Abstract
Brucella, a gram-negative intracellular bacterium, causing Brucellosis, a zoonotic disease with a range of clinical manifestations, from asymptomatic to fever, fatigue, loss of appetite, joint and muscle pain, and back pain, severe patients have developed serious diseases affecting various organs. The mRNA vaccine is an innovative type of vaccine that is anticipated to supplant traditional vaccines. It is widely utilized for preventing viral infections and for tumor immunotherapy. However, research regarding its effectiveness in preventing bacterial infections is limited. In this study, we analyzed the epitopes of two proteins of brucella, the TonB-dependent outer membrane receptor BtuB and the LPS assembly protein LptD, which is involved in nutrient transport and LPS synthesis in Brucella. In order to effectively stimulate cellular and humoral immunity, we utilize a range of immunoinformatics tools such as VaxiJen, AllergenFPv.1.0 and SignalP 5.0 to design proteins. Finally, five cytotoxic T lymphocyte (CTL) cell epitopes, ten helper T lymphocyte (HTL) cell epitopes, and eight B cell epitopes were selected to construct the vaccine. Computer simulations are also used to verify the immune response of the vaccine. The codon optimization, in silico cloning showed that the vaccine can efficiently transcript and translate in E. coli. The secondary structure of mRNA vaccines and the secondary and tertiary structures of vaccine peptides were predicted and then docked with TLR-4. Finally, the stability of the developed vaccine was confirmed through molecular dynamics simulation. These analyses showed that the design the multi-epitope mRNA vaccine could potentially target extracellular protein of prevalent Brucella, which provided novel strategies for developing the vaccine.
Similar content being viewed by others
Introduction
Brucellosis is a zoonotic bacterial infection caused by Brucella1.This gram-negative coccus lacks flagella, does not produce spores, and grows slowly2. The World Health Organization(WHO) reports more than 500,000 new cases of human brucellosis annually3.At the same time, the WHO considers brucellosis to be one of the seven neglected zoonotic diseases4. It poses a threat to public health in the Mediterranean region, the Middle East, Central Asia, Southeast Asia, sub-Saharan Africa, and some parts of Latin America5. In 2019, there were 44,036 confirmed cases, resulting in an incidence rate of 3.2513 per 100,000 people in China 6.There are six common species of Brucella: Brucella melitensis, Brucella abortus, Brucella ovis, Brucella suis, Brucella canis, and Brucella neotomae7. The most virulent causes of human infection are Brucella melitensis, Brucella abortus, Brucella suis, and Brucella canis8.
Brucella can enter the body through multiple pathways owing to its potent pathogenicity and subsequently generate endotoxin, precipitating bacteremia and toxemia9. Brucella is able to survive in the human immune system is attributed to its array of invasive virulence factors and diverse strategies for evading cellular immunity, allowing it to replicate within intracellular niches10. Patients with brucellosis present a spectrum of symptoms, which can vary from being asymptomatic to experiencing fever, fatigue, loss of appetite, and joint, muscle, and back pain11.Severe individuals may manifest multiorgan dysfunction10. However, the increasing drug resistance rate of Brucella brought great challenges for clinical treatment12,13. Therefore, it is necessary to develop a new therapeutic approach against Brucella in early prevention. Vaccines are one of the most important tools in public health and play an important role in preventing and controlling infectious diseases14. Unfortunately, a vaccine for brucellosis in human beings has not been approved yet15 . Several types of vaccines are presently under development to address brucellosis, such as attenuated live, multi-epitope, DNA, recombinant protein, and mRNA vaccines. The mRNA vaccines have been applied in Achromobacter xylosoxidans16 、Citrobacter freundii (C. freundii)17 and so on. mRNA vaccines are an emerging approach to vaccine development, encoding only the target antigen to enhance the safety and efficacy of the vaccine18.
In order to expand the scope of vaccine application and provide cross-protection against Brucella melitensis, Brucella abortus, and Brucella suis, we selected the BtuB and LptD proteins, which are present in all three strains19. The outer membrane proteins BtuB, which transport vitamin B12, has been proved to have good immunogenicity in previous experiments and is suitable as a vaccine design candidate protein20,21. LptD is one of the outer membrane proteins of Brucella and has good immunogenicity19. The difference is that LptD is to transport lipopolysaccharides (LPS) which lead to the occurrence of an inflammatory response22. Therefore, vaccines based on BtuB and LptD may reduce the inflammatory reaction and slow down the progression of the disease.
