Abstract
Alzheimer’s disease (AD), a neurodegenerative disease that mostly affects the elderly, slowly impairs memory, cognition, and daily tasks. AD has long been one of the most debilitating chronic neurological disorders, affecting mostly people over 65. In this study, we investigated the use of Vision Transformer (ViT) for Magnetic Resonance Image processing in the context of AD diagnosis. ViT was utilized to extract features from MRIs, map them to a feature sequence, perform sequence modeling to maintain interdependencies, and classify features using a time series transformer. The proposed model was evaluated using ADNI T1-weighted MRIs for binary and multiclass classification. Two data collections, Complete 1Yr 1.5T and Complete 3Yr 3T, from the ADNI database were used for training and testing. A random split approach was used, allocating 60% for training and 20% for testing and validation, resulting in sample sizes of (211, 70, 70) and (1378, 458, 458), respectively. The performance of our proposed model was compared to various deep learning models, including CNN with BiL-STM and ViT with Bi-LSTM. The suggested technique diagnoses AD with high accuracy (99.048% for binary and 99.014% for multiclass classification), precision, recall, and F-score. Our proposed method offers researchers an approach to more efficient early clinical diagnosis and interventions.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is distinguished by the accumulation of aberrant protein deposits in the brain, known as plaques and tangles, which result in the death of nerve cells and the degeneration of brain tissue. Neural degeneration reduces cognitive function and causes mood and behavior changes1,2. AD is typically categorized in three stages3,4. The first stage is the preclinical stage, characterized by brain, blood, and cerebrospinal fluid (CSF) abnormalities without outward signs5. It is believed that AD pathology begins at least 20 years before symptoms appear6. The second stage of the disease is referred to as mild cognitive impairment (MCI), which involves cognitive impairment confined to a single cognitive domain, usually memory. Dementia, the final stage of the disease, is defined as a cognitive disturbance in more than one domain, often memory and executive function, with substantial interference with daily life activities.
With the recent approval of new drugs for early AD intervention, early detection of AD and differentiation of MCI have become of primary importance for successful disease treatment and management7,8, and to slow disease progression and improve the quality of life for those with AD9,10. AD classification is one of the most challenging problems neurologists face1. Advances in computer-aided diagnosis (CAD) systems based on neuroimaging data tools have improved classification. CAD systems can be divided into conventional and deep learning-based techniques. Most traditional approaches to image analysis employ a four-stage pipeline of pre-processing, segmentation, feature extraction, and classification9. Deep learning (DL) algorithms have an advantage over conventionally based methods because they require little or no image pre-processing. They can automatically infer an optimal data representation from raw images without requiring prior feature selection, resulting in a more objective and less biased process10,11,12,13,14. CNN-based architectures are used extensively for medical image analysis. They have been applied to 2D and 3D ultrasound and MRI images15 and are the deep models used most frequently to detect AD15,16,17,18. However, a 3D MRI brain image consists of stacked 2D data slices. 3D-CNN model to learn spatiotemporal features would be optimal, which is impossible with 2D CNN. But, because it requires many parameters and a high amount of computation, the 3D model cannot be used to construct deep models15.
Although transformer architecture dominates natural language processing, its use in medical imaging has been limited19. However, Vision Transformer (ViT) has recently gained popularity due to its impressive results in various medical imaging tasks, including image classification, object detection, and semantic segmentation20. ViT took note of the scaling success of Transformers in NLP and applied a standard Transformer to images with minimal modifications. Transformer-based architectures have also been used in medical image analysis21,22,24. ViT has recently demonstrated superior performance in many computer-vision tasks, making it a viable alternative to CNN as a network architecture23. CNNs collect features gradually from local to global by adding more convolutional layers. ViT, on the other hand, uses a multi-headed self-attention mechanism to capture long-range dependencies. For this approach, the model equally weights all elements in the input sequence for superior performance. ViT extracts features across the entire image without degrading image resolution, preventing spatial loss from information skipping. Thus, ViT is ideal for brain imaging analysis. The self-attention strategy of ViT has the capacity to accurately capture the interdependencies between various dispersed networks of brain regions21.
