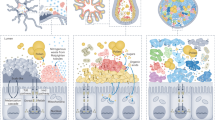
Abstract
The health of honey bee queens is crucial for colony success, particularly during stressful periods like overwintering. To accompany a previous longitudinal study of colony and worker health, we explored niche-specific gut microbiota, host gene expression, and pathogen prevalence in honey bee queens overwintering in a warm southern climate. We found differential gene expression and bacterial abundance with respect to various pathogens throughout the season. Biologically older queens had larger microbiotas, particularly enriched in Bombella and Bifidobacterium. Both Deformed Wing Virus A and B subtypes were highest in the fat body tissue in January, correlating with colony Varroa levels, and Deformed Wing Virus titers in workers. High viral titers in queens were associated with decreased vitellogenin expression, suggesting a potential trade-off between immune function and reproductive capacity. Additionally, we found a complex and dynamic relationship between these viral loads and immune gene expression, indicating a possible breakdown in the coordinated immune response as the season progressed. Our study also revealed a potential link between Nosema and Melissococcus plutonius infections in queens, demonstrating that seasonal opportunism is not confined to just workers. Overall, our findings highlight the intricate interplay between pathogens, metabolic state, and immune response in honey bee queens. Combined with worker and colony-level metrics from the same colonies, our findings illustrate the social aspect of queen health and resilience over the winter dearth.
Similar content being viewed by others
Introduction
The honey bee (Apis mellifera) is a domesticated insect valued for its contributions to honey production and pollination services1. An observed trend of the last decade has amateur and commercial beekeeping operations in North America and Europe losing 30–50% of their hives annually2. These losses are attributed to a complex interplay of biotic and abiotic factors, often involving various pathogens and environmental stressors3,4,5. Among the leading causes of such losses are queen failures. Historically queens survived 2–5 years, but more recently, queen losses and supersedures occur much earlier and more frequently6,7,8,9,10. Now it’s common practice by many beekeepers to prophylactically replace queens on an annual basis, as younger queens are generally considered healthier and more productive2,11,12. The health and vitality of a honey bee colony depends on the well-being of its queen, who, as the sole egg-laying individual in the hive, plays a critical role in determining the colony's overall success. Amidst the myriad of challenges that honey bee colonies face, the overwintering period stands out as a time of heightened vulnerability. Overwintering is a particularly stressful period for honey bee colonies, marked by reduced foraging opportunities and increased susceptibility to pathogens. In the US, colonies that overwinter in warm southern climates like Southern California, Arizona, Texas, and Florida deal with shorter and warmer winters. Colonies may struggle with unsustainable brood rearing, forage dearth, and spikes of extreme cold weather. The queen's health during this period is crucial, as it sets the stage for the colony's survival and productivity in the subsequent foraging season.
While the queen is generally protected from pathogens by several social immune mechanisms like grooming by her retinue of nurse bees, the secretion of antimicrobial substances in royal jelly13, and the isolation within the hive, she is not entirely immune to infections. Viruses that infect honey bees have evolved strategies to also infect queens horizontally through worker contact14 and vertically through mating15. Thus, most of the pathogens that infect workers can also be found in queens16,17,18,19, however most infections appear to be innocuous with no overt symptomology20. While it’s true that some pathogens have been linked with colony loss21,22,23,24, the direct effects on queen health are far less understood. Among the viruses that infect queens, Deformed Wing Virus (DWV) is the most widespread and well-known. In the absence of the varroa mite (Varroa destructor) DWV normally persists at low levels within a colony with no apparent detrimental effects. It can be found in all life stages and castes, including glandular secretions used to feed developing larvae and members of the hive14,16,25,26. When Varroa is abundant, there is a dramatic rise in DWV, which is aided in transmission by being directly injected into worker and drone hemolymph during feeding. While Varroa do not feed directly on the queen, high mite populations can cause high DWV viral titers in queen tissues. The combination of high viral titers and Varroa load is linked to immunosuppression in workers27, with the number of workers with overt symptomology (deformed/crippled wings) serving as a predictive marker for colony loss28. In extreme cases, newly emerged queens can present with the telltale sign of crippled wings29, but subclinical infections on queen health, fecundity, and overall colony dynamics warrants further investigation.
The gut microbiota plays a significant role in nutrient absorption and overall immune function in honey bees. Previous studies have shown that the composition of the queen gut microbiota can change with the age30,31, possibly affecting her health and susceptibility to disease. Like workers, the queen microbiota is highly structured and represented by few species which belong to the genera Bombella, Lactobacillus, Apilactobacillus, Commensalibacter (Alpha 2.1), Bombilactobacillus, and Bifidobacterium. Worker-centric genera like Snodgrassella, Gilliamella, and Frischella are sometimes found as either dominance environments (dysbiosis) or relatively low abundance which may indicate they do not serve essential functions in the queen and may even be detrimental. Other honey-bee associated bacteria like Bartonella and Melissococcus, the causative agent in the brood disease European Foulbrood have not been reported in queens.
Queen health and immunity is intrinsically linked with vitellogenin expression, a molecule that is constitutively expressed throughout the queens’ body, but mostly by the fat body tissue32,33. Vitellogenin is an egg-yolk protein, co-opted by honey bees to serve multiple purposes not seen in other animal systems such as immunity, oxidative stress, nutrition, and longevity33,34,35. The fat body in honey bees serves multiple essential functions, akin to the mammalian liver and adipose tissue. It is also a critical site for the production of antimicrobial peptides (AMPs) in response to pathogen associated molecular patterns (PAMPS) of bacterial and fungal origin. Understanding how gene expression in the fat body and other tissues change in response to pathogen pressure is vital for a comprehensive view of queen health. Among the inducible immune-effectors of the honey bee immune response are Peptidoglycan Recognition Proteins (PGRPs), which can recognize and bind to bacterial cell walls, thereby triggering immune responses. A single membrane-bound PGRP in bees (PGRP-LC) acts as a transmembrane receptor upstream of the IMD pathway and is activated by the DAP-type peptidoglycan of Gram-negative bacteria or certain Gram-positive bacteria36,37. There are also short PGRPs that recognize lysine-containing peptidoglycan, triggering a proteolytic cascade that activates the Toll pathway. Honey bees also have defenses against viruses through the a Jak/STAT homologue pathway and RNA interference (RNAi)38,39. Thus, honey bee immune response is capable of action against bacteria and viruses but can also respond to fungi like Varimorpha (Nosema) ceranae; a microsporidian midgut parasite and the leading cause of dysenteric colony losses worldwide.