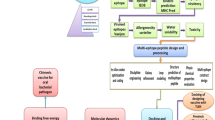
In this study, we will design the mRNA vaccine based on LptD and BtuB. The immunological characteristics of the selected protein were predicted using an online tool. The prediction of cytotoxic T lymphocyte (CTL) epitopes, helper T lymphocyte (HTL) epitopes, and B cell epitopes was conducted. After predicting T-cell and B-cell epitopes, it is essential to identify antigenic, non-allergenic and non-toxic epitopes for subsequent construction of the vaccine23.Additionally, we will conduct molecular docking with MHC alleles to further explore their interactions. Finally, the mRNA vaccine was developed and its physical and chemical properties were assessed. The Immune simulation was then conducted. The nucleotide sequence of the vaccine was optimized through codon analysis, followed by in silico cloning and simulated gel electrophoresis. The secondary structure of the constructed mRNA vaccine was analyzed, along with the secondary and tertiary structures of the multi-epitope fusion proteins. The mRNA vaccine was also docked with TLR-4 molecules. To assess the stability of atoms and molecules in the vaccine, a molecular dynamics model was used.
Results
Physicochemical properties of proteins
The protein sequences of LptD and BtuB obtained from NCBI will be included in the supplementary information. Conducting online software analysis, it has been confirmed that BtuB and LptD have antigenicity scores of 0.811642 and 0.860866, respectively. With both surpassing the 0.5 threshold and demonstrating strong antigenicity. Additionally, the AlgPred server has indicated that the BtuB and LptD proteins are likely non-allergenic. The amino acid number of BtuB and LptD are 620aa and 792aa respectively. The molecular weight of BtuB and LptD are 66,892.00 and 88553.50 respectively. Both BtuB and LptD are outer membrane proteins. Further predictions regarding the bioinformatics analysis of these two proteins will be presented in Table 1.
Prediction and excision of signal peptides
SignalP 5.0 and LiPOP 1.0 were used to predict the signal peptides. The SignalP 5.0 server indicated that it is likely that the signal peptide of BtuB is located within the 1–24 peptide range (Fig. 1A). The LiPOP1.0 suggested that the 13–28 peptide may function as the signal peptide (Fig. 1B). According to the SignalP5.0 website, the 1–42 peptide of the LptD protein is highly likely to be the signal peptide (Fig. 1C). Additionally, the LiPOP 1.0 tool suggested that the signal peptide could be located between amino acids 26–39(Fig. 1D). Finally, we combined the results from two software programs and selected the peptide sequence that completely does not containa signal peptide in order to enhance the the effectiveness of the cell epitope. We choose the epitope after the 28 peptide of BtuB, and we choose the epitope after the 42 peptide of LptD.
(A–D) Signal peptide of proteins using SignalP-5.0 and LiPOP1.0 analysis. SP (Sec/SPI) and Spl: type of signal peptide predicted; CS: the cleavage site; Other: the probability that the sequence does not have any kind of signal peptide. (A) The signal peptide prediction of BtuB by SignalP5.0. (B) The signal peptide prediction of BtuB by LiPOP1.0. (C) The signal peptide prediction of LptD by SignalP5.0.(D) The signal peptide prediction of LptD by LiPOP1.0.
Prediction and assessment of B cell epitopes
After obtaining the predicted top ten epitopes from Ellipro and ABCpred respectively, we selected two cross-result epitopes. The next steps involved verifying their antigenicity, toxicity( See supplementary document 1), and allergenicity using VaxiJen, AllerTop V.2.0, and ToxinPred in turn. We screened all the predicted epitopes to determine whether they have homology with Homo sapiens (Taxid:9606), and we excluded peptides with E value less than 0.05. We selected two epitopes in BtuB protein, and six epitopes in LptD protein in Table 2.
Prediction and assessment of T cell epitopes
We used the IEDB and the NetCTLpan1 server to predict CTL epitopes. We selected the top ten epitopes, two of which are consistent in both predictions. We also utilized IEDB and the NetMHC-IIpan-4.0 server to predict HTL epitopes. We selected the top ten epitopes, including two cross-result epitopes. It is worth mentioning that all the selected epitopes have been verified as antigenic, non-toxic, non-homologous, and non-allergenic. When predicting and evaluating CTL cell epitopes, five epitopes were selected: three from the BtuB protein and two from the LptD protein in Table 3. HTL cell epitopes were predicted and assessed. Six epitopes of the BtuB protein and four epitopes of the LptD protein were selected, as shown in Table 4. In the end, we selected 8 B cell epitopes, 5 CTL cell epitopes, and 10 HTC cell epitopes for the construction of subsequent mRNA vaccines.
