a total of 113.40 G clean bases were generated, with respective lowest, highest, and average clean base values of 6.47 G, 8.85 G, and 7.56 G. Quality control data exhibited a Phred quality score Q30 ranging from 92 to 93.22%, coupled with an average GC content of 54.32%. Importantly, all samples demonstrated total mapping and uniquely mapping rates of 91.38% and 84.12%, respectively (average values), affirming the high quality and suitability of the sequencing data for subsequent gene expression analysis (Table S1). A comprehensive assembly yielded 118095 expressed genes (91852 known and 26243 new) and 192506 transcripts (113961 known and 78545 new). Subsequently, 91313 genes and 113446 transcripts were annotated by referencing KEGG, Swiss-Prot, Pfam, GO, COG, and NR databases. Principal component analysis (PCA) distinguished the 12 samples into two groups, Fielder and Ardito, effectively capturing differences between NP and LP treatments. PC1 and PC2 explained 18.34% and 39.70% variations, respectively (Fig. S2). The results ensured accurate replication within each group and highlighted distinct differences between genotypes and treatments. To validate RNA-Seq gene expression accuracy, qRT-PCR was conducted on 20 randomly selected DEGs (Table S2) using identical cDNA templates. The results exhibited a highly significant positive correlation with transcriptome data, affirming the reliability of the RNA-Seq results (Fig. 2).
Identification of differentially expressed genes in response to LP stress
Pairwise comparisons of genotype and treatment identified genes associated with Pi starvation. Fielder exhibited 3029 DEGs (1591 upregulated and 1438 downregulated), while Ardito had 1430 DEGs (1176 upregulated and 254 downregulated) compared to NP treatment. Surprisingly, Fielder had more DEGs than Ardito. P efficient genotype Fielder had 6704 DEGs (3328 upregulated and 3376 downregulated) compared to P inefficient genotype Ardito under LP stress (Fig. 3 and Table S3).
GO annotations and KEGG enrichment analysis
We performed GO annotation and KEGG pathway enrichment analyses of the DEGs identified in wheat under LP stress. In total, 3029 DEGs were grouped in 45 terms in Fielder, 1493 DEGs were grouped in 44 terms in Ardito, and 6704 DEGs were grouped in 47 terms in the comparison Fielder with Ardito were discovered in the GO database. Metabolic process (GO: 0008152), cellular process (GO: 0009987), and response to stimulus (GO: 0050896) were the most abundant GO terms annotated in the biological process (BP) in the three comparison groups (Table S4). Furthermore, membrane part (GO: 0044425), cell part (GO: 0044464), and organelle (GO: 0043226) were the most abundant GO terms in the cellular component (CC) (Table S4). Finally, binding (GO: 0005488) and catalytic activity (GO: 0003824) were the most enriched GO terms in the molecular function (MF) category (Fig. 4 and Table S4).
The next step is to understand the functions of these significantly enriched DEGs. The KEGG pathway enrichment analysis showed that under LP stress conditions, the top 20 enriched KEGG pathways in Fielder, Ardito, and Fielder vs. Ardito are presented in Fig. 5. DEGs from three comparisons were enriched in two identical pathways, 94 genes in Fielder, 82 genes in Ardito, and 169 genes in Fielder vs. Ardito under LP stress were significantly enriched in phenylpropanoid biosynthesis (ko00940) pathway; 13 genes in Fielder, 11 genes in Ardito, and 21 genes in Fielder vs. Ardito were enriched in arginine and proline metabolism (ko00330) pathway. In addition, for Fielder, protein processing in the endoplasmic reticulum (ko04141, 117 genes), diterpenoid biosynthesis (ko00904, 17 genes), and nitrogen metabolism (ko00910, 14 genes) were the three main pathways. For Ardito, tryptophan metabolism (ko00380, 20 genes), Cysteine and methionine metabolism (ko00270, 20 genes), and diterpenoid biosynthesis (ko00904, 16 genes) were the three main pathways with many DEGs. For the comparison of Fielder with Ardito, plant-pathogen interaction (ko04626, 107 genes), glutathione metabolism (ko00480, 57 genes), MAPK signaling pathway-plant (ko04016, 52 genes), and starch and sucrose metabolism (ko00500, 52 genes) were the enriched pathways with the highest number of DEGs (Fig. 5). The differential expression of genes in response to LP stress-activated varied molecular mechanisms in various lines, as revealed by enriched KEGG pathways.
DEGs related to pulper acid phosphatase and phosphate transporter
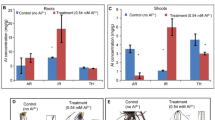
The transcriptome data revealed the identification of seven differentially expressed genes (DEGs) that encode pulper acid phosphatase (PAP) under LP stress (Fig. 6 and Table S5). Among these, five PAP genes, including one PAP2, three PAP27, and one PAP29, were significantly upregulated in Fielder; two PAP7 and one PAP29 genes were upregulated in Ardito under LP stress. One PAP1, one PAP27, and one PAP29 genes were upregulated, and one PAP1 gene was downregulated in Fielder compared with Ardito under LP stress. Four genes encoding phosphate transporter (PHT), including two PHT1-8 and one PHO1-3, were upregulated, and one PHO1-2 was downregulated in Fielder, but no response in Ardito. In addition, one SPX1 and two SPX6 genes were upregulated, and one SPX3 gene was downregulated in Fielder compared with Ardito under LP stress (Fig. 6 and Table S5).
DEGs involved in purple acid phosphatase and phosphate transporter were identified in the RNA-seq analysis that showed changes in expression under LP stress in wheat. The numbers in the black rectangles show Log2(fold-changes) of the comparisons. The comparisons show that each gene in the picture is significantly differentially expressed. PAP purple acid phosphatase, PHT, PHO phosphate transporter, SPX SPX-domain-containing protein.