ViT is based on the concept of Transformers from natural language processing (NLP) applied to medical images. It uses a standard Transformer architecture, with minimal modifications, to process MRI images instead of text. Other neural network models, which process the image sequences sequentially (RNN) or in parallel (CNN), require more time to train and infer the results, and fail to control for long-term dependencies among the image layers24. The joint transformer handles long-range dependencies, avoids recursion, and allows parallel computation to reduce training time and avoid performance drops due to long-range dependencies24.
ViT has outperformed CNN in several computer-vision tasks, giving it an appropriate network architectural option. We were motivated to use ViT's benefits to diagnose AD patients using 3D MRIs. The lack of large-scale datasets in this field is one of the major obstacles to training deep models from scratch. The model can adapt to the smaller target dataset by using transfer learning to learn from a larger dataset. The 3D data from a plane was divided into a 2D slice array in order to benefit from transfer learning using a pre-trained ViT. Furthermore, it should be possible for slice-based methods to track the dependencies of related features across slices. The sequence classification task uses a time series classification with a transformer to get around this issue. We have combined time series transformers and pre-trained ViT to create a deep learning-based classification system for AD.
The goal of this article is twofold with respect to the proposed ViT: (1) evaluate the predictive performance of ViT combined with a transformer neural network; and (2) capture long-range dependencies and the global context of MRIs, allow parallel computation to reduce training time, and avoid performance drops due to long-range dependencies. We propose testing the hypothesis that ViT with a time series transformer performs better for AD patient classification based on MRI by using the self-attention mechanism to capture long-range dependencies and contextual relationships from MRI images.
Methods
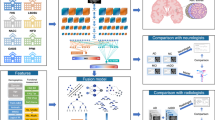
We used ViT to derive T1-weighted MRI slice attributes and a transformer neural network model for sequential feature classification, maintaining inter-association between the slices. The transformer neural network architecture and the ViT architecture for the sequential feature classification model are explained in Supplementary Sections 1 and 1.1. The summary of the ADNI dataset and steps of the proposed method are described in Sections 2.1–2.4, and the pipeline of the proposed method’s architecture is shown in Fig. 1.
The proposed pipeline of the ViT-TST. MRI images were pre-processed using CAT12 (image registration to standardize the images and skull stripping to reduce biases by ensuring consistent voxel intensities). ViT was used from each plane to derive slice attributes. Finally, the time series transformer was used to classify the feature sequences.
Dataset
Data used in this study was obtained from the Alzheimer’s disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public–private partnership led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD).
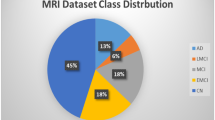
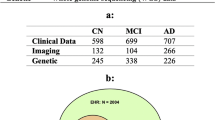
We performed binary and multiclass classification using T1-weighted 3D MRI scans from ADNI25,26. We trained and tested the models on subjects who had scans taken at screening and at 6- and 12-month visits (ADNI1: Complete 1Yr 1.5T data) and on subjects who had scans taken at screening, and at 6 months, 1 year, and 18 months (MCI only), and 2 and 3 years (normal and MCI only) (ADNI1: Complete 3Yr 3T data). We tested the model performance in three types of MRI scans (from the top down, axial plane; from front to back, coronal plane; and side to side, sagittal plane). We randomly split each data set into 60% training, 20% testing, and 20% validation sets. We performed extensive experiments on binary (NC/AD) and multi-classification tasks (NC/MCI/AD) to assess the proposed method. The details of model variants are listed in Supplementary Table S1. The configuration of training parameters is summarized in Supplementary Table S2. Both binary and multi-classifications were performed for all sagittal, coronal, and axial planes. We also implemented different baseline architectures to make comparisons with the proposed method. The details of baseline model variants are listed in Supplementary Table S3. The Complete 1Yr 1.5T data results are provided in Supplementary Section 3. The descriptive statistics of the ADNI data are provided in Table 1.
MRI Pre-processing
T1-weighted MRI scans were standardized in Montreal Neurological Institute (MNI) space. For comparison across subjects each skull was stripped using Statistical Parametric Mapping 12 (SPM12)27 and Computational Anatomy Toolbox (CAT12; http://www.neuro.uni-jena.de/cat/) in MATLAB (see Fig. 2).