Molecular docking
To assess the structural relationship between HLA alleles and T cell epitopes. The HDOCK server provided a list of the top ten docking models, and we chose the first model for evaluation. Molecular docking analysis of CTL epitopes and HLA alleles resulted in a docking score of -251.12 and a ligand RMSD of 48.75 Å (Fig. 2A). The HTL epitopes and HLA allele molecular docking analysis yielded a docking score of -249.48 and a ligand RMSD of 192.84 Å (Fig. 2B).
Vaccine design
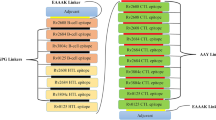
The proposed construction of mRNA vaccine is shown in Fig. 3 MRNA vaccine we designed consists of the following from the N-terminal to C-terminal, that is 5’m7GCap- 5’UTR–Kozak sequence-tPA (Signal peptide)-GPGPGGLNWQATASTSLDMAGPGPGVGLNWQATASTSLDMGPGPGEVGLNWQATASTSLDGPGPGVTGLEATLSHRFNEQGPGPGKVTGLEATLSHRFNEGPGPGNPDLQPEKSRSVEVGGPGPGWPILFSTTSSTHILEGPGPGEYDFRIAGIHQLKPEGPGPGTYEIQGYNAQTQVSKGPGPGGDAIRVDTNFDPANAkkPECIESTNDEPENDENkkQATASTSLDMALYQTRkkMAYIQTRNPGDEKASHkkYSYTMPEPVYGGELNFkkTYEIQGYNAQTQVSKIkkMALRGDAIRVDTNFDPkkPGFSGTNLRFTSEAEWkkYLPSDKYEDEHPNDDSSRAAYFLQGSFNFALAAYLQTYSGISVKAAYNIYMRGMSSKAAYALAPNYDLTLAAYLVYDRDVNTV-GGGSHHHHHH-Stop codon-3’ UTR-Poly (A) tail. (Fig. 3).
Vaccine physicochemical properties and immunological characterization evaluation
The molecular formula of the vaccine is C1991H3033N553O635S9. The molecular weight is 45,164.85 kDa, and the number of amino acid residues is 421. The vaccine had a solubility of 0.978002, indicating that the protein antigen was soluble. The instability index (II) was computed to be 28.54, which is less than the threshold of 40, indicating that the vaccine protein was stable. The gravy was − 0.727. The indication shows that the vaccine was a hydrophilic protein. In addition, the vaccine's antigenicity is 1.1082, which is greater than the threshold value of 0.4, indicating that the protein is highly antigenic. According to the allergenicity prediction results, the vaccine is likely non-allergenic. In conclusion, the vaccine design is feasible. Finally, Table 5 presents all the final results.
Simulated immune response
After using the C-ImmSim server to simulate the immune response to three vaccine injections, the results showed that the LptD-BTuB mRNA vaccine could induce both cellular and humoral immune responses. B cells were identified to have a part in moderating the immune response, gradually escalating subsequent to each vaccination, and ultimately reaching its zenith (Fig. 4A). Two subsets of T cells, namely TH cells (Fig. 4B) and TC cells, displayed an upward trend and reached a peak following three simulated immune injections. However, the upward trend of TH cells was marginally lower than that of B cells. Contrary to expectations, the levels of TC cells (Fig. 4C), NK cells (Fig. 4D), DC cells (Fig. 4E), and EP (Fig. 4F) remained stable and consistent. Additionally, the data revealed a continuous increase in the concentration of IgM + IgG (Fig. 4G) until it reached its peak. Finally, the vaccine also resulted in alterations of various cytokines (Fig. 4H), particularly IFN and IL-2 cytokines.
The results of C-ImmSim server immune simulation reaction: (A): The B-cell isotypes during each stage. (B): The TH isotypes during each stage. (C): The TC isotypes during each stage. (D): The NK cells during each stage. (E): The DC cells during each stage. (F): The EP cells during each stage. (G): Antibodies of various types during each stage. (H): The Cytokines and interleukin during each stage.