The primary aim of this study is to explore variation in queen gut microbiota, host gene expression, and pathogen prevalence associated with overwintering in a warm southern climate within a large-scale commercial beekeeping operation. We seek to understand how factors at the worker and colony level interact, and what implications they may have for queen health. To this end, we expand on a longitudinal study performed in the Imperial Valley of California from September 2016 through January 2017. In previous work it was revealed that colony or worker level factors Nosema, Varroa, and DWV increased from early to late winter, while the same time period was characterized by decreased Vitellogenin (Vg) and increased AMP expression in workers40. Over the same time period, queens showed a sharp increase in mandibular pheromone compliment homovanillic alcohol (HVA) suggesting a shift in queen quality41. We suggest that changes in colony level metrics and worker gene expression form the foundation for queen health documented in this study. By collecting interconnected data points on workers, queens, and colonies over time, we seek to produce a comprehensive framework that can better predict changes in colony and queen health.
Methods
Experimental design
Queens were obtained from colonies under the management of Ashurst Bee Co. Inc. (Westmoreland, CA), following the methodology previously established by Ricigliano et al., 2018. Briefly, these colonies were distributed across three sites in Southern California: Site 1 (Best 115 E and W, coordinates 33° 01′ 38.8′ ′N 115° 31′ 07.8′′ W), Site 2 (Young and River, coordinates 33° 07′ 58.8′′ N 115° 34′ 05.3′′ W), and Site 3 (Jacobsen 111, coordinates 33° 04′ 30.5′′ N 115° 31′ 14.9′′ W). Each site hosted a balanced mixture of both young and mature Italian queens, all originating from the same commercial breeding source. Young queens were approximately 4 months old and older queens were approximately 16 months old. A carbonyl assay of fat body tissue was also used to confirm biological age. A subset of queens of each age were randomly sampled from each site across the span of September 2016, November 2016, and January 2017 resulting in 96 queen samples over the 5-month period.
Dissections, protein oxidation assay, and nucleic acid extractions
All 96 queens were collected into sterile 2.0-ml tubes and immediately frozen on dry ice and stored at − 80 °C for nucleic acid extraction. Queens were pinned through the thorax in 70% ethanol to wash and aid in dissection. Micro-dissection scissors were used to cut through the sides of the abdomen to access the digestive tract which was removed and floated in ethanol to separate the gut tissues. The abdominal fat body, along with the attached dorsal sclerites, were extracted as a single unit for use in gene expression and protein oxidation assays, aimed at quantifying biological aging.
The fat body, midgut, and ileum tissues were bead-beaten in 1× TE buffer for 2 min at 30-s intervals and centrifuged to recover the supernatant. The fat body supernatant fraction used in a protein oxidation assay quantifies the accumulation of protein carbonyl groups associated with oxidative stress and aging42. This well-defined assay, previously deployed in related studies, was applied in accordance with the methods described in Copeland et al.43. To extract nucleic acids (DNA and RNA simultaneously) we used Qiagen AllPrep PowerViral DNA/RNA Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol and methodology also reported in Copeland et al.43.
Using the DNA, we quantified total bacterial abundance for the midgut and ileum tissues using a quantitative PCR (qPCR) assay of the 16 rRNA gene44. We created a standard curve using a tenfold serial dilution series of a plasmid standard containing a full-length Escherichia coli 16S rRNA gene. We amplified a 466 bp fragment in the V3–V4 region of the 16S rRNA gene using universal primer pair (5′-CCTACGGGDGGCWGCA-3′ and 5′-GGACTA CHVGGGTMTCTAATC-3′). PCR reactions were performed in triplicate on a BioRad CFX96 (Biorad, Hercules, California, US) as follow: 12 μl reactions containing 9 μl of iTaq Universal SYBR Green Supermix (BioRad, Hercules, California, US), 0.5 μl forward primer,0.5 μl reverse primer, and 2 μl of DNA template. The cycling conditions were 95 °C for 3 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 60 s. The qPCR results were expressed as the total number of 16S rRNA gene copies per DNA extraction (200 μl volume elution). To characterize the microbial communities of the queen midgut and ileum, we amplified the V3–V4 region of the 16S rRNA gene using PCR primers (341F 5′-CCTACGGGNGGCWGCAG-3′; 805R 5′-GACTACHVGGGTATCTAATCC-3′). DNA library preparation was performed following Illumina MiSeq DNA library preparation protocol. Sequencing was performed at the University of Arizona Genetics Core (UAGC) on a MiSeq following the manufacturer’s guidelines. The sequence data for this study have been deposited in GenBank, Sequence Read Archive no. as PRJNA1061232.
Immune gene expression and quantification of pathogen titers
To quantify mRNA levels, a cDNA template was generated from the purified RNA fraction. Briefly, RNA was converted into cDNA with Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, Massachusetts, United States) following manufacturer’s instructions. PCR reactions were performed using gene-specific primers and the following protocol: initial denaturation at 95 °C for 5 min; 40 cycles with denaturation at 95 °C for 15 s; and a combined annealing and extension step at 58 °C for 30 s. The reactions were carried out using iTaqTM Universal SYBR® Green Supermix (Biorad, Hercules, California, US) in triplicate on an CFX96TM Real-Time PCR Detection System (Biorad, Hercules, California, US).
RNA fractions were used to create cDNA and examine gene expression, Nosema intensity, virus abundance, and viral titers qPCR. To confirm the absence of contaminant DNA and primer dimers, we used no-template controls made of water and analyzed melt-curves for each qPCR plate. Relative gene expression was calculated using the 2–∆∆Ct method45 using both β-actin and RPS18 as reference genes46. Genes were chosen to cover a range of processes associated with innate immunity including; B-Gluc2, DOPA Carboxylate, abaecin, apidaecin, defensin1, defensin2, hopscotch, hymenoptaecin, IRP30, lysozyme1, lysozyme2, lysozyme3, PGRP-LC, PGRP-S1, PGRP-S2, PGRP-S3, PPO, vitellogenin, AmEater, argonaut2, and dicer (Table S1). After normalization, we log-transformed expression levels to approximate normality and tested for differences by month and site using the parametric test, ANOVA with Tukey’s HSD in JMP 14. We also pooled cDNA to quickly screen for honey bee viral pathogens: Acute Bee Paralysis Virus (ABPV), Black Queen Cell Virus (BQCV), Chronic Bee Paralysis Virus (CBPV), Deformed Wing Virus (DWV-A and DWV-B), Israeli Acute Paralysis Virus (IAPV), Kashmir Bee Virus (KBV), Lake Sinai Virus (LSV), Sacbrood Virus (SBV), and Slow Bee Paralysis Virus (SBPV).
Virus titer standards were generated from PCR products that were purified using the GeneJET PCR Purification Kit (Thermo Scientific, Waltham, MA), ligated using pGEM®-T Easy Vector Systems (Promega, Madison, WI) to yield plasmid vectors, which were then transformed into DH5α™ Competent Cells. Transformed colonies were selected and grown overnight in broth. Plasmids were isolated using the GeneJET Plasmid Miniprep Kit (Thermo Scientific, Waltham, MA) and quantified using an Implen nanophotometer P300. The known mass of the plasmid plus PCR insert was used to calculate DWV plasmid-standard copies per µL. Primers for immune genes and pathogens can be found in Table S126,47,48,49,50.