DEGs related to plant hormones signal transduction
The KEGG enrichment analysis showed that 52 DEGs enriched in the MAPK signaling pathway were identified under LP stress. Thirty-nine DEGs involved in auxin, abscisic acid (ABA), brassinosteroid (BR), ethylene (ETH), gibberellin (GA), jasmonic acid (JA), salicylic acid (SA), and zeatin (ZT) signaling pathways were identified to be associated with plant hormone signal transduction (Fig. 7 and Table S6). In the auxin signaling pathway, one ARF, one GH3, and two IAA genes were upregulated, but one ARF and three SAUR genes were downregulated in Fielder compared with Ardito under LP stress. One ARF, one GH3, and two SAUR genes were upregulated in Ardito, but one SAUR gene was downregulated in Fielder. In the abscisic acid signaling pathway, one PYL and one PP2C genes were upregulated in Fielder and Fielder vs. Ardito, respectively, while three PP2C and one ABF genes were downregulated. In the brassinosteroid signaling pathway, two C92C genes were downregulated, and one BR gene was downregulated in Fielder compared with Ardito. One ERF1 and one EIN2 genes were upregulated in the ethylene signaling pathway in Ardito and Fielder vs. Ardito. In contrast, two EIN and two ERF1 genes in Fielder, three EIN genes in Ardito, and one ERF1 gene in Fielder vs. Ardito were downregulated. In the gibberellin signaling pathway, two CPS4, one GA3, and one KSL4 genes were upregulated in Fielder and Ardito, while one GA2 gene was downregulated in Fielder. In the jasmonic acid signaling pathway, four JAZ genes were upregulated in Fielder compared with Ardito. In the salicylic acid signaling pathway, one SABP2 gene was upregulated, and one SABP2 gene was downregulated in Fielder compared with Ardito. In the zeatin signaling pathway, four U73C and one CZOG genes in Fielder compared with Ardito and one CKX1 gene in Ardito were upregulated under LP stress. Two U73C genes were significantly downregulated in Fielder and Ardito, and one CKX1, one CZOG, and one U73C3 genes in Fielder compared with Ardito were downregulated under LP stress (Fig. 7 and Table S6).
DEGs involved in plant hormone signal transduction in the RNA-seq analysis showed changes in expression during LP stress in wheat. ARF auxin response factor, GH indole-3-acetic acid-amido synthetase GH3, SAU auxin-responsive protein SAU, ABF abscisic acid factor, PP2C protein phosphatase 2C, PYL abscisic acid receptor PYL, BR brassinosteroid, C92C cytochrome P450, EIL ethylene insensitive 3-like protein, ERF ethylene-responsive transcription factor, CPS syn-copalyl diphosphate synthase, GA gibberellin, KSL terpene synthase, JAZ jasmonate ZIM-domain, SABP salicylic acid binding protein, CZOG cis-zeatin O-glucosyltransferase, CKX cytokinin dehydrogenase, U37 UDP-glycosyltransferase.
DEGs related to secondary metabolic synthesis
Twenty-nine critical DEGs involved in secondary metabolic synthesis were significantly expressed in three comparisons, Fielder_LP vs. NP, Ardito_LP vs. NP, and Fielder vs. Ardito under LP stresses (Fig. 8 and Table S7). In the pathway of phenylpropanoid metabolism, four PAL, one 4CL5, and two 4CL7 genes were upregulated, while one PAL, two 4CL7, and one C73A10 genes were downregulated in Fielder vs. Ardito under LP stress, respectively. One 4CL7 gene in Fielder, two PAL, and two C73A10 genes in Ardito were upregulated under LP stress. Most structural genes implicated in the flavonoid metabolism pathways were significantly upregulated. For instance, four genes encoding shikimate o-hydroxycinnamoyltransferase (EC 2.3.1.133), four CYP75A genes, three genes encoding chalcone-flavanone isomerase (EC 5.5.16), two CHS, one CAMT, and one F3H genes were upregulated. Moreover, one CHS1 gene was downregulated in Fielder and Fielder vs. Ardito. One FLS and one C75A5 genes were downregulated in Fielder and Ardito. In the pathway of Anthocyanin metabolism, one BZ1 gene was downregulated in Fielder and Fielder vs. Ardito under LP stress (Fig. 8 and Table S7).
DEGs involved in secondary metabolic synthesis in the RNA-seq analysis showed changes in expression during LP stress in wheat. (A) The pathway of phenylpropanoid metabolism. (B) Flavonoid metabolism. (C) Anthocyanin metabolism. PAL phenylalanine ammonia-lyase, CYP73A, CYP75A cytochrome P450, 4CL 4-coumarate-CoA ligase, E2.3.1.133 Shikimate o-hydroxycinnamoyltransferase, ACT agmatine coumaroyl transferase, PHT putrescine hydroxycinnamoyl transferase, TBT1 tryptamine benzoyl transferase, PCMT caffeoyl-CoA O-methyltransferase, CHS chalcone synthase, E5.5.1.6 chalcone-flavanone isomerase, F3H naringenin,2-oxoglutarate 3-dioxygenase, FLS flavonol synthase/flavanone 3-hydroxylase, BZ anthocyanidin 3-O-glucosyltransferase.
DEGs related to carbohydrate metabolism
Twenty-eight crucial DEGs involved in carbohydrate metabolism were significantly expressed in three comparisons under LP stress (Fig. 9 and Table S8). In the starch and sucrose metabolism pathway, three SUS genes were upregulated in Fielder and Ardito, and one SUS4 gene was downregulated in Fielder under LP stress. One UGP2 gene was upregulated in Fielder vs. Ardito under LP stress. One GBE1 gene was significantly downregulated in Fielder under LP stress. Two BAM3 genes were upregulated, and one BAM gene was downregulated. In the glycolysis pathway, one GALM gene was upregulated in Fielder and Ardito and Fielder vs. Ardito under LP stress, respectively. One HK4 gene in Fielder vs. Ardito was downregulated, and one HK8 gene in Fielder was upregulated. One PFP gene was upregulated in Fielder and Ardito under LP stress. One ALDO3 gene was upregulated, and one ALDO1 gene was downregulated in Fielder vs. Ardito. One GAPDH2 and one PGK genes were downregulated in Fielder vs. Ardito. One ENO1 and one PPDK1 genes were upregulated in Ardito. Genes involved in the citrate cycle (TCA cycle) pathway were significantly expressed in the comparison Fielder with Ardito, but no response in Fielder and Ardito comparison LP with NP, including the upregulation of one ACLY gene and one ACO gene and the downregulation of the MDH2 gene and SDHA gene.
DEGs involved in carbohydrate metabolism in the RNA-seq analysis showed changes in expression during LP stress in wheat. (A) The pathway of starch and sucrose metabolism. (B) Glycolysis/gluconeogenesis. (C) Citrate cycle (TCA cycle). SUS sucrose synthase, UGP UTP-glucose-1-phosphate uridylyltransferase, BAM beta-amylase, GBE 1,4-alpha-glucan-branching enzyme, GALM galactose mutarotase, HK hexokinase, PFP pyrophosphate–fructose 6-phosphate, ALDO fructose-bisphosphate aldolase, GAPDH glyceraldehyde-3-phosphate dehydrogenase, PGK phosphoglycerate kinase, ENO enolase, PPDK pyruvate, phosphate dikinase, SDHA succinate dehydrogenase, MDH malate dehydrogenase, ACLY ATP-citrate synthase beta chain protein, ACO aconitate hydratase.