Handling 3D MRI using 2D ViT
ViT models are pre-trained using a vast number of 2D data (mageNet21K28 and 21,843 classes at a resolution of \(224\times 224\) pixels. By taking advantage of transfer learning with a pre-trained 2D network model, we split the standardized 3D MRI into 2D slices. In the 2D slices, the sizes of each slice in the axial, coronal, and sagittal planes were \(113\times 137\), \(113\times 113\), and \(137\times 113\) pixels, respectively.
Each slice was then converted to an image of \(224\times 224\) pixels, and each slice was divided into \(14\times 14\) patches where each patch was \(16\times 16\) pixels (see Fig. 3).
These patches, which were flattened into a vector as a sequence of 196 patches, were considered to be an input token for the model, called the self-attention model. Later, we applied a multiple-layer self-attention model and feed-forward neural network to process the sequence of patched pixels and perform the feature selection. The proposed model comprised multiple transformer blocks, each applying the multi-head attention layer as a self-attention mechanism to the patch sequence. Finally, the output of the transformer encoder was processed via a classifier head to produce the final class probability output. Figure 4 provides a visual summary of the proposed model.
Sequence classification using a time series transformer
After features extraction using ViT, a transformer model24 was used as a tensor for time-series classification to maintain the relationships between the slices. Due to the self-attention mechanisms of transformer-based time series classifications, long-term dependencies between time steps (features of each slice) were captured more effectively. We then used feature embedding to map each token sequence to a meaningful numeric vector. Since we used ViT to map the slices of each MRI to a sequence of dimensional numerical features, this transformer did not need an embedding module. As each slice provides dimensional numerical features, the MRI classification problem was changed to a multi-dimensional time series classification. The summary of the model is illustrated in Fig. 5.
By utilizing transfer learning with the ViT, we adeptly address the challenges posed by the voluminous nature of 3D MRI datasets. Splitting the 3D MRIs into 2D slices allows for the application of pre-trained models, which are predominantly designed and optimized for 2D image data. This strategy not only circumvents the need for large 3D medical imaging datasets but also uses the power of models trained on extensive 2D image datasets.
Furthermore, our method extends the benefits of transfer learning beyond mere feature extraction from individual slices. By applying transfer learning to the time series classification of these extracted features, we innovatively create a two-tiered application of transfer learning. This approach capitalizes on the inherent temporal information within MRI scans, treating the progression of slices as a sequence that can provide valuable insights into the underlying medical conditions.
This dual application of transfer learning—first, to the feature extraction from 2D slices, and second, to the temporal analysis of these features—ensures a comprehensive utilization of available data. It effectively applies deep learning advances to medical imaging issues, specifically managing 3D MRI data with limited resources and data privacy concerns.
Results
The analysis was performed in two main steps: feature extraction and sequence modeling tasks on ADNI1: Complete 3Yr 3T and ADNI1: Complete 1Yr 1.5T MRI data. First, we compared our classification result with CNN alongside Bi-LSTM, CNN alongside Transformer, and ViT alongside Bi-LSTM. Then, we compared the classification performance, model accuracy, precision, F-score, and recall. The results for ADNI1: Complete 1Yr 1.5T are shown in the Supplement Section 2.1.
Experiments on ADNI1: complete 3Yr 3T
Binary classification
The results from the four architectures were similar, with CNN-TST and ViT-TST achieving the highest accuracy and precision scores of 98.81% and 0.99, respectively. Meanwhile, CNN-Bi-LSTM and ViT-Bi-LSTM had slightly lower scores but still performed well, with accuracy scores of 97.14% and 97.38%, respectively. To measure the ability of our proposed model to capture the positive instances, we calculated the recall score, and to measure the balance between the precision and the recall, we calculated the F-Score. The F-scores and recall scores for all architectures were very similar, ranging from 0.97 to 0.99, indicating good performance in these metrics (see Table 2).
The architectures CNN-Bi-LSTM and ViT-TST had the highest accuracy, at 98.333%, while CNN-TST had the lowest accuracy, at 97.381%. Based on precision, F-score, and recall, the CNN-Bi-LSTM and ViT-TST architectures achieved the highest performance, with precision and recall scores of 0.98 and an F-score of 0.98. The CNN-TST also had a strong performance, with a precision and recall of 0.98 and an F-score of 0.97. The ViT-Bi-LSTM model had the lowest precision score, 0.97, but still achieved a strong F-score and recall score of 0.98 and 0.97, respectively.