16S rRNA gene community analysis
16S rRNA gene sequence data were processed using MOTHUR v.1.44.34351 according to previously published protocols43. Briefly, paired end reads were joined using the make.contigs command. We used the SED command in UNIX to remove the first and last five nucleotides from each sequence. Sequences were then screened to remove ambiguous bases, using the screen.seqs command. Unique sequences were generated using the unique.seqs command. A count file containing group information was generated using the count.seqs command. Sequences were aligned to the BEExact database52 using the align.seqs command. Sequences were filtered to remove overhangs at both ends and gaps using filter.seqs. The unique.seqs command was ran again to remove new redundancies from filtering. A precluster step using pre.cluster was performed followed by the chimera.uchime command53 to remove chimeric sequences. Sequences not of bacterial origin (fungi, archaea, mitochondria, and chloroplasts) were removed using the remove.seqs command. All unique sequences with one or two members (single/doubletons) were removed using the AWK command in UNIX. A distance matrix was constructed for the aligned sequences using the dist.seqs command. Sequences were classified at the unique level with the BEExact database using classify.seqs command. Unique sequences were then merged at the species-level with the merge.otus command.
We curated the data to remove sources of contamination associated with low abundance DNA environments54. Queen libraries, especially the midgut may contain very little DNA and contamination skews results. Following criteria previously established55, we used three exclusion criteria to identify contaminant OTUs: (1) strong negative Pearson’s correlations with read number and microbiome size based on 16S rRNA sequences (BactQuant), (2) intercorrelations with other known or suspected contaminants, and (3) association with contamination from other works54. Additionally, we corroborated our contamination with kit controls and the R program decontam56.
Statistical analyses
The 13 most abundant operational taxonomic units (OTUs) and a sum of remaining OTUs were normalized by qPCR BactQuant absolute abundances by first calculating the proportion of each OTU by dividing the raw read count into the total number of sequences per sample. Each ratio was multiplied by the total BactQuant 16S rRNA gene copies qPCR for each sample. Next, each OTU was corrected for 16S gene copies per bacterial cell; 16S rRNA gene copy number were assigned based on the exact match or closest taxonomic representative57. OTUs without a close taxonomic representative in the database was assigned 4.2 gene copies, the mean 16S rRNA gene copy number58. Next, the data were CLR-transformed using the software CoDaPack59.
A two-way multivariate analysis of variance (MANOVA) was performed on CLR-transformed data with OTUs 1–15 as dependent variables. The MANOVA examined sampling month and site as independent variables, and post hoc pairwise analyses were conducted using Tukey honestly significant difference (HSD) test. Normalized absolute abundance data, gene expression, and viral titers were also analyzed using ANOVA with Tukey HSD by both sampling month and site. When appropriate, a false discovery rate (FDR) was employed to account for multiple comparisons. We examined carbonyl as a continuous variable that serves as a proxy for biological age. These values were used in a bivariate analysis against bacterial cell numbers and gene expression. These were analyzed with ANOVA and corrected with FDR for multiple comparisons.
We also used Pearson’s correlations as a statistical measure to quantify the strength and direction of a linear relationship between two continuous variables. In our case, we explore patterns between OTU abundance, gene expression, viral titers, and microbiome size. The coefficient ranges from − 1 to 1, with values closer to 1 indicating a strong positive linear relationship, values closer to − 1 indicating a strong negative linear relationship, and values around 0 suggesting little to no linear correlation. To further explore the distribution of these correlations, we categorized them into different levels of significance based on the value of the Pearson's coefficient R. For positive correlations, 'Strong Positive' was defined as R ≥ 0.8, 'Moderate Positive' as 0.8 > R ≥ 0.5, and 'Mild Positive' as 0.5 > R > 0. Similarly, for negative correlations, 'Strong Negative' was defined as R ≤ − 0.8, 'Moderate Negative' as − 0.8 < R ≤ − 0.5, and 'Mild Negative' as − 0.5 < R < 0. Correlations close to zero and not significant were categorized as 'Not Significant'. We visualized these data as stacked bar charts, tallying the number of correlations falling into each category for different tissues (midgut, ileum, and fat body) and across different months (September, November, and January). To statistically assess the variations in these distributions across months, we performed Chi-Square Tests for Independence. This allowed us to determine whether the distribution of significance levels was consistent across the different months for each tissue type. We calculated the expected frequencies for each cell in the contingency table and used these to compute the Chi-Square statistic and associated p-value. Cells contributing significantly to the Chi-Square statistic were further identified using standardized residuals, calculated as \(\frac{(Observed-Expected)}{\sqrt{Expected}}\). Standardized residuals greater than 1.96 or less than − 1.96 were considered to significantly contribute to the Chi-Square value, indicating that the observed frequency was significantly different from the expected frequency under the assumption of independence between months and significance levels.
Results
Sequence read curation and microbial community analysis
Next generation sequencing provided 9,532,084 raw reads (400 bp) across 192 libraries; half were queen midguts and the other queen ileums (Table S2). The BEExact classifier matched many sequences to the species-level52, however, most Bombella sequences were classified as Bombella_unclassified, while Bo_apis was sparse with only 33% of samples having any reads even though it was the 7th most abundant OTU. For downstream analyses we combined these into Bombella sp. After quality filtering and removal of laboratory contamination (Table S3), 8,002,199 reads were conserved accounting for 98.23% of sequences and 14 operational taxonomic units (OTUs) (Table S4): Bombella sp., Lactobacillus paniaspium, Commensalibacter sp. (Alpha 2.1), Lactobacillus apis, Snodgrassella alvi, Lactobacillus helsingborgensis, Frischella perrara, Apilactobacillus kunkeei, Bombilactobacillus mellis, Lactobacillus kimbladii, Lactobacillus sp., Bifidobacterium asteroides, Bombilactobacillus mellifer, and Lactobacillus melliventris. The remaining 144,429 reads and 1.77% of sequences were conserved and summed as the 15th OTU. The average read depth was 43,359 per library, (29,296 for midguts and 57,277 for ileums). The average size of the midgut based on 16S rRNA gene copies was 5.33E+04. Queen ileum tissues averaged 1.15E+06 16S rRNA gene copies. The size of the queen microbiota was driven largely by the top 4 OTU, Bombella sp., L. panisapium, Alpha 2.1, and L. apis which accounted for 84% of total sequences in the curated dataset.
Queen microbiota
Queen midgut tissues were dominated by Bombella sp. (Fig. 1) but differed significantly by month in the MANOVA model (Table S5). The MANOVA uses CLR-adjusted values to interpret significant differences in the relative abundance of bacteria strains. L. helsingborgensis, A. kunkeei, and B. mellifer were significant under the MANOVA model. Microbiota size, Bombella sp., L. apis, and B. asteroides all increased with biological age measured through fat body carbonyl contents (Table S6). Absolute abundance generally saw patterns of highest abundance in November for 6 of the 15 OTUs (Fig. 1). A. kunkeei was the only bacteria that decreased over the sampling period, a pattern also observed in the relative abundance MANOVA (Fig. 1, Table S5). Notably, five queen midguts harbored a relative abundance of Melissococcus plutonius at greater than 25%.