DEGs related to transcription factors (TFs)
GO analysis results showed that numerous DEGs involved in transcription factors (TFs) were annotated in the GO database in the present study. A total of 426 TFs from 29 families were significantly expressed in three comparisons under LP stress. MYB, AP2, and bHLH were the top three leading TF families, containing 82, 50, and 44 differentially expressed genes (Table 3 and Table S9). Three TFs MYB (TraesCS7D02G295400), bHLH (TraesCS5A02G049600), and NAC (TraesCS2A02G101900) families were expressed in three comparisons which showed a downward trend. However, numerous TFs showed different trends in the two cultivars under LP stress. For instance, for the WRKY family, two genes were upregulated, and one gene was downregulated in Fielder; four genes were upregulated, and one gene was downregulated in Ardito; but in Fielder comparison with Ardito, nineteen genes were upregulated, and three genes were downregulated under LP stress. For the HSF family, twenty-three genes were downregulated, and one gene was upregulated in Fielder; six genes were upregulated, and two genes were downregulated in Fielder vs. Ardito, but no significantly responded in Ardito under LP stress. In addition, two genes from the SPB family, two from the RAV family, and one from the Nin-like family were exclusively downregulated in Fielder vs. Ardito under LP stress. One gene from NF-YA was exclusively upregulated, and one gene from ZF-HD was exclusively downregulated in Fielder under LP stress, respectively. One gene from E2F/DP was upregulated in Ardito under LP stress (Table 3 and Table S9). These transcription factors showed specialty in three comparisons.
Phytohormone profiles in wheat roots
To confirm the transcriptomic findings, we performed targeted metabolomics analysis on 41 plant hormones in the roots, detecting 34 of them, including 7 AUXs (IAA, IAAla, IAAsp, IALeu, IAPhe, IATrp, and IPA), ABA, 6 GAs (GA1, GA3, GA4, GA7, GA8, and GA9), 2 GA synthesis inhibitors (UCZ and PP333), 3 JAs (JA, MeJA, and OPDA), SA, 10 CKs (mT, cZ, cZR, tZ, tZR, DZ, iP, iPR, 6-BA, and DPU), and 4 other plant growth regulators (MH, 1-NAD, 4-CPA, and 5-NG) (Table 4). The results were consistent with the transcriptome data. LP stress significantly increased the total auxin, total JA, and SA contents but decreased the total ABA and total CK contents in Fielder. Additionally, the total GA and SA increased, while the total IAA, CK, JA, and ABA content decreased in Ardito under LP stress. Furthermore, compared to Ardito, the total IAA, ABA, GA, JA, and SA content increased in Fielder under LP stress (Table 4). Regarding auxin, the IAA and IALeu content significantly increased, while the IAPhe content decreased in Fielder. The content of IAA has been increased by more than 2-fold in Fielder under LP stress. In contrast, the IAA, IAAsp, IAAleu, IAPhe, and IATrp content significantly decreased but increased the IAAla content in Ardito. As for GA, the GA1 and GA8 contents significantly increased, but the GA3 content decreased in Fielder. In Ardito, the GA7, GA8, and GA9 contents significantly increased, while the GA1, GA3, and GA4 content decreased. In terms of JA, the JA and OPDA content significantly increased, while MeJA decreased in Fielder. Conversely, the MeJA content decreased in Ardito, and the JA and OPDA content showed no significant influence. The total CK content accounts for about 80% of the total plant hormone content in the wheat root, with the highest being cZR. The CZ and DZ content increased, while the mT, CZR, and tZ content decreased in Fielder. In Ardito, the tZ content increased, while the cZ, cZR, tZR, and 6-BA content decreased. LP stress significantly decreased the MH and 4-CPA content in Fielder. Interestingly, 1-NAD was detected in Ardito under LP stress (Table 4).
Discussion
PAP and PHT regulated P assimilation and remobilization
To respond to LP stress, plants have evolved lots of strategies known as phosphate starvation responses (PSR) regulated by phosphorus-starvation-induced (PSI) genes (PHT1, PAP, SPX)32. In our study, six PAP genes were upregulated. They had a higher expression level in P efficient genotype Fielder under LP stress, which can support the high release of phosphorus from organic phosphorus under LP stress. Plants increase the secretion of purple acid phosphatases (PAPs) into the rhizosphere to scavenge organic phosphorus for plant use under low phosphorus stress33. It found that Cm-PAP10.1 and Cm-PAP10.2, genes encoding purple acid phosphatases, were upregulated in melon under LP stress, the activity of PAPs secreted by plant roots under LP stress was positively correlated with the degree of P deficiency34. In rice, OsPAP10c overexpression increased acid phosphatase (APase) activity by almost fivefold in both roots and leaves under low phosphorus. Meanwhile, genes encoding phosphorus transporter proteins were screened in transcriptome data, among which four DEGs of high-affinity phosphorus transporter systems (PHT1 and PHO1 families) were screened; three of these were significantly upregulated in P efficient genotype but not response in P inefficient genotype Ardito treated with LP. The high-affinity transporter TaPHT1;9-4B and its transcriptional regulator TaMYB4-7D contributed to efficient Pi acquisition and plant growth under Pi-limiting conditions35. In Zygophyllum xanthoxylum, PHO1 and PHT1 genes were upregulated in the roots, suggesting that these phosphorus transporter protein genes may play a vital role in regulating the distribution, transport, and maintenance of dynamic homeostasis of P in the plant body during P deficiency36. Our results that Fielder has a higher total phosphorus content both in the stem and at the root suggests that Fielder may have activated and mobilized phosphorus by secreting more PAPs and enhanced phosphorus absorption by PHTs. SPX-domain-containing proteins (SPXs) play a crucial role in the sensing, signaling, and transport of inorganic phosphate (Pi) in eukaryotes. AtSPX1 is a phosphate-dependent inhibitor of PHR1 in Arabidopsis; OsSPX1, OsSPX4, and OsSPX6 are involved in Pi starvation signaling and acting as a negative regulator of PHR in rice9,10,37,38. Transgenic plants overexpressing SPX6 exhibited decreased Pi concentrations and suppression of phosphate starvation-induced (PSI) genes37. In our study, four SPX genes were expressed in different trends under LP stress; further research is needed on the function of SPX in response to LP stress.