The table shows the performance of four different deep-learning architectures on a particular task. The ViT-TST architecture performed the best among all the models, achieving the highest accuracy of 99.048% and the highest scores for precision, F-score, and recall. The ViT-Bi-LSTM architecture also performed well, achieving an accuracy of 98.571% and competitive precision, F-score, and recall scores. The CNN-TST architecture attained an accuracy of 98.333% and high precision, F-score, and recall scores. Overall, the ViT-TST and ViT-Bi-LSTM architectures were better suited for the given task, followed by CNN-TST. The CNN-Bi-LSTM architecture was the least effective, although the differences were minor.
The results on sagittal, coronal, and axial planes for binary classification (NC and AD) are listed in Table 2.
To calculate the performance of the proposed model for binary classification, we calculated the confusion matrix based on the proposed method and compared it with the other methods. The performance of the proposed method for binary disease classification using different MRI planes is shown in Fig. 6. The diagonal cells indicate the prediction performance of the models when used to identify the true positive cases. ViT with time series transformer performed better than the other models for all the planes.
Multiclass classification
ViT-TST architecture achieved the highest accuracy and precision scores for multiclass classification based on sagittal planes. The CNN-Bi-LSTM model achieved an accuracy of 98.028%, with a precision of 0.98, an F-score of 0.98, and a recall of 0.98. The ViT-TST model achieved an accuracy of 98.310%, with a precision of 0.98, an F-score of 0.98, and a recall of 0.98. All four architectures had the best precision score of 0.98, with the ViT-TST architecture achieving the highest accuracy and precision scores (see Table 3).
For the coronal plane, CNN-Bi-LSTM achieved an accuracy of 96.479%, with a precision of 0.96, an F-score of 0.96, and a recall of 0.96. ViT-TST achieved the highest accuracy, 99.014%, with a precision of 0.99, an F-score of 0.99, and a recall of 0.99. Overall, the CNN-Bi-LSTM architecture had lower scores than the other architectures, especially for accuracy.
For the axial plane, the ViT-Bi-LSTM and CNN-TST architectures also performed well, with accuracy, precision, F-score, and recall values above 0.98. The CNN-Bi-LSTM architecture had the lowest precision and F-score values but achieved high accuracy and recall value. These results suggest that the ViT-TST architecture is the most efficient for this task.
The results on sagittal, coronal, and axial planes for multiclass classification (NC, MCI, and AD) are listed in Table 3.
The ViT-TST algorithm excelled in multiclass classification, demonstrating superiority in true positive (TP), true negative (TN), false positive (FP), and false negative (FN). It consistently demonstrated exceptional performance in accurately classifying instances belonging to various classes, with high TP and TN values across all classes. This demonstrated its ability to accurately identify positive instances and differentiate them from negatives. Additionally, the algorithm minimized false negatives (FN), reducing the risk of misclassifying positive occurrences as negatives, crucial in medical diagnostic applications such as AD classification. The ViT-TST also excelled in managing false positives (FP), ensuring that instances from other classes were correctly identified as non-relevant. Overall, the superiority of ViT-TST in TP, TN, FN, and FP further solidified its position as a highly effective and reliable choice for multiclass disease classification tasks.
The performance of the proposed method based on a confusion matrix for multiclass classification on different planes is shown in Fig. 7.
We also compared our proposed model with conventional deep models. Tables 4 and 5 show numerical results from different deep learning model for the binary and multiclassification tasks on ADNI datasets and compared with our proposed model. For binary classification, the proposed method outperformed other traditional models. Moreover, as shown in Table 5, the proposed method ranked first among all competitors.
The results on sagittal, coronal, and axial planes for multiclass disease patient classification (NC, MCI, and AD) are shown in Table 5.
The ADNI1: Complete 1Yr 1.5T results are shown in Supplement Section 2.2.1.
Discussion
In this study, we proposed use of a vision transformer with sequential transformer architecture for binary and multiclass MRI classification. We also compared the performance of the proposed model with other traditional image analysis architectures such as convolutional neural networks and long short-term memory networks on medium and large-sized ADNI datasets.