Relative and absolute abundance of midgut and ileum microbiotas. (A) The relative abundance of the honey bee queen midgut and ileum microbiotas by month. Color-coded bars represent relative abundance that has been corrected by species-specific 16S rRNA gene. (B) Absolute abundance statistical test with ANOVA Tukey HSD, ordered letters report. A is always highest abundance/expression. Levels not connected by same letter are significantly different. Gray = did not significantly differ.
In queen ileums we found a significant effect by month and site, but not as an interaction effect in the MANOVA model (Table S5). For month, the dependent variables that were significant were Bombella sp., A. kunkeei, and B. asteroides and despite site being significant in the overall model, after FDR correction no dependent variables met the significance threshold of p < 0.05. Microbiota size, Bombella sp., L. panisapium, and B. asteroides increased with biological age measured through fat body carbonyl contents (Table S6). The absolute abundance differences in the ileum generally matched the midgut, with most OTUs having the highest abundance in November, however 10 out of 15 OTUs differed significantly (Fig. 1). Interestingly, we also found that Bombella sp. absolute abundance increased through the sampling period with the highest levels in January.
Gene expression
We quantified gene expression in the midgut, ileum, and fat body (Fig. 2, Figure S1). We found differential gene expression in all tissues, but the largest differences were in the midgut and fat body. Starting with the midgut, nearly all genes peaked in expression levels during the month of November. This included genes argonaut2 and hopscotch which are involved in RNA interference and immune signaling, respectively. Antimicrobial peptides defensin1, hymenoptaecin, and prophenol oxidase (PPO) which is involved in the melanization pathway. Several lysozymes and peptidoglycan recognition proteins (PGRPs) were also upregulated in November, which are typically innate immune responses to the peptidoglycan layer found in bacterial cell walls. Pearson’s correlations of gene expression in the midgut were extremely positively intercorrelated (Fig. 2; Table S7). During November when gene expression levels are highest, the number of significant positive intercorrelations are severely suppressed in November and January. We used a Chi-Square Tests for Independence to determine if there is a significant association between two categorical variables: the month (MG Sep, MG Nov, MG Jan) and the number of significant Pearson's correlations (Strong Positive, Moderate Positive, etc.). The midgut had a high Chi-Square statistic (817.31, df = 12, p value < 0.0001) reinforcing the result of a strong departure from independence between the two variables (Table S8) with the number of positive correlations contributing most to the statistic. The remaining strong positive Pearson’s correlations (R > 0.8) in November are related to the genes Beta-glucanase2 and DOPA carboxylase. Not only are these genes highly intercorrelated (R = 0.8039, p value < 0.0001), they also both form strong positive correlations with Lysozyme1, Lysozyme3, PGRPS1, and PPO. By January there is only one strong correlation remaining: PRPLC and Lysozyme1 (R = 0.847 p value < 0.0001). The number of negative correlations also increased throughout the sampling period, for example by January there’s a moderate correlation between DOPA carboxylase and hymenoptaecin expression (R = − 0.6034, p value = 0.0002).
Gene expression and correlations. (A) Gene expression reported by month using ANOVA Tukey HSD, ordered letters report. A is always highest abundance/expression. Levels not connected by same letter are significantly different. Gray = did not significantly differ. (B) Stacked bar chart showing the number of statistically significant Pearson’s correlations by tissue and month.
Ileum gene expression was largely static between months outside of argonaut2, defensin1, hopscotch, and PGRPS1 which showed variable differential patterns (Fig. 2, Figure S1). Argonaut2 expression increased over the sampling months, with highest levels in January. Defensin1 peaked in November, which was like the expression pattern for the midgut. Hopscotch and PGRPS1 expression were static in September and November but dropped significantly in January. Despite low differential expression, the highest number of significant correlations was found in November (Fig. 2), while both September and January had low correlations matching the later months of the midgut (Table S7). The ileum also showed a substantial Chi-Square statistic (435.71, df = 12, p value < 0.0001), with the number of non-significant correlations between the months contributing the most to the statistic (Table S8). Ileums in November also had the most moderate negative correlations in the dataset, with some nearly meeting our threshold for strong negative correlation. These moderate negative correlations (R < − 0.7, p value < 0.0001) are with PGRPS3 and several viral signaling and immune genes: argonaut2, dicer, hopscotch, and Lysozyme3. PGRPS2R also had moderate negative correlations (R < − 0.5, p value < 0.005) with the same genes. Correlations in September and January are much sparser. In September, all of the strong positive correlations disappear, and dicer has moderate negative correlations (R < − 0.5, p value < 0.0011) with DOPA carboxylase, PGRPS1 and PGRPS2R.
In contrast to the midgut and ileum, the fat body showed more variable gene expression patterns, with the overall pattern showing decreased expression in November relative to September and January for six out of eight significant genes (Fig. 2, Figure S1). The six genes which were suppressed in September were DOPA carboxylase, an enzyme that catalyzes the decarboxylation of DOPA (3,4-dihydroxyphenylalanine) to dopamine, a neurotransmitter, hopscotch, lysozyme3, PGRPLC, and PPO. Beta-glucanase2, an enzyme that breaks down beta-glucans, complex sugars found in the cell walls of fungi, bacteria, and some plants decreased over the sampled season. Similar to the midgut, September had the most significant correlations which decreases over the sampled time period (Fig. 2, Table S9). The fat body also had a substantial Chi-Square statistic (508.85, df = 12, p value < 0.0001), with the changes in the number of positive correlations between the months contributing most to the statistic. Nearly all strong positive correlations disappear from September to November, except hymenoptaecin with PGRPS2R (R = 0.8383, p value < 0.0001) and PGRPS3 (R = 0.7734, p value < 0.0001) which seemingly persist through January. January also added strong positive correlations: argonaut2 with hymenoptaecin (R = 0.7831, p value < 0.0001) and PGRPS2 (R = 0.8571, p value < 0.0001).
DWV, Nosema, and European Foulbrood
Our initial screening for viruses yielded mostly negative results for Acute Bee Paralysis Virus (ABPV), Chronic Bee Paralysis Virus (CBPV), Israeli Acute Paralysis Virus (IAPV), Kashmir Bee Virus (KBV), Lake Sinai Virus (LSV), Sacbrood Virus (SBV), and Slow Bee Paralysis Virus (SBPV). We found variable levels of Black Queen Cell Virus (BQCV) just above our limit of detection, so we excluded this and all negative viruses from downstream analysis due to low prevalence and abundance. Deformed Wing Virus (DWV-A and DWV-B) was found in all tissues tested: midgut, ileum, and fat body (Fig. 3). Over the course of the sampling period, DWV-A increased by several orders of magnitude in all tissues. DWV-B matched DWV-A levels for September for both September and November, but in January increased by several orders of magnitude in all tissues. Individual viral titers were similar across all tissues, for example high titer samples were high across all three tissues. Carbonyl accumulation was not a factor in DWV titers.