Codon optimization and in silico cloning
The amino acid sequence of the fused epitope protein was successfully converted to nucleotide sequence using the online server EMBOSS Backtranseq. The nucleotide sequence was then optimized with the JCat tool, excluding XHOI and BamHI restriction endonuclease sites. E. coli served as the host for the optimization process. Finally, the optimized codon adaptation index (CAI) value is 0.98, with an average percentage of GC content percentage of 53%(Fig. 5A).The 5' and 3' ends of the target gene were modified with BamHI and XhoI restriction sites. Design the primers with the following specifications: The forward primer sequence is 5’GGATCCACATTTGCTTCTGACACAACTG, with a length of mer, a Tm value of 62°C, and GC content of 46%. The downstream primer sequence is 5’CTCGAGTGCTGCCCACTCAGACTT, with a length of 24-mer, a Tm value of 64°C, and GC content of 58%.The target gene was amplified using simulated PCR in SnapGene and inserted into the multiple cloning site (MCS) region of the plasmid after removing the previously designed primer restriction site. The plasmid sequence of pVAX1 is 2999 bp, while that of the recombinant plasmid is 4448 bp (Fig. 5B) The target gene sequence is 1511 bp (Fig. 5C). Finally, a 1% agarose was selected for the simulated electrophoresis reaction (Fig. 5D) (see Supplementary Fig. 1).
(A): Adaptation of optimized codons. (B): The constructed recombinant(red) was inserted into the pVAX1 vector(black) in silico cloning. The XHOI and BamHI is restriction endonuclease sites. (C): The amplified target gene sequence is 1511bp.D: Simulated agarose gel experiment (1.0% agarose ). As shown in the figure, “1” stands for LptD-BtuB mRNA vaccine, “2” stands for pVAX1, and “3” stands for recombinant illustrated in the fgure, “1” stands for the MEV, “2” stands for pET-28a ( +), and “3” stands for recombinant plasmid.
mRNA Vaccine’s and peptides’s structure
RNAfold predicts the RNA's secondary structure, producing two structures as output, which are the minimum corresponding free energy (MFE) and the centroid structure. The centroid structure that contains a minimal base-pair distance to all structures in the thermodynamic ensemble. After inputting the optimized vaccine codon sequence, the optimal secondary structure is obtained, guaranteeing the minimum corresponding free energy (MFE) of − 495.13 kcal/mop (Fig. 6A). The other is a secondary centroid structure with a stability value of − 401.50 kcal/mol (Fig. 6B). The smaller value indicates greater stability. Predicting the secondary structure of mRNA is significant in enhancing its stability to withstand endonuclease cleavage and chemical degradation.
The peptide's two-dimensional structure, as predicted by SOPMA, is depicted in Fig. 7A, consisting of 13.30% alpha-helix, 1.66% β-turn, 65.32% random coil, and 19.71% extended strand. The ratio of The alpha-helix, random coil, extended strand andβ-turn the secondary structure aligns with that of the tertiary structure. This suggests that the tertiary structure (Fig. 7B) predicted by Robetta is accurate. We selected model one out of the five models, and its confidence level is 0.40. The three-level structure was optimized using GalaxyRefine (Fig. 8). Next, we used Discover Studio to display the PDB format of the vaccine. The pink areas in the model represent the donor, while the green areas represent the recipient (Fig. 7C). Following up with an analysis and application of the optimized three-level structure model.
Evaluation of tertiary structure quality
Using ProSA-web server to evaluate the predicted the model of tertiary structure (Fig. 9A), The light blue region in the figure represents the source of X-rays in the predicted tertiary structure, and the dark blue region represents the source of nuclear magnetic resonance (NMR) in the predicted tertiary structure. Verification of the model was performed with the ERRAT quality factor of 80.110 (see Supplementary Fig. 2 )while Verify 3D score was 92.40%(see Supplementary Fig. 3 ). The Z-score of the LptD-BTuB mRNA vaccine was -6.36, indicating the protein has the correct structure. In order to ensure the accuracy of structural prediction again, we use SWISS-MODEL online service and predicted using the Ramachandran plots. The allowable area as the dark green region and restricts the maximum allowable area to the light green region (Fig. 9B). The blank area is prohibited. Ramachandran plot analysis showing 94.75% in favored, 5.15% in allowed, and 0.72% in disallowed regions of protein residues. Based on the figure, a significant number of amino acids fall within the specified range, leading to a conclusion that the three-level structure prediction model for the LptD-BTuB mRNA vaccine is highly credible.