Plant hormones signal transduction in wheat root
LP stress disrupts plant hormone synthesis and distribution, consequently impacting root growth5. Our study reveals significant alterations in the expression of genes involved in plant hormone signal transduction, underscoring the pivotal role of hormones in wheat root development under low phosphorus stress. Under Pi starvation, heightened auxin signaling at the root tip and lateral root primordia inhibits primary root growth while stimulating lateral root formation31. The growth hormone-responsive transcription factor SAUR modulates root morphology and fosters lateral root development39,40. Similarly, increasing cytokinin (CK) is crucial in maintaining the root-to-shoot ratio, thereby reducing CK concentrations under low phosphorus conditions30. Previous research indicates that the transcription factor MYB62 regulates phosphate starvation response (PSR) by modulating GA metabolism and signaling41, suggesting the necessity for MYB TFs and GA signaling regulation in wheat to cope with low P stress. Jasmonate (JA) and salicylic acid (SA) are additional hormones crucial for integrating environmental cues and influencing root growth under LP stress32,42. Our study demonstrates the upregulation of genes involved in auxin, CK, GA, JA, SA, and brassinosteroid (BR) signaling in Fielder compared to Ardito under low phosphorus stress, indicating their involvement in regulating wheat root morphology in response to low-P stress. Targeted metabolome results corroborate the transcriptome data, showing increased total IAA, ABA, GA, JA, and SA in Fielder than in Ardito under LP stress (Table 4). Under LP stress, Fielder exhibited a significant increase in total auxin, total JA, and SA contents while experiencing a decrease in total ABA and total CK contents. On the other hand, Ardito showed an increase in total GA and SA contents but a decrease in total IAA, total JA, total CK, and ABA contents under LP stress. Root morphological indicators were significantly higher in Fielder compared to Ardito, indicating Fielder’s ability to expedite root development in response to stress through upregulation of gene expression and elevation of hormone contents. Brassinosteroids (BRs) are plant hormones that promote cell elongation and division, crucial for plant growth and development43. Recent studies have implicated brassinosteroid signaling in regulating phosphate starvation-induced malate secretion in plants44. Additionally, ABA and ETH signaling were observed to be downregulated in both Fielder and Ardito under low phosphorus stress. While ABA and ETH signals may negatively regulate low phosphorus stress in roots, further investigation is required to ascertain the precise mechanism.
Secondary metabolic synthesis regulated to LP stress
Secondary metabolites, such as phenylpropanoids, flavonoids, and anthocyanins, can scavenge reactive oxygen species, delay microbial degradation of organic acids, and enhance mobilization of rhizosphere phosphorus45,46,47. Numerous DEGs were enriched in the phenylpropanoid and flavonoid metabolism pathways, suggesting that P starvation might affect the metabolic product content of secondary metabolic synthesis in wheat roots. In our study, PAL, CYP73A, and 4CL in the phenylpropanoid metabolism pathway were upregulated under LP stress, and E2.3.1.133, PCMT, and CYP75A were significantly upregulated in the comparison Fielder with Ardito, F3H was exclusively upregulated in Fielder and E5.5.1.6 exclusively upregulated in Ardito suggesting the increase of genes related to flavonoid metabolism may be enhanced the low P tolerance in Fielder. Similar to our results, flavonoids were highly accumulated in P-efficient cotton genotype Jimain169 roots under low phosphorus due to the upregulation of the genes responsible for flavonoids48. In style, a set of genes involved in flavonoid synthesis were found to be upregulated by Pi starvation accompanied by the flavonoid metabolites, phenolic acids, and phenyl amides contents were increased in roots, which might facilitate P solubilization and cooperate with beneficial microorganisms in the rhizosphere, and thus contributing to P acquisition and utilization49. The above results indicate that Pi starvation altered the gene expression related to flavonoid biosynthesis. Genes and metabolites that regulate flavonoid metabolism might regulate phosphorus signaling and improve wheat tolerance to low phosphorus stress. However, the flavonoid content decreased significantly in P-resistant maize roots under low phosphorus stress, which may be attributed to the levels of phosphorus supply50. This may be related to the supply of phosphorus levels. In soybeans, there is more anthocyanin production in LP and more isoflavonoid production in NP51. Anthocyanin is also one of the most observed flavonoids in the roots and shoots under LP stress. Among various functions of flavonoids in plants, the higher accumulation of anthocyanin in leaves has a vital role in the photo-protection of the leaf. However, the function of anthocyanin in the roots is still to be elucidated52.
Changes in carbohydrate biosynthesis under LP stress
Plants produce ATP and CO2 through photosynthetic product breakdown (e.g., glucose) by respiration to promote root growth and development or maintain root activity for nutrient and water uptake and translocation53. Higher root biomass and physiology are required for higher production and are important traits supporting normal plant growth under stress conditions. P‐deficiency was reported to increase carbohydrate translocation via the phloem to roots to favor root growth for better acquisition of Pi from soil54. Fielder has a higher root morphology than Ardito under low phosphorus stress but at the cost of an increased response to root-to-shoot ratio. In the previous study, the P efficient genotype had a more remarkable ability to maintain phosphorylated sugars (i.e., glucose-6-P and fructose-6P) by upregulating the genes involved in glycolysis, starch, and sucrose synthesis that are important for glycolysis as well as the biosynthesis of sugars and starch55,56. The above results suggest that wheat roots maintain ATP supply by continuously altering the transcription levels of genes encoding key enzymes in the glycolysis/gluconeogenesis pathway. Under low P conditions, SUS, UGP, GALM, HK, PFP, ALDO, and BAM were upregulated in Fielder, suggesting that Fielder better maintains the phosphorylated sugars required for sugar and starch biosynthesis to regulate the metabolic processes required for energy and carbon skeleton production to supply root growth57.
Plants secrete organic acids from TCA cycle derivatives (Major citrate and malate) to help solubilize sparingly soluble inorganic phosphate and mobilize Pi from the soil58. Citrate exudation from roots increased P uptake from Fe-P in soils59. Similarly, in the case of GhmMDH1 from cotton (Gossypium hirsutum L.), overexpressed cotton showed higher malate exudation and stimulation of P uptake from sparingly soluble forms60. It found that transgenic rice of maize PEPC showed a more significant increase of oxalate exudation and accumulated P than the wild type61. ATP-citrate lyase (ACLY) could catalyze the transformation among acetyl-CoA, oxaloacetate, and citrate, the key node of pyruvate metabolism going into the TCA cycle under low phosphorus. Here, ACLY showed a higher expression in the Fielder roots than Ardito and perhaps played a key role in promoting the TCA cycle. ACO catalyzes the transformation among isocitrate, cis-Aconitate, and citrate, also upregulated in Fielder vs. Ardito, suggesting that wheat roots mainly mobilize P absorption through the secretion of citric acid under LP stress in Fielder. However, the MDH2 was downregulated compared to Fielder with Ardito, perhaps due to energy balance.