We also checked Pearson’s correlations of DWV-A and -B with gene expression and total brood area. When DWV-A levels begin to rise in November, the midgut had the strongest immune response across all tested gene expression (Fig. 2, Table S11). Hymenoptaecin had a moderate positive correlation (R = 0.6692, p value < 0.0001) that becomes more potent in January (R = 0.7604, p value < 0.0001) along with RNAi-pathway gene argonaut2 (R = 0.7041, p value < 0.0001). The pattern with these genes exists in the fat body as well, but includes moderate positive correlations (R > 0.5, p value < 0.024) for the antimicrobial peptides apidaecin and defensin1 and PGRPs PGRPS2R and PGRPS3. Correlations with DWV-B during the spike in January are sparse with most only being mild correlations, (R < 0.5). There were mild negative correlations (R > 0.5, p value < 0.035) of DWV-A and -B with total brood when sampled and the mean total brood across the sampling period (Table S12).
Nosema intensity was measured in the midgut across the season (Fig. 4). Overall Nosema intensity was low and not different between the months, however there were five out of six samples with the highest Nosema burden that all came from Site 1 and were sampled in September and November. We examined gene expression and did not find any significant correlations between Nosema intensity and the expression levels of the immune-related genes we analyzed in these five samples. The microbiota of these five queens also had high (< 25%) relative abundance of Melissococcus plutonius, the causative agent in European Foulbrood. The sixth queen with higher Nosema burden also came from Site 1 in November but did not carry M. plutonius greater than 1% relative abundance.
Discussion
Queen health is a critical measure of a successful colony, especially during stressful events such as disease, dearth, or overwinter. However, it is too costly to sacrifice the queen, such that her health is typically inferred from the way workers treat the queen, egg laying capability, brood patterns, and other colony level metrics. We designed this experiment to reveal changes in the gut microbiota, host gene expression, and pathogen pressure associated with overwintering in a warm southern climate in a large-scale commercial beekeeping operation. Our results support the hypothesis that overwintering has a systemic effect, allowing queen health to be predicted based on worker characteristics, pathogen levels, and colony metrics.
Although queen age was not a factor in this study, we used oxidative damage accumulated in the fat body tissue as a proxy for chronological age. Each sampling month had an equal number of young and old queens, and overall differences in microbiota were minimal. When we regressed carbonyl accumulation against our top OTUs we found that biologically older queens had larger microbiotas and more Bombella sp. and Bifidobacterium in both the midgut and ileum tissues. Lactobacillus apis also increased with age in the midgut, while Lactobacillus panisapium increased with age in the ileum highlighting strain specificity and diversity within the tissues. The increase in abundance of Lactobacillus spp. and Bifidobacterium align with previously reported studies on queen microbiota aging30,31,60. Generally, the relative abundance of species rarely differed significantly between months. Apilactobacillus kunkeei is one standout from midguts which decreased over the sampling period. A. kunkeei is a fructophilic lactic acid bacteria that prefers fructose as its primary carbon source61. Its decrease over the sampling period could be indicative of changes in the dietary intake of the queens, a shift in the microbial community dynamics within the midgut, or a change in the queens metabolism as a result of a low sugar diet.
The absolute abundance of many OTU peaked in the ileum during November, and these tissues were dominated by top OTUs Bombella, L. panisapium, Alpha 2.1, and L. apis. The ileum appeared more stable in terms of relative abundance, but there were some dominance libraries of Snodgrassella alvi and Frischella perrara which are broadly considered worker bacteria. In workers, S. alvi interfaces with the ileum epithelial tissue, consumes available oxygen, and forms a biofilm with Gilliamella and L. apis62,63. S. alvi appears to be tolerated by queens to some degree, apparently replacing Alpha 2.130,31. F. perrara causes a scab formation in the worker pylorus64, but we did not detect any tissue abnormalities during dissection.
Queens are protected by several social immune mechanisms, such as the retinue of workers that groom and feed her with the diet of royal jelly. Despite the highly antimicrobial diet and the protection from mostly living in the hive, she is still vulnerable to pathogens through horizontal transmission17 and vertical transmission during mating nuptial flights14,15. Multiple virus infections in queens are common16, yet it’s unclear how these infections effect health, behavior, and overall physiology of queens as overt symptomology often remains elusive. Out of three tissues tested, the fat body had the highest DWV viral titers in January which correspond to hive Varroa levels and nurse DWV intensity from the same hives and workers associated with these queens40. DWV titers correlated between tissues, such that queens with high titers in the fat body were also high in the ileum and midgut suggesting systemic infection. In honey bees, the fat body provides many of features of the mammalian liver and adipose tissue, fulfilling essential metabolic functions such as the production of antimicrobial peptides and vitellogenin33. Vitellogenin is synthesized in the fat body and transported to the ovaries, where it is incorporated into developing oocytes65. Beyond its role in reproduction, vitellogenin also has immunomodulatory functions and is involved in lifespan regulation and stress resistance in honey bees35,66. Given that DWV replication occurs in the cytoplasm and plasma membrane of fat body cells67, this could impair the queen’s immune function and fecundity, potentially leading to the uptick in yearly queen failures experienced by commercial beekeepers. One large study surveyed honey bee queens and found coinfections of both DWV-A and -B, but determined they had little impact on the health and functional status of the queen despite instances of degenerated ovaries18. However, even with the overall patterns of depressed gene expression in the fat body during November, queens with the highest viral loads expressed greater antimicrobial peptides and PGRPs, and the lowest expression of vitellogenin hinting that covert DWV infections may impact queen health. Workers overwintering experience a reduced expression of immune genes and higher DWV loads68, suggesting queens could be responding in a similar manner. DWV-B is considered the more virulent strain69 replacing A across the US70, but in our data DWV-B lagged behind DWV-A.
In a separate queen cohort study sampled during the same overwintering time points from the same beekeeping operation, Carroll et al. found that queen fat body protein contents were lowest in September and November, while lipid contents were highest in November and January41. These findings suggest a complex interplay between viral infections, metabolic status, and immune response. The elevated expression of antimicrobial peptides and PGRPs in queens with high viral loads may represent a compensatory immune response to the viral infection. However, the concomitant low expression of vitellogenin could indicate a trade-off between immune function and reproductive capabilities. This is supported by the negative correlations between brood area and DWV viral titers. The data from Carroll et al. further complicates the picture, showing that protein and lipid reserves in the queen fat body fluctuate during the overwintering period, potentially affecting both immune responses and reproductive output. These metabolic changes could be influenced by or be a response to the viral infections and may serve as an additional stressor that impacts queen health.