Reducing false negatives (FN) is crucial in medical diagnosis and healthcare, since models can inaccurately categorize positive occurrences as negative, causing failure to identify or detect cases. In AD categorization, false negative outcomes may misclassify individuals as being in good health, preventing timely identification and administration of appropriate medical interventions. The ViT-TST algorithm appears to be the best choice for binary AD classification. It provides a balanced combination of accuracy and sensitivity, which is crucial for medical applications where both false positives and false negatives can have significant implications. Since minimizing FN is a top priority in AD classification, ViT-TST would be the most suitable choice.
Our proposed method, the ViT-TST, outperformed the ViT-Bi-LSTM, CNN-TST, and CNN-Bi-LSTM models when classifying MCI patients. The ViT-TST achieved higher accuracy, sensitivity, or recall, resulting in an increase in the number of TP predictions and a decrease in the number of FN. This suggests that the ViT-TST accurately identified more MCI patients, reducing the chances that they would be misclassified as healthy or as belonging to other classes. The viability of the ViT-TST for multiclass disease classification tasks, particularly in the diagnosis of AD, is supported by its promising results.With the approval of new drugs for early intervention in AD7,8, early detection of AD and differentiation of MCI have become critical. Classifying MCI vs. NC or AD in a multiclassification framework is challenging because it is a heterogeneous condition with many subtypes and causes. Many studies have overlooked this problem and have experimented only with binary classification. However, our experiments were performed on binary (NC/AD) and multiclass disease classification tasks (NC/MCI/AD), and the performance of each model was evaluated using accuracy, precision, F-score, and recall.
When averaging the accuracy rates over all planes for binary disease classification on the ADNI1: Complete 3Yr 3T data, ViT-TST achieved the highest average accuracy of 98.65% (see Table 2), followed closely by CNN-TST with 98.17%. ViT-Bi-LSTM and CNN-Bi-LSTM also performed well, with average accuracies of 97.85% and 97.22%, respectively (see Table 2). These results suggest that all four architectures are effective for binary disease patient classification on this dataset, with CNN-Bi-LSTM being the most effective.
When averaging the accuracy rates over all planes for multiclass disease classification on the ADNI1: Complete 3Yr 3T, ViT-TST achieved the highest average accuracy of 98.77%, followed by ViT-Bi-LSTM with 97.74%. CNN-TST and CNN-Bi-LSTM also performed well, with average accuracies of 97.64% and 97.08%, respectively (see Table 3). These results suggest that all four architectures are effective for multiclass classification on this dataset, with ViT-TST being the most effective.
Evaluation of all the results obtained revealed the ViT-TST architecture to be consistently among the top-performing architectures across all datasets and classification tasks. Therefore, the ViT-TST architecture may be a good choice when designing a classification model for similar datasets and tasks if computational resources and other practical considerations are allowed.
Thus, we have devised a way to classify AD based on deep learning by combining pre-trained ViT and time series transformers. Many tasks have limited data, making training a model from scratch difficult. In small or unbalanced target datasets, transfer learning lets the model learn from a large dataset and adapt to the smaller target dataset. Transfer learning in ViT improves generalization, training speed, and adaptability to datasets and tasks; the proposed method generalizes well when trained on insufficient amounts of data. To take advantage of transfer learning with a pre-trained ViT, the 3D data from a plane were split into a 2D slice array. The problem with slice-based approaches such as CNN is that they fail to retain the dependencies of associated features between slices. To overcome this problem, the sequence classification task uses a time series classification with a transformer. Another alternative is to use 3D deep models; however, transfer learning with a pre-trained 3D is not currently available. Therefore, these approaches do not generalize well when trained on insufficient data.
Conclusions
Overall, our results show that all four architectures achieved high levels of accuracy and performance on binary and multiclass disease classification tasks. Based on accuracy scores, all models performed well. However, we conclude that the Vi-TST and ViT-Bi-LSTM models perform better than the CNN-TST and CNN-Bi-LSTM models in terms of long-term dependencies among the spatial and temporal patterns in dynamic MRI sequences. In addition, they capture the global context using the self-attention mechanism to ensure that the relevant information from the entire or sequence of images is considered during classification, which reduces overfitting. Transfer learning with the ViT can efficiently handle large 3D MRI datasets by splitting them into 2D slices and applying pre-trained models. This approach not only minimizes the need for large datasets but also utilizes models trained on extensive 2D image datasets. The method also extends transfer learning beyond feature extraction to time series classification, providing valuable insights into underlying medical conditions. We gain deeper understanding of the underlying medical conditions by leveraging the attention mechanism in both feature extraction and classification, demonstrating a sophisticated combination of advanced AI techniques for medical image analysis.