Transcription factors responded to LP tolerance in wheat
The transcriptional regulation of PSR involves several transcription factors (TFs), including members of the MYB, WRKY, and bHLH families. These TFs positively or negatively regulate Pi signaling and Pi homeostasis in plants11. In our study, the identification of 426 transcription factors suggested complex regulation in wheat root response to LP stress (Table 3). Among those identified transcription factors, 82 DEGs belonging to the MYB transcription factor family were significantly expressed by LP stress. For instance, a MYB gene (TraesCS7D02G295400) showed a significantly different expression in three comparisons (Table S9-1), indicating that MYB transcription factors are crucial in responding to low phosphorus stress. OsMYB4P- and OsMYB2P-1-overexpression upregulated the expression of the Pi transporter genes, leading to higher Pi accumulation in shoots and roots62,63. In addition, MYB genes have also been shown to respond to low phosphorus levels by modulating gibberellin (GA) metabolism and signaling and regulation of the miR399f promoter41,64.
Forty-four transcription factor genes belonging to the bHLH family were significantly expressed by LP stress. One bHLH gene (TraesCS5A02G049600) was downregulated in Fielder, Ardito, and Fielder vs. Ardito under low phosphorus stress, suggesting that wheat tends to downregulate bHLH in response to low phosphorus stress. It has also been found in other plants. For instance, AtbHLH32 is a negative regulator of several Pi-starvation responses in Arabidopsis. The bHLH TF OsPTF1 (Pi starvation-induced transcription factor 1) was reported to be involved in the PSR in rice. OsPTF1-overexpression enhanced tolerance to Pi starvation in transgenic rice by modulating root architecture rather than triggering increased expression of phosphate transporters65.
Additionally, 30 transcription factors belonging to the WRKY family were identified in this study. Most WRKY genes showed significant upregulation, especially in the comparison between Felder and Ardito, suggesting the WRKY may have a positive regulatory effect on low phosphorus tolerance in Fielder. WRKY45 and WRKY75 positively regulate the expression of PHT1;1. RNA interference (RNAi) lines of both WRKY45 and WRKY75 displayed impaired Pi uptake66,67. In rice, transgenic seedlings overexpressing OsWRKY74 display increased Pi uptake, longer roots, increased biomass, and higher iron accumulation levels68, WRKY21 and WRKY108 activate the expression of OsPHT1;1 under Pi-sufficient conditions to promote Pi accumulation69. The vast and complex regulatory mechanisms indicate that TFs play an important role in responding to low phosphorus stress, and further research is needed on these TFs.
Materials and methods
Plant materials and treatment
Two wheat genotypes with different responses to low phosphorus (LP), including Fielder (P efficient) and Ardito (P inefficient), were used in this study. Wheat seeds were surface-sterilized with 0.5% NaClO (v/v) solution for 20 min, rinsed with sterile water, and allowed to germinate in the wet paper towel for 4–5 days at 4 °C before the seedlings were transplanted into continuously aerated 1/5 Hoagland hydroponic solution (pH 4.2) for a 2-day acclimation to low pH in a walk-in growth chamber with a stable temperature of 23 °C, a 14 h photoperiod of 400 μmol m−2·s−1 illumination level, and relative humidity of 60–80%. For different Pi-level treatments, germinated seedlings were moved into the solutions with two concentrations of NaH2PO4: 200 μmol/L (normal Pi-level, NP) or 2 μmol/L (low Pi-level, LP) and grow for 4 days. The experimental samples are entirely random and arranged with at least three replicates.
Determination of phenotypic indicators
The plant height of seedlings was measured with a ruler, and Electronic vernier calipers were used to measure stem thickness. The relative chlorophyll content in fully developed leaves, as indicated by SPAD (Soil and Plant Analyzer Development) value, was determined with a portable Konica SPAD-502Plus instrument (Konica Minolta Holdings Inc., Tokyo, Japan). Fresh plant roots from different treatment groups were collected and completely expanded onto the scanner platform (Epson Expression 1000XL, Seiko Epson Corporation, Nagano, Japan). Analyze the scanned root photos using WinRhizo Pro software (Regent Instruments Inc., Quebec, Canada) to obtain the roots’ total length, diameter, surface area, and volume. Afterward, dry the stem and root samples at 75 ° C for 4 days and further determine the dry mass of the stem and roots using an electronic balance.
P concentration determination
Oven-dried shoot and root samples were ground into fine powder for P concentration analysis. A 50-microgram sample was weighed out and digested with 9 mL nitric acid and 1 mL H2O2 using a microwave digestion instrument, and phosphorus concentration was measured by ICP-AES (Juguang Co., Beijing, China). Total phosphorus in the shoot (TPL) was calculated based on shoot phosphorus concentration (LPC) and shoot weight. Total phosphorus in roots (TPR) was calculated based on root phosphorus concentration (RPC) and root weight70.
RNA extraction, library preparation, sequencing, and read mapping
Follow the instructions of the TRIzol kit (Invitrogen Co., Carlsbad, CA, USA) to extract total RNA from a 1 cm root tip. RNA quality and integrity were measured using a 5300 bioanalyzer (Agilent Co., Santa Clara, CA, USA) and quantified using ND-2000 (Thermo Fisher NanoDrop, Waltham, MA, USA). Only high-quality RNA samples (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, > 1 μg, RIN ≥ 6.5, 28S: 18S ≥ 1.0) were used to construct sequencing library and verified by qRT-PCR. Entrust Shanghai Majorbio Biopharmaceutical Biotechnology Co., Ltd. (Shanghai, China) to perform RNA purification, reverse transcription, and library construction according to the manufacturer’s instructions (Illumina, San Diego, CA, USA), followed by sequencing analysis. 12 RNA-seq transcriptome libraries (2 genotypes × 2 treatments × 3 biological replicates) were prepared and sequenced. The FASTP71 soft was used to remove low-quality readings from the originally paired ends, and HISAT272 (Ver. 2.2.1) was used to map clean readings onto the wheat genome for comparison. The StringTie73 (Ver. 1.3.6) was then used to assemble the mapped reads in a reference-based approach. All genes were annotated against public databases, including NR, Pfam, COG, Swiss-Prot, KEGG, and GO.
Differential expression and functional enrichment analysis
Express the level of each transcript using Fragments Per Kilobases Per Millionreads (FPKM). RSEM74 was used to quantify gene abundances. Differential expression analysis was performed using the DESeq275. DEGs with |log2FC| ≥ 1 and FDR ≤ 0.05 were considered significantly different expressed genes. Cluster heat diagrams were drawn using Toolkit for Biologists (TBtools Ver. 2.030) with default settings76. Those data were added to the Comprehensive Gene Expression Database with the accession number PRJNA1033153. In addition, Goatools and KOBAS6377 were used to analyze these DEGs’ GO and KEGG functions. When the Bonferroni corrected, the p-value was ≤ 0.05, and GO terms, and KEGG metabolic pathways were significantly enriched compared to the entire transcriptome background.