The relationship between DWV titers and gene expression of the midgut from September to January was also complex and dynamic. DWV did not reach the same levels of infection as it did in the fat body, but replication in the guts of queens is documented67. In September, when DWV titers are lowest, there were 164 significant positive correlations between different genes, suggesting a coordinated and possibly effective immunity prior to rampant Varroa pressure and high levels of DWV infection over the season. As DWV titers increase in November and peak in January, the number of significant correlations among immune genes drops dramatically to 42 and 34, respectively. This indicates a disruption in the immune system's coordinated response with several potential explanations. The immune system might be reallocating resources as the viral infection progresses, leading to a more complex or less straightforward relationship between the expression of different immune genes. In response to co-infections, the immune system allocates resources to fight multiple pathogens simultaneously. In such cases, the immune system often prioritizes one pathogen over another, leading to a more complex or less straightforward relationship between the expression of different immune genes. This has been observed in co-infections with parasites and bacteria71, or between different viral infections72. It's also possible that the virus has evolved mechanisms to disrupt the host's immune response73, especially as the viral load increases25. Lastly, there may be a threshold level of DWV that, once exceeded, disrupts the normal, coordinated immune response in queens, however this has not yet been established. Unfortunately, our design did not distinguish between these alternative hypotheses.
Lysozymes and peptidoglycan recognition proteins (PGRPs) are mainly geared towards bacterial defense and are not directly involved in combating viral infections like DWV. However, there are several indirect ways these molecules could be relevant in the context of a DWV infection. For instance, a DWV infection could weaken the bee's immune system, making it more susceptible to bacterial infections. Indeed, the midgut gene expression seems to indicate an active response to a perceived pathogen during November, which suggests bacterial opportunism. In Drosophila melanogaster, immune response to pathogens is highly regionalized in the gut with the Toll pathway active in the foregut and hindgut, and IMD regulating the midgut37. This may explain the robust immune response seen in queen midguts through the Toll and IMD pathways, as most gene expression of the ileum was not significantly different. Recent research suggests that Toll and IMD pathways may also contribute to antiviral defenses. For example, some studies on Drosophila have suggested these pathways can be activated by specific viral infections and may help curb viral replication74,75. The increased expression of both PGRP-LC and short PGRPs partly explain the high activation of immune pathways. The midgut, as the primary site of food digestion and absorption, is frequently exposed to potential pathogens, so it's not uncommon for it to have a robust immune response. In immune trade-offs, a robust response requires substantial energy, whereas the queen may allocate resources to the midgut at the expense of other tissues, including the fat body. In Drosophila, the midgut acts as a sentinel tissue, detecting pathogens first and conveying the signal to the rest of the body76 which may explain the delay in fat body gene expression. There’s also a possibility of an active infection of worker nestmates affecting honey bee queen immunity through exposure to cuticular hydrocarbons (CHCs). Immune-challenged workers present immunological cues through CHCs that increase queen expression of AMPs77. Varroa mites and DWV infection also influence CHC expression78,79 and high viral titers and Varroa load present with immunosuppressed workers27. It’s also possible that if the infection is localized primarily in the midgut, other tissues like the fat body might not be stimulated to the same degree. DWV can actively suppress host immunity by modulating the host immune response, downregulating the expression of immune-related genes in certain tissues to aid their own survival73. It’s also possible that different tissues respond to infections in different ways, such that the fat body is mounting a different type of immune response that does not involve the upregulation of the particular genes we explored. In such scenarios, lysozymes and PGRPs might be upregulated as part of a broader immune response. Additionally, while not antiviral themselves, the expression of these molecules could be influenced by the overall activation of the immune system in response to the viral infection or opportunism. A DWV infection could also disrupt the gut microbiome, which might trigger a response from bacterial defense mechanisms like lysozymes and PGRPs. We found significant differences in the absolute abundance of several core microbial members in November midguts, however, studies on queen host-microbe interactions are limited and these results should be interpreted cautiously.
Colonies heavily infected by Nosema ceranae often experience excessive levels of queen supersedures17,49,80. Queens are partially resistant to N. ceranae17, and all of our queens had relatively low Nosema infection rates, even though relative worker Nosema loads rose precipitously in January40. However, our queens with the highest Nosema loads also revealed heavy loads of M. plutonius suggesting either a co-infection or secondary infection had taken place. Our findings indicate a potential link between Nosema and M. plutonius infections in queens, raising the possibility of synergistic or facilitative interactions between these pathogens that could exacerbate their individual effects. This is supported by the fact that both pathogens target the midgut, a key site for nutrient absorption and immune function. During a N. ceranae infection, queens present similar lesions in the epithelial cells of the midgut as workers and with high enough spore counts can succumb to death17. M. plutonius causes the brood disease European Foulbrood (EFB)81. EFB infects unsealed brood when freshly hatched larva ingest food contaminated with M. plutonius. M. plutonius proliferates rapidly in the midgut causing it to twist, shrivel, and decompose into a brown/black scale. Prevalence of M. plutonius can be high in asymptomatic colonies—one study using qPCR found 90% of colonies within an apiary containing EFB symptomatic apiaries had adult bees that carried M. plutonius82. Even though adult workers can carry M. plutonius, they do not show symptoms of disease, and queens may be similarly unaffected. However, this is the first study to find M. plutonius at such high relative abundances in queens to our knowledge. In general, the coincidence of Nosema and M. plutonius in queen guts may reflect the environmental similarities of queen and larval guts, as both appear to support aerobes in the presence of pure royal jelly, a substance packed with broad-spectrum antimicrobial activity13. Members of the microbiome shared by queen and larval guts may also facilitate the transmission of pathogens like Nosema and M. plutonius between distinct host environments within the colony.
Conclusion
Our work highlighted the complex interplay between the microbiota, Deformed Wing Virus (DWV), gene expression, and immune responses. We suggest that high DWV loads could disrupt the queen’s coordinated immune response, potentially leading to immune exhaustion or resource reallocation. Its currently unknown whether queens can overcome DWV infections. It should also be noted that if there are high levels of colony viral infection, likewise the queen probably suffers too. These findings are particularly relevant given the high prevalence of multiple viral infections in queens and their potential impact on queen health and hive stability. Furthermore, our results indicate a potential link between Nosema ceranae and Melissococcus plutonius infections in queens, suggesting that pathogen opportunism is not confined to just workers. These relationships are complex and are dependent on many additional factors like season, location, hive social immunity, and individual queen fitness. Future research should aim to extend these findings through longer-term studies to determine if DWV viral titers recede and how other pathogens like Nosema and M. plutonius can parasitize queens. Overall, the multifaceted interactions between the microbiota, pathogens, immune responses, and metabolic status presented here highlight the utility of a holistic colony-level approach to understanding queen health and resilience.
Data availability
The dataset generated for this study can be found in the GenBank, Sequence Read Archive as PRJNA1061232.