Data availability
No datasets were generated or analysed during the current study.
References
Hazarika, R. A., Kandar, D. & Maji, A. K. An experimental analysis of different deep learning based models for Alzheimer’s disease classification using brain magnetic resonance images. J. King Saud Univ. Comput. Inf. Sci. 34(10), 8576–8598. https://doi.org/10.1016/J.JKSUCI.2021.09.003 (2022).
Jain, R., Jain, N., Aggarwal, A. & Hemanth, D. J. Convolutional neural network based Alzheimer’s disease classification from magnetic resonance brain images. Cogn. Syst. Res. 57, 147–159. https://doi.org/10.1016/J.COGSYS.2018.12.015 (2019).
Blennow, K., Zetterberg, H. & Fagan, A. M. Fluid biomarkers in Alzheimer disease. Cold Spring Harbor Perspect. Med. 2(2012), a006221. https://doi.org/10.1101/cshperspect.a006221 (2012).
Khojaste-Sarakhsi, M., Haghighi, S. S., Ghomi, S. M. T. F. & Marchiori, E. Deep learning for Alzheimer’s disease diagnosis: A survey. Artif Intell Med 130, 102332. https://doi.org/10.1016/J.ARTMED.2022.102332 (2022).
Alzheimer’s Association. 2019 Alzheimer’s Disease Facts and figures (Wiley Online Library, 2012).
Alberdi, A., Aztiria, A. & Basarab, A. On the early diagnosis of Alzheimer’s disease from multimodal signals: A survey. Artif. Intell. Med. 71, 1–29. https://doi.org/10.1016/J.ARTMED.2016.06.003 (2016).
McDade, E. et al. Lecanemab in patients with early Alzheimer’s disease: Detailed results on biomarker, cognitive, and clinical effects from the randomized and open-label extension of the phase 2 proof-of-concept study. Alzheimers Res. Ther. 14(1), 191. https://doi.org/10.1186/S13195-022-01124-2 (2022).
Sims, J. R. et al. Donanemab in early symptomatic Alzheimer disease: The TRAILBLAZER-ALZ 2 randomized clinical trial. JAMA https://doi.org/10.1001/JAMA.2023.13239 (2023).
Loddo, A., Buttau, S. & Di Ruberto, C. Deep learning based pipelines for Alzheimer’s disease diagnosis: A comparative study and a novel deep-ensemble method. Comput. Biol. Med. 141, 105032. https://doi.org/10.1016/J.COMPBIOMED.2021.105032 (2022).
Zhao, B., Lu, H., Chen, S., Liu, J. & Wu, D. Convolutional neural networks for time series classification. J. Syst. Eng. Electron. 28(1), 162–169. https://doi.org/10.21629/JSEE.2017.01.18 (2017).
Wen, Q. et al. Transformers in time series: A survey (2022). https://doi.org/10.48550/arxiv.2202.07125.
Yue, L. et al. Hierarchical feature extraction for early Alzheimer’s disease diagnosis. IEEE Access 7, 93752–93760. https://doi.org/10.1109/ACCESS.2019.2926288 (2019).
Silva, I. R. R., Silva, G. S. L., de Souza, R. G., dos Santos, W. P. & de Fagundes, R. A. A. Model based on deep feature extraction for diagnosis of Alzheimer’s disease. In Proceedings of the International Joint Conference on Neural Networks Vol. 2019 (2019). https://doi.org/10.1109/IJCNN.2019.8852138.
Zhang, F. et al. Multi-modal deep learning model for auxiliary diagnosis of Alzheimer’s disease. Neurocomputing 361, 185–195. https://doi.org/10.1016/J.NEUCOM.2019.04.093 (2019).
Jang, J. & Hwang, D. M3T: Three-dimensional medical image classifier using multi-plane and multi-slice transformer. 20718–20729 (2022).