Metabolome detection
The frozen wheat root apices were dispatched to Shanghai Majorbio Biopharmaceutical Biotechnology Co., Ltd. (Shanghai, China) for phytohormone extraction and analysis. Precision weighing was conducted on 100 mg root samples, which were then placed into 2 mL grinding tubes. Subsequently, 498 μL of 80% methanol and 2 μL of SA-D4 internal standard solution (2 μg/mL) were added to each tube. The mixture was then ground at low temperature for 3 minutes and subjected to ultrasound extraction at low temperature for 1 hour. To the extracted sample, 25 mg of EN15662 (consisting of magnesium sulfate 4 g, sodium chloride 1 g, sodium citrate 2 hydrate 1 g, disodium hydrogen citrate 1.5 hydrate 0.5 g) was added as a salt packet. The mixture was immediately shaken and vigorously vibrated on an oscillator for 10 minutes. Following this, it was centrifuged at 10 °C for 10 min, and 100 μL of supernatant was extracted. To this, 60 μL of water was added, and the solution was thoroughly swirled before transferring it into a sample vial. Qualitative and quantitative detection of the target substances in the samples was carried out using LC-ESI-MS/MS (UHPLC-Qtrap). The specific parameters included a WatersBEHC18 (2.1 × 100 mm, 1.7 μm) liquid chromatography column with a column temperature of 30 °C and a sample size of 10 μL. The mobile phase consisted of mobile phase A (0.1% formic acid water) and mobile phase B (0.1% formic acid acetonitrile), with a balance time of 3 min and a collection time of 10 min. In Sciex quantitative software OS, default parameters were utilized to automatically identify and integrate each ion fragment and assist in manual inspection. A standard linear regression curve was established with the mass peak area of the analyte as the ordinate and the analyte concentration as the abscissa. For sample concentration calculation, the mass spectrum peak area of the sample analyte was substituted into the linear equation to determine the concentration result.
Quantitative real-time PCR (qRT-PCR) validation
Using the identical RNA/cDNAs for RNA-seq as templates, qRT-PCR was performed on a Bio-Rad CFX96 (Bio-Rad Laboratories, Hercules, CA, USA) to verify the authenticity of transcriptomic profile expression patterns. The 10 μL reaction system contained TB Green® Premix Ex Taq™ II (Takara standard Co., Osaka, Japan) 5 μL, 10 μM primers each 0.2 μL, cDNA template 1 μL, and ddH2O 3.6 μL. The amplification procedure was initially 95 °C for 30 s, 40 cycles of 95 °C for 10 s, and 60 °C for 30 s (two-step thermal cycling). Twenty candidate DEGs involved in various processes were randomly picked up as target genes, and the housekeeping gene TaActin was used as an internal control. The primers used for qRT-PCR are listed in Table S1. The expression level of the gene was determined using the 2−ΔΔCT formula78.
Statistical analysis
Analysis of variance (ANOVA) followed by Duncan’s multiple range test (DMRT) was conducted to compare means among treatments using SPSS software (version 19.0). Before ANOVA, the data underwent normality testing using Chi-square analysis79.
Ethics declarations
The authors declare no competing interests. This study does not include human or animal subjects. All experimental research and studies on plants (wheat cultivars) comply with relevant institutional, national, and international guidelines and legislation, as well as the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Data availability
The original contributions presented in the study are available in the article or Supplementary. The RAN-seq raw data can be found on the NCBI repository, accession number PRJNA1033153. Supplementary material associated with this article can be found in the online version.
References
Chiou, T. & Lin, S. I. Annual Review of Plant Biology. Vol. 62 (eds. Merchant, S. S., Briggs, W. R. & Ort, D.). 185–206 (2011).
Chen, Z. et al. Improving phosphorus acquisition efficiency through modification of root growth responses to phosphate starvation in legumes. Front. Plant Sci. https://doi.org/10.3389/fpls.2023.1094157 (2023).
Lizbeth Lopez-Arredondo, D., Antonio Leyva-Gonzalez, M., Isabel Gonzalez-Morales, S., Lopez-Bucio, J. & Herrera-Estrella, L. Annual Review of Plant Biology. Vol. 65 (ed. Merchant, S. S.). 95–123 (2014).
Niu, Y. F. et al. Responses of root architecture development to low phosphorus availability: A review. Ann. Bot. 112, 391–408. https://doi.org/10.1093/aob/mcs285 (2013).
Zhang, Z., Liao, H. & Lucas, W. J. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 56, 192–220. https://doi.org/10.1111/jipb.12163 (2014).
Gu, M., Chen, A., Sun, S. & Xu, G. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing?. Mol. Plant 9, 396–416. https://doi.org/10.1016/j.molp.2015.12.012 (2016).
Mlodzinska, E. & Zboinska, M. Phosphate uptake and allocation—A closer look at Arabidopsis thaliana L. and Oryza sativa L.. Front. Plant Sci. https://doi.org/10.3389/fpls.2016.01198 (2016).
Nilsson, L., Mueller, R. & Nielsen, T. H. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 30, 1499–1512. https://doi.org/10.1111/j.1365-3040.2007.01734.x (2007).
Puga, M. I. et al. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. PNAS 111, 14947–14952. https://doi.org/10.1073/pnas.1404654111 (2014).
Wang, Z. et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. PNAS 111, 14953–14958. https://doi.org/10.1073/pnas.1404680111 (2014).
Wang, F., Deng, M., Xu, J., Zhu, X. & Mao, C. Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 74, 114–122. https://doi.org/10.1016/j.semcdb.2017.06.013 (2018).
Yang, M. et al. Chloroplastic Sec14-like proteins modulate growth and phosphate deficiency responses in Arabidopsis and rice. Plant Physiol. https://doi.org/10.1093/plphys/kiad212 (2023).
Walkowiak, S. et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 588, 277–283. https://doi.org/10.1038/s41586-020-2961-x (2020).
Chen, W. et al. Research advances in the mutual mechanisms regulating response of plant roots to phosphate deficiency and aluminum toxicity. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23031137 (2022).
Madsen, C. K. & Brinch-Pedersen, H. Molecular advances on phytases in barley and wheat. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20102459 (2019).
Miao, J. et al. Characterization of the promoter of phosphate transporter TaPHT1.2 differentially expressed in wheat varieties. J. Genet. Genomics 36, 455–466. https://doi.org/10.1016/s1673-8527(08)60135-6 (2009).