References
Otto, C. R. V. et al. Past role and future outlook of the Conservation Reserve Program for supporting honey bees in the Great Plains. Proc. Natl. Acad. Sci. 115, 7629–7634 (2018).
Bruckner, S. et al. A national survey of managed honey bee colony losses in the USA: Results from the Bee Informed Partnership for 2017–18, 2018–19, and 2019–20. J. Apic. Res. 1, 1–15 (2023).
Cox-Foster, D. L. et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287 (2007).
Genersch, E. Honey bee pathology: Current threats to honey bees and beekeeping. Appl. Microbiol. Biotechnol. 87, 87–97 (2010).
vanEngelsdorp, D., Tarpy, D. R., Lengerich, E. J. & Pettis, J. S. Idiopathic brood disease syndrome and queen events as precursors of colony mortality in migratory beekeeping operations in the eastern United States. Prev. Vet. Med. 108, 225–233 (2013).
Winston, M. L. The Biology of the Honey Bee (Harvard University Press, 1987).
Szabo, T. Length of life of queens in honey bee colonies. Am. Bee J. 20, 20 (1993).
Tarpy, D. R., vanEngelsdorp, D. & Pettis, J. S. Genetic diversity affects colony survivorship in commercial honey bee colonies. Naturwissenschaften 100, 723–728 (2013).
Seeley, T. D. Honeybee Ecology: A Study of Adaptation in Social Life (Princeton University Press, 2014).
Withrow, J. M., Pettis, J. S. & Tarpy, D. R. Effects of temperature during package transportation on queen establishment and survival in honey bees (Hymenoptera: Apidae). J. Econ. Entomol. 112, 1043–1049 (2019).
Spleen, A. M. et al. A national survey of managed honey bee 2011–12 winter colony losses in the United States: Results from the Bee Informed Partnership. J. Apic. Res. 52, 44–53 (2013).
Kulhanek, K. et al. A national survey of managed honey bee 2015–2016 annual colony losses in the USA. J. Apic. Res. 56, 328–340 (2017).
Vojvodic, S. et al. The transcriptomic and evolutionary signature of social interactions regulating honey bee caste development. Ecol. Evol. 5, 4795–4807 (2015).
Chen, Y., Evans, J. & Feldlaufer, M. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invert. Pathol. 92, 152–159 (2006).
Amiri, E. et al. Quantitative patterns of vertical transmission of deformed wing virus in honey bees. PLoS One 13, e0195283 (2018).
Chen, Y., Pettis, J. S. & Feldlaufer, M. F. Detection of multiple viruses in queens of the honey bee Apis mellifera L. J. Invert. Pathol. 90, 118–121 (2005).
Higes, M., Martín-Hernández, R., García-Palencia, P., Marín, P. & Meana, A. Horizontal transmission of Nosema ceranae (Microsporidia) from worker honeybees to queens (Apis mellifera). Environ. Microbiol. Rep. 1, 495–498 (2009).
Gauthier, L. et al. Viruses associated with ovarian degeneration in Apis mellifera L. Queens. PLoS One 6, e16217 (2011).
Francis, R. M., Nielsen, S. L. & Kryger, P. Patterns of viral infection in honey bee queens. J. Gen. Virol. 94, 668–676 (2013).
McMenamin, A. J., Brutscher, L. M., Glenny, W. & Flenniken, M. L. Abiotic and biotic factors affecting the replication and pathogenicity of bee viruses. Curr. Opin. Insect Sci. 16, 14–21 (2016).
Highfield, A. C. et al. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol.ogy 75, 7212–7220 (2009).
Hristov, P., Shumkova, R., Palova, N. & Neov, B. Factors associated with honey bee colony losses: A mini-review. Vet. Sci. 7, 1–16 (2020).
Insolia, L. et al. Honey bee colony loss linked to parasites, pesticides and extreme weather across the United States. Sci. Rep. 12, 20787 (2022).
Muz, D. & Muz, M. N. Acute bee paralysis virus field isolates from apiaries suffering colony losses in Türkiye. J. Apic. Res. 20, 1–7 (2023).
de Miranda, J. R. & Genersch, E. Deformed wing virus. J. Invert. Pathol. 103, S48-61 (2009).
Posada-Florez, F. et al. Varroa destructor mites vector and transmit pathogenic honey bee viruses acquired from an artificial diet. PLoS One 15, e0242688 (2020).
Yang, X. & Cox-Foster, D. L. Impact of an ectoparasite on the immunity and pathology of an invertebrate: Evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. 102, 7470–7475 (2005).
Dainat, B., Evans, J. D., Chen, Y. P., Gauthier, L. & Neumann, P. Dead or alive: Deformed Wing Virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 78, 981–987 (2012).
Williams, G. R. et al. Deformed wing virus in western honey bees (Apis mellifera) from Atlantic Canada and the first description of an overtly-infected emerging queen. J. Inverteb. Pathol. 101, 77–79 (2009).
Anderson, K. E. et al. The queen’s gut refines with age: Longevity phenotypes in a social insect model. Microbiome. 6, 1–16 (2018).
Copeland, D. C., Anderson, K. E. & Mott, B. M. Early queen development in honey bees: Social context and queen breeder source affect gut microbiota and associated metabolism. Microbiol. Spectr. 20, e00383-e422 (2022).
Corona, M., Hughes, K. A., Weaver, D. B. & Robinson, G. E. Gene expression patterns associated with queen honey bee longevity. Mech. Ageing Dev. 126, 1230–1238 (2005).
Corona, M. et al. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. PNAS 104, 7128–7133 (2007).
Amdam, G. V. et al. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp. Gerontol. 39, 767–773 (2004).
Seehuus, S.-C., Norberg, K., Gimsa, U., Krekling, T. & Amdam, G. V. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. 103, 962–967 (2006).
Royet, J. & Dziarski, R. Peptidoglycan recognition proteins: Pleiotropic sensors and effectors of antimicrobial defences. Nat. Rev. Microbiol. 5, 264–277 (2007).
Buchon, N., Silverman, N. & Cherry, S. Immunity in Drosophila melanogaster—from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810 (2014).
Evans, J. D. et al. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 15, 645–656 (2006).
Brutscher, L. M., Daughenbaugh, K. F. & Flenniken, M. L. Antiviral defense mechanisms in honey bees. Curr. Opin. Insect Sci. 10, 71–82 (2015).
Ricigliano, V. A. et al. Honey bees overwintering in a southern climate: Longitudinal effects of nutrition and queen age on colony-level molecular physiology and performance. Sci. Rep. 8, 1–11 (2018).
Carroll, M. J., Brown, N. J., Ruetz, Z., Ricigliano, V. A. & Anderson, K. E. Honey bee retinue workers respond similarly to queens despite seasonal differences in Queen Mandibular Pheromone (QMP) signaling. PLoS One 18, e0291710 (2023).