Gunawardena, K. A. N. N. P., Rajapakse, R. N. & Kodikara, N. D. Applying convolutional neural networks for pre-detection of Alzheimer’s disease from structural MRI data. In 2017 24th International Conference on Mechatronics and Machine Vision in Practice, M2VIP 2017 vol. 2017 1–7 (2017). https://doi.org/10.1109/M2VIP.2017.8211486.
Choi, H. & Jin, K. H. Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav. Brain Res. 344, 103–109. https://doi.org/10.1016/J.BBR.2018.02.017 (2018).
Esmaeilzadeh, S., Belivanis, D. I., Pohl, K. M., Adeli, E. End-to-end Alzheimer’s disease diagnosis and biomarker identification. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 11046 LNCS 337–345 (2018). https://doi.org/10.1007/978-3-030-00919-9_39/COVER.
Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale (accessed 28 March 2023); https://github.com/
Kim, J., Shim, K., Kim, J. & Shim, B. Vision transformer-based feature extraction for generalized zero-shot learning (2023). arXiv:2302.00875.
Lyu, Y., Yu, X., Zhu, D. & Zhang, L. Classification of Alzheimer’s disease via vision transformer: Classification of Alzheimer’s disease via vision transformer. In ACM International Conference Proceeding Series 463–468 (2022). https://doi.org/10.1145/3529190.3534754.
Kushol, R., Masoumzadeh, A., Huo, D., Kalra, S. & Yang, Y. H. Addformer: Alzheimer’s disease detection from structural MRI using fusion transformer. In Proceedings - International Symposium on Biomedical Imaging vol. 2022 (2022). https://doi.org/10.1109/ISBI52829.2022.9761421.
Li, J. et al. Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives. Med. Image Anal. 85, 102762. https://doi.org/10.1016/J.MEDIA.2023.102762 (2023).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 5999–6009 (2017).
“ADNI | Alzheimer’s Disease Neuroimaging Initiative (accessed 03 April 2023). https://adni.loni.usc.edu/
Jack, C. R. et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27(4), 685–691. https://doi.org/10.1002/JMRI.21049 (2008).
Ashburner, J. & Friston, K. J. Unified segmentation. Neuroimage 26(3), 839–851. https://doi.org/10.1016/J.NEUROIMAGE.2005.02.018 (2005).
Ridnik, T., Ben-Baruch, E., Noy, A. & Zelnik-Manor, L. ImageNet-21K pretraining for the masses. 2021 (accessed 05 April 2023). arXiv:2104.10972v4
Lian, C., Liu, M., Zhang, J. & Shen, D. Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI. IEEE Trans. Pattern Anal. Mach. Intell. 42(4), 880–893. https://doi.org/10.1109/TPAMI.2018.2889096 (2020).
Liu, M. et al. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. Neuroimage 208, 116459. https://doi.org/10.1016/J.NEUROIMAGE.2019.116459 (2020).
Li, J. et al. 3-D CNN-Based multichannel contrastive learning for Alzheimer’s disease automatic diagnosis. IEEE Trans. Instrum. Meas. 71, 1–11. https://doi.org/10.1109/TIM.2022.3162265 (2022).
Zhu, J. et al. Efficient self-attention mechanism and structural distilling model for Alzheimer’s disease diagnosis. Comput. Biol. Med. 147, 105737. https://doi.org/10.1016/J.COMPBIOMED.2022.105737 (2022).
Karasawa, H., Liu, C. L. & Ohwada, H. Deep 3D convolutional neural network architectures for Alzheimer’s disease diagnosis. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) vol. 10751 LNAI, 287–296 (2018). https://doi.org/10.1007/978-3-319-75417-8_27.
Basaia, S., Agosta, F., Wagner, L., Canu, E., et al. Automated classification of Alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks. Elsevier (accessed 29 March 2023). https://www.sciencedirect.com/science/article/pii/S2213158218303930
Lin, W. et al. Convolutional neural networks-based MRI image analysis for the Alzheimer’s disease prediction from mild cognitive impairment. Front. Neurosci. 12, 777. https://doi.org/10.3389/FNINS.2018.00777/BIBTEX (2018).