Liu, X. et al. TaPht1;4, a high-affinity phosphate transporter gene in wheat (Triticum aestivum), plays an important role in plant phosphate acquisition under phosphorus deprivation. Funct. Plant Biol. 40, 329–341. https://doi.org/10.1071/fp12242 (2013).
Guo, C. et al. TaPT2, a high-affinity phosphate transporter gene in wheat (Triticum aestivum L.), is crucial in plant Pi uptake under phosphorus deprivation. Acta Physiol. Plant 36, 1373–1384. https://doi.org/10.1007/s11738-014-1516-x (2014).
Wang, P. et al. TaPHT1;9–4B and its transcriptional regulator TaMYB4-7D contribute to phosphate uptake and plant growth in bread wheat. New Phytol. 231, 1968–1983. https://doi.org/10.1111/nph.17534 (2021).
Guo, C. et al. Function of wheat phosphate transporter gene TaPHT2;1 in Pi translocation and plant growth regulation under replete and limited Pi supply conditions. Planta 237, 1163–1178. https://doi.org/10.1007/s00425-012-1836-2 (2013).
Ouyang, X. et al. Knock out of the PHOSPHATE 2 gene TaPHO2-A1 improves phosphorus uptake and grain yield under low phosphorus conditions in common wheat. Sci. Rep. https://doi.org/10.1038/srep29850 (2016).
Wang, J. et al. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann. Bot. 111, 1139–1153. https://doi.org/10.1093/aob/mct080 (2013).
Qu, B. et al. A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 167, 411–423. https://doi.org/10.1104/pp.114.246959 (2015).
Zheng, X. et al. The MYB transcription factor TaPHR3-A1 is involved in phosphate signaling and governs yield-related traits in bread wheat. J. Exp. Bot. 71, 5808–5822. https://doi.org/10.1093/jxb/eraa355 (2020).
Liu, Z. et al. TaMIR1139: A wheat miRNA responsive to Pi-starvation, acts a critical mediator in modulating plant tolerance to Pi deprivation. Plant Cell Rep. 37, 1293–1309. https://doi.org/10.1007/s00299-018-2313-6 (2018).
Li, P. et al. Integrated analysis of metabolome and transcriptome reveals insights for low phosphorus tolerance in wheat seedling. Int. J. Mol. Sci. https://doi.org/10.3390/ijms241914840 (2023).
Cuyas, L., Jing, L., Pluchon, S. & Arkoun, M. Unraveling metabolic profile of wheat plants subjected to different phosphate regimes. J. Soil Sci. Plant Nutr. 23, 974–990. https://doi.org/10.1007/s42729-022-01097-z (2023).
Zheng, L. et al. Comparative physiological and proteomic response to phosphate deficiency between two wheat genotypes differing in phosphorus utilization efficiency. J. Proteom. 280, 104894–104894. https://doi.org/10.1016/j.jprot.2023.104894 (2023).
Satheesh, V., Tahir, A., Li, J. & Lei, M. Plant phosphate nutrition: Sensing the stress. Stress biol. 2, 16–16. https://doi.org/10.1007/s44154-022-00039-0 (2022).
Silva-Navas, A. et al. Role of cis-zeatin in root responses to phosphate starvation. New Phytol. 224, 242–257. https://doi.org/10.1111/nph.16020 (2019).
Bhosale, R. et al. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. https://doi.org/10.1038/s41467-018-03851-3 (2018).
Chien, P., Chiang, C., Leong, S. J. & Chiou, T. Sensing and signaling of phosphate starvation: From local to long distance. Plant Cell Physiol. 59, 1714–1722. https://doi.org/10.1093/pcp/pcy148 (2018).
Fei, Y., Xue, Y., Du, P., Yang, S. & Deng, X. Expression analysis and promoter methylation under osmotic and salinity stress of TaGAPC1 in wheat (Triticum aestivum L). Protoplasma 254, 987–996. https://doi.org/10.1007/s00709-016-1008-5 (2017).
Fita, A. et al. Diversity in expression of phosphorus (P) responsive genes in Cucumis melo L.. PLoS ONE https://doi.org/10.1371/journal.pone.0035387 (2012).
Wang, Y., Chen, Y. & Wu, W. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 63, 34–52. https://doi.org/10.1111/jipb.13053 (2021).
Hu, X. et al. Transcriptome analysis of Zygophyllum xanthoxylum adaptation strategies to phosphate stress. Front. Plant Sci. https://doi.org/10.3389/fpls.2021.723595 (2021).
Zhong, Y. et al. Rice SPX6 negatively regulates the phosphate starvation response through suppression of the transcription factor PHR2. New Phytol. 219, 135–148. https://doi.org/10.1111/nph.15155 (2018).
Lv, Q. et al. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26, 1586–1597. https://doi.org/10.1105/tpc.114.123208 (2014).
Du, Y. & Scheres, B. Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69, 155–167. https://doi.org/10.1093/jxb/erx223 (2018).
Castrillo, G. et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513. https://doi.org/10.1038/nature21417 (2017).
Devaiah, B. N., Madhuvanthi, R., Karthikeyan, A. S. & Raghothama, K. G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2, 43–58. https://doi.org/10.1093/mp/ssn081 (2009).
Ham, B., Chen, J., Yan, Y. & Lucas, W. J. Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 49, 1–9. https://doi.org/10.1016/j.copbio.2017.07.005 (2018).
Planas-Riverola, A. et al. Brassinosteroid signaling in plant development and adaptation to stress. Development https://doi.org/10.1242/dev.151894 (2019).
Liu, T. et al. Brassinosteroid signaling regulates phosphate starvation-induced malate secretion in plants. J. Integr. Plant Biol. 65, 1099–1112. https://doi.org/10.1111/jipb.13443 (2023).
Wu, Q. et al. Endophytic Bacillus sp. AP10 harboured in Arabis paniculata mediates plant growth promotion and manganese detoxification. Ecotox. Environ. Safe https://doi.org/10.1016/j.ecoenv.2023.115170 (2023).
Chen, Y. et al. Enhancement of polyphenolic metabolism as an adaptive response of lettuce (Lactuca sativa) roots to aluminum stress. Environ. Pollut. https://doi.org/10.1016/j.envpol.2020.114230 (2020).
Tomasi, N. et al. Flavonoids of white lupin roots participate in phosphorus mobilization from soil. Soil Biol. Biochem. 40, 1971–1974. https://doi.org/10.1016/j.soilbio.2008.02.017 (2008).