Reznick, A. Z. & Packer, L. Oxidative damage to proteins: Spectrophotometric method for carbonyl assay. Methods Enzymol. 233, 357–363 (1994).
Copeland, D. C., Maes, P. W., Mott, B. M. & Anderson, K. E. Changes in gut microbiota and metabolism associated with phenotypic plasticity in the honey bee Apis mellifera. Front. Microbiol. https://doi.org/10.3389/fmicb.2022.1059001 (2022).
Liu, C. M. et al. BactQuant: An enhanced broad-coverage bacterial quantitative real-time PCR assay. BMC Microbiol. 12, 25 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001).
Jeon, J. H., Moon, K., Kim, Y. & Kim, Y. H. Reference gene selection for qRT-PCR analysis of season- and tissue-specific gene expression profiles in the honey bee Apis mellifera. Sci. Rep. 10, 13935 (2020).
Evans, J. D. Beepath: An ordered quantitative-PCR array for exploring honey bee immunity and disease. J. Invert. Pathol. 93, 135–139 (2006).
Simone, M., Evans, J. D. & Spivak, M. Resin collection and social immunity in honey bees. Evolution 63, 3016–3022 (2009).
Alaux, C. et al. Pathological effects of the microsporidium Nosema ceranae on honey bee queen physiology (Apis mellifera). J. Invert. Pathol. 106, 380–385 (2011).
Traynor, K. S. et al. Multiyear survey targeting disease incidence in US honey bees. Apidologie 47, 325–347 (2016).
Schloss, P. D. et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Daisley, B. A. & Reid, G. BEExact: A metataxonomic database tool for high-resolution inference of bee-associated microbial communities. Systems 6, 25 (2021).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Anderson, K. E. et al. A high-throughput sequencing survey characterizing European foulbrood disease and Varroosis in honey bees. Sci. Rep. 13, 1162 (2023).
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. K. & Schmidt, T. M. rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598 (2015).
Větrovský, T. & Baldrian, P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8, e57923 (2013).
Comas, M., & Thió-Henestrosa, S. CoDaPack 2.0: A stand-alone, multi-platform compositional software. In Proceedings of the 4th International Workshop on Compositional Data Analysis, 1–10 (2011).
Li, W.-L. et al. Gut microbiota-driven regulation of queen bee ovarian metabolism. Microbiol. Spectrum 10, e02145-e2223 (2023).
Endo, A. et al. Characterization and emended description of Lactobacillus kunkeei as a fructophilic lactic acid bacterium. Int. J. Syst. Evol. Microbiol. 62, 500–504 (2012).
Martinson, V. G., Moy, J. & Moran, N. A. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 78, 2830–2840 (2012).
Zheng, H., Powell, J. E., Steele, M. I., Dietrich, C. & Moran, N. A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. 20, 5 (2017).
Engel, P., Bartlett, K. D. & Moran, N. A. The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. mBio https://doi.org/10.1128/mBio.00193-15 (2015).
Raikhel, A. S. & Dhadialla, T. S. Accumulation of yolk proteins in insect oocytes. Annu. Rev. Entomol. 37, 217–251 (1992).
Amdam, G. V. & Omholt, S. W. The regulatory anatomy of honeybee lifespan. J. Theoret. Biol. 216, 209–228 (2002).
Fievet, J. et al. Localization of deformed wing virus infection in queen and drone Apis mellifera L. Virol. J. 3, 16 (2006).
Steinmann, N., Corona, M., Neumann, P. & Dainat, B. Overwintering is associated with reduced expression of immune genes and higher susceptibility to virus infection in honey bees. PLoS One 10, e0129956 (2015).
McMahon, D. P. et al. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. R. Soc. B Biol. Sci. 283, 20160811 (2016).
Ryabov, E. V. et al. Recent spread of Varroa destructor virus-1, a honey bee pathogen, in the United States. Sci. Rep. 7, 17447 (2017).
Graham, A. L. Ecological rules governing helminth–microparasite coinfection. Proc. Natl. Acad. Sci. 105, 566–570 (2008).
Sulkowski, M. S. Hepatitis C virus infection in HIV-infected patients. Curr. Infect. Dis. Rep. 3, 469–476 (2001).
Nazzi, F. et al. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog. 8, e1002735 (2012).
Zambon, R. A., Nandakumar, M., Vakharia, V. N. & Wu, L. P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. 102, 7257–7262 (2005).
Sabin, L. R., Hanna, S. L. & Cherry, S. Innate antiviral immunity in Drosophila. Curr. Opin. Immunol. 22, 4–9 (2010).
Bosco-Drayon, V. et al. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 12, 153–165 (2012).
Hernández López, J., Riessberger-Gallé, U., Crailsheim, K. & Schuehly, W. Cuticular hydrocarbon cues of immune-challenged workers elicit immune activation in honeybee queens. Mol. Ecol. 26, 3062–3073 (2017).
Salvy, M., et al. Modifications of the cuticular hydrocarbon profile of Apis mellifera worker bees in the presence of the ectoparasitic mite Varroa jacobsoni in brood cells. Parasitology [Internet]. 2001. http://www.journals.cambridge.org/abstract_S0031182001007181. Accessed 25 May 2023.
Baracchi, D., Fadda, A. & Turillazzi, S. Evidence for antiseptic behaviour towards sick adult bees in honey bee colonies. J. Insect Physiol. 58, 1589–1596 (2012).
Higes, M. et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10, 2659–2669 (2008).
Forsgren, E., Locke, B., Sircoulomb, F. & Schäfer, M. O. Bacterial diseases in honeybees. Curr. Clin. Microbiol. Rep. 5, 18–25 (2018).
Erban, T. et al. Bacterial community associated with worker honeybees (Apis mellifera) affected by European foulbrood. PeerJ. 5, 1–28 (2017).
Acknowledgements
We thank Ashurst Bee Co. Inc. (Westmoreland, CA), and especially Bryan Ashurst for the donation and maintenance of the bee colonies and queens that made this research possible. The authors would also like to thank Tiffany Ann Wolff and Kaiju P. Chan for their valuable input.
Funding
The study was supported by the ARS-USDA, Carl Hayden Bee Research Center, CRIS project plan Anderson 2022-21000-021-00D. The ARS is an equal opportunity employer and provider.
Author information
Authors and Affiliations
Contributions
Experimental design by V.A.R., B.M.M., and K.E.A. D.C.C., V.A.R., O.L.K., R.J.E. performed laboratory work with the input of K.E.A. and B.M.M. D.C.C. and J.G.O. analyzed the data. D.C.C., O.L.K., and R.J.E., designed the figures. D.C.C., V.A.R., and K.E.A. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Copeland, D.C., Ricigliano, V.A., Mott, B.M. et al. A longitudinal study of queen health in honey bees reveals tissue specific response to seasonal changes and pathogen pressure. Sci Rep 14, 8963 (2024). https://doi.org/10.1038/s41598-024-58883-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-58883-1
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.