Billones, C. D., Demetria, O. J. L. D., Hostallero, D. E. D., Naval, P. C. DemNet: A convolutional neural network for the detection of Alzheimer’s disease and mild cognitive impairment. In IEEE Region 10 Annual International Conference, Proceedings/TENCON 3724–3727 (2017). https://doi.org/10.1109/TENCON.2016.7848755.
Hosseini-Asl, E., Keynton, R. & El-Baz, A. Alzheimer’s disease diagnostics by adaptation of 3D convolutional network. 2016 (accessed 29 2023 March). https://ieeexplore.ieee.org/abstract/document/7532332/?casa_token=Neb5n7ikTZMAAAAA:PkEGLJT7qw9U49OS9KRibb0AFV1ImpMxt_SViSMvquUaRK5BjceVLgbe3YznJAG0Tw20np3KSMNT0Pk
Valliani, A. & Soni, A. Deep residual nets for improved Alzheimer’s diagnosis. In: Proceeding of the 8th ACM International Conference, and Undefined 2017 615 (2017). https://doi.org/10.1145/3107411.3108224.
Gunawardena, K., Rajapakse, R. N., Kodikara, N. D. Applying convolutional neural networks for pre-detection of Alzheimer’s disease from structural MRI data. 2017 (accessed 29 March 2023). https://ieeexplore.ieee.org/abstract/document/8211486/?casa_token=0Vm5OBjwvlYAAAAA:PKVbNMAMIsDz-HLaNNWN_khu_UcFL6wO3FEtTxOxVE5tuCet49yXSNB-smS9lU5C5knfNN1GTZT9RWs
Vu, T. D., Ho, N. H., Yang, H. J., Kim, J. & Song, H. C. Non-white matter tissue extraction and deep convolutional neural network for Alzheimer’s disease detection. Soft Comput. 22(20), 6825–6833. https://doi.org/10.1007/S00500-018-3421-5 (2018).
Jain, R., Jain, N., Aggarwal, A. & Hemanth, D. J. Convolutional neural network based Alzheimer’s disease classification from magnetic resonance brain images. Elsevier 2019 (accessed 29 March 2023). https://www.sciencedirect.com/science/article/pii/S1389041718309562
Wang, H. et al. Ensemble of 3D densely connected convolutional network for diagnosis of mild cognitive impairment and Alzheimer’s disease. Neurocomputing 333, 145–156. https://doi.org/10.1016/J.NEUCOM.2018.12.018 (2019).
Goenka, N. & Tiwari, S. AlzVNet: A volumetric convolutional neural network for multiclass classification of Alzheimer’s disease through multiple neuroimaging computational approaches. Biomed. Signal Process. Control 74, 103500. https://doi.org/10.1016/J.BSPC.2022.103500 (2022).
Odusami, M., Maskeliūnas, R. & Damaševičius, R. An intelligent system for early recognition of Alzheimer’s disease using neuroimaging. Sensors 22(3), 740. https://doi.org/10.3390/S22030740 (2022).
Acknowledgements
Data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the National Institutes of Health (Grant U01 AG024904), and the DOD ADNI Department of Defense (award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Co.; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Co.; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development, LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research provides funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. SAC is a Muslow, MD Endowed Chair in Healthcare Informatics.
Replication of results
The codes and data used are available on request to enable the method proposed in the manuscript to be replicated by readers.
Funding
Three National Institutes of Health grants supported this work: R01HL145753, R01HL145753-01S1, and R01HL145753-03S1; in addition, the work was supported by LSUHSC-S CCDS Finish Line Award, COVID-19 Research Award, and LARC Research Award to MSB; and Institutional Development Award (IDeA) from the National Institutes of General Medical Sciences of the NIH under grant number P20GM121307 and R01HL149264 to CGK.
Author information
Authors and Affiliations
Contributions
M.A.N.B. conceptualized and designed the study; S.A. and T.A. took part in data accumulation and data analysis; M.A.N.B., S.A., T.A., M.S.B., E.D., S.C., J.V., and C.G.K. wrote the manuscript; and all the authors have read, edited, and approved the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alp, S., Akan, T., Bhuiyan, M.S. et al. Joint transformer architecture in brain 3D MRI classification: its application in Alzheimer’s disease classification. Sci Rep 14, 8996 (2024). https://doi.org/10.1038/s41598-024-59578-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-59578-3
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.