Iqbal, A. et al. Integrative physiological, transcriptome and metabolome analysis reveals the involvement of carbon and flavonoid biosynthesis in low phosphorus tolerance in cotton. Plant Physiol. Biochem. 196, 302–317. https://doi.org/10.1016/j.plaphy.2023.01.042 (2023).
Luo, J. et al. Metabolic alterations provide insights into Stylosanthes roots responding to phosphorus deficiency. BMC Plant Biol. https://doi.org/10.1186/s12870-020-2283-z (2020).
Luo, B. et al. Metabolite profiling and genome-wide association studies reveal response mechanisms of phosphorus deficiency in maize seedling. Plant J. 97, 947–969. https://doi.org/10.1111/tpj.14160 (2019).
Nezamivand-Chegini, M. et al. Integration of transcriptomic and metabolomic analyses provides insights into response mechanisms to nitrogen and phosphorus deficiencies in soybean. Plant Sci. (Amsterdam, Neth.) https://doi.org/10.1016/j.plantsci.2022.111498 (2023).
Zeng, X., Chow, W. S., Su, L., Peng, X. & Peng, C. Protective effect of supplemental anthocyanins on Arabidopsis leaves under high light. Physiol. Plant 138, 215–225. https://doi.org/10.1111/j.1399-3054.2009.01316.x (2010).
Hammond, J. P. & White, P. J. Sucrose transport in the phloem: Integrating root responses to phosphorus starvation. J. Exp. Bot. 59, 93–109. https://doi.org/10.1093/jxb/erm221 (2008).
Iqbal, A. et al. Phosphorus and carbohydrate metabolism contributes to low phosphorus tolerance in cotton. BMC Plant Biol. https://doi.org/10.1186/s12870-023-04100-6 (2023).
Nguyen, V. L., Palmer, L. & Stangoulis, J. Higher photochemical quenching and better maintenance of carbon dioxide fixation are key traits for phosphorus use efficiency in the wheat breeding line, RAC875. Front Plant Sci 12, 816211. https://doi.org/10.3389/fpls.2021.816211 (2021).
Kumar, S. et al. Characterization of contrasting rice (Oryza sativa L.) genotypes reveals the Pi-efficient schema for phosphate starvation tolerance. BMC Plant Biol. https://doi.org/10.1186/s12870-021-03015-4 (2021).
Amtmann, A., Hammond, J. P., Armengaud, P. & White, P. J. Advances in Botanical Research: Incorporating Advances in Plant Pathology. Vol. 43 (ed. Callow, J. A.). 209–257 (2006).
Wang, J. et al. Ethylene inhibits malate accumulation in apple by transcriptional repression of aluminum-activated malate transporter 9 via the WRKY31-ERF72 network. New Phytol. 239, 1014–1034. https://doi.org/10.1111/nph.18795 (2023).
Wang, Y. et al. Dual effects of transgenic Brassica napus overexpressing CS gene on tolerances to aluminum toxicity and phosphorus deficiency. Plant Soil 362, 231–246. https://doi.org/10.1007/s11104-012-1289-1 (2013).
Wang, Z. et al. The mitochondrial malate dehydrogenase 1 gene GhmMDH1 is involved in plant and root growth under phosphorus deficiency conditions in cotton. Sci. Rep. https://doi.org/10.1038/srep10343 (2015).
Begum, H. H. et al. The function of a maize-derived phosphoenolpyruvate carboxylase (PEPC) in phosphorus-deficient transgenic rice. Soil Sci. Plant Nutr. 51, 497–506. https://doi.org/10.1111/j.1747-0765.2005.tb00058.x (2005).
Yang, W. T. et al. Overexpression of OsMYB4P, an R2R3-type MYB transcriptional activator, increases phosphate acquisition in rice. Plant Physiol. Biochem. 80, 259–267. https://doi.org/10.1016/j.plaphy.2014.02.024 (2014).
Dai, X., Wang, Y., Yang, A. & Zhang, W. OsMYB2P, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol. 159, 169–183. https://doi.org/10.1104/pp.112.194217 (2012).
Baek, D. et al. Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol. 161, 362–373. https://doi.org/10.1104/pp.112.205922 (2013).
Yi, K. K. et al. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 138, 2087–2096. https://doi.org/10.1104/pp.105.063115 (2005).
Wang, H. et al. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER 1; 1 expression in response to phosphate starvation. Plant Physiol. 164, 2020–2029. https://doi.org/10.1104/pp.113.235077 (2014).
Devaiah, B. N., Karthikeyan, A. S. & Raghothama, K. G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. https://doi.org/10.1104/pp.106.093971 (2007).
Dai, X., Wang, Y. & Zhang, W. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. https://doi.org/10.1093/jxb/erv515 (2016).
Zhang, J. et al. OsWRKY21 and OsWRKY108 function redundantly to promote phosphate accumulation through maintaining the constitutive expression of OsPHT1;1 under phosphate-replete conditions. New Phytol. 229, 1598–1614. https://doi.org/10.1111/nph.16931 (2021).
Dai, B. et al. Physiological, biochemical, and transcriptomic responses of Neolamarckia cadamba to aluminum stress. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21249624 (2020).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890. https://doi.org/10.1093/bioinformatics/bty560 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357-U121. https://doi.org/10.1038/nmeth.3317 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290. https://doi.org/10.1038/nbt.3122 (2015).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. https://doi.org/10.1186/1471-2105-12-323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Chen, C. et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 (2020).
Xie, C. et al. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. https://doi.org/10.1093/nar/gkr483 (2011).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Luo, D. et al. Physiological and transcriptomic analyses reveal commonalities and specificities in wheat in response to aluminum and manganese. Curr. Issues Mol. Biol. 46, 367–397. https://doi.org/10.3390/cimb46010024 (2024).
Acknowledgements
This research was funded by the Guangxi Natural Science Foundation, grant number 2023GXNSFAA026445, and the National Natural Science Foundation of China, grant number 31960633. The Authors are highly grateful to Jiping Liu from Robert W. Holley Center, USDA-ARS/Cornell University for providing wheat seed.
Author information
Authors and Affiliations
Contributions
Conceptualization: D. D., Y.X., and Y.L.; conducting experiment and formal analysis: D.L. and M.U.; data curation: F.P.; statistical expertise: W.Z. and F.P.; writing-original draft preparation: D.L., M.U. and D.D.; writing-review and editing: D.D. and Y.L.; visualization: Y.Q. and Q.L.; funding acquisition: D.D. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Luo, D., Usman, M., Pang, F. et al. Comparative transcriptomic and physiological analyses unravel wheat source root adaptation to phosphorous deficiency. Sci Rep 14, 11050 (2024). https://doi.org/10.1038/s41598-024-61767-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-61767-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.