Molecular docking
The HDOCK website utilizes a semi-flexible docking approach while taking into account non-bonding interactions, including electrostatic interaction and van der Waals force, to forecast its binding mode and affinity. The website provided a list of the top ten docking models, and we selected the first model for evaluation. The results of the molecular docking analysis indicate a docking score of − 364.59, a ligand RMSD of 93.69 Å and a confidence score of 0.9865 in Fig. 10A.We utilized PyMOL for three-dimensional visualization. The purple dotted line in the Fig. 10B represents hydrogen bonding.
Molecular dynamic simulation
The IMOD server conducts molecular dynamics simulations of TLR4 complexes under specific conditions. The binding of mRNA vaccine and TLR4 is shown in Fig. 11A. A service provided by Cluspro. Within the dynamic region, the covariance matrix illustrates the relationship between amino acid dimers in Fig. 11B. The red section represents related residues, the white denotes anti-related residues, and the blue indicates irrelevant residues. In Figs. 11C, the elastic network model is represented by ordered pairs of atoms connected by springs, with the gray area in the figure indicating the hard region. Therefore, the constructed vaccine constructed can be considered to be harder and more stable. The B-factor graph illustrates the correlation between the Normal Mode Analysis and PDB regions of the complex in Fig. 11D. The flexible regions of the structure represent amino acids with high deformability, as reflected in the peak map of deformability shown in Figs. 11E.The eigenvalues of the two docking complexes are shown in the Fig. 11F.
Discussion
Brucella, a Gram-negative coccidian, has the potential to cause zoonotic diseases and lead to public health incidents, economic losses, and casualties worldwide. Therefore, it is crucial to understand and prevent the spread of this pathogen1. For humans, the most pathogenic species are Brucella melitensis, Brucella abortus and Brucella suis. Brucellosis is potentially harmful with variable clinical manifestations , and no effective treatment is available 24. Vaccination remains the most effective preventive measure25. Development of highly effective vaccines against human Brucella remains a public health priority26. In our study, we designed the multi-epitope mRNA vaccine, which provided novel strategies for developing the vaccine.
The primary attenuated live vaccines for brucellosis in China include Brucella melitensis strain M5, Brucella suis strain S2, and Brucella abortus strain A1927. However, there are many shortcomings, such as persistent infection in vaccinated animals, risk of toxicity reversal, lack of lasting protection, interference with brucellosis diagnosis and so on28.Developing a new vaccine needs to verify its effectiveness and safety, which is a time-consuming and expensive process. Therefore, using immunoinformatics approaches to design new kind of vaccines could be a magnificently additive in the way forward of preventing Brucella. Many studies focused on design of novel vaccines using bioinformatics tools29. Subunit vaccines are developed utilizing fragments of pathogens like proteins30. Subunit vaccines encompass a range of types such as include recombinant protein vaccine, epitope vaccine, DNA vaccine, gene marker vaccine, conjugate vaccine, recombinant vector vaccine, nanoparticle vaccine, bacterial shadow vaccine, mRNA vaccine and so on31. Compared to other vaccines, mRNA vaccines have obvious advantages. First, they are safe, as there is no potential risk of infection or insertion of mutant genes, and mRNA is degraded through normal cell processes. Secondly, using various modifications can make mRNA more stable and highly translatable. Thirdly, the in vitro transcription reaction yields a high amount of mRNA, which gives mRNA vaccines the potential for rapid, cost-effective, and large-scale production32.
A qualified vaccine should also contain effective epitopes of cytotoxic T lymphocytes (CTL), helper T lymphocytes (HTL), and B cells33, thus has the capability to induce the activation of both cellular and humoral immune responses. In this study, the epitopes of the LptD and BtuB proteins were analyzed and predicted. Before making the prediction, we removed the signal peptide from the protein. The signal peptide can impact the initial stage of protein translation and even affect the subsequent folding and transportation of the protein34. In this study, we found 5 CTL epitopes and 10 HTL epitopes using IEDB NetCTLpan1 and NetMHC-IIpan-4.0. T cell epitopes and HLA molecules are well matched. The incidence of brucellosis in Xinjiang is much higher than that of most provinces in China35. Therefore, our focus is on the high-frequency alleles of the Xinjiang population to predict T cell epitopes. We selected the alleles with high frequency in Xinjiang (HLA-A*1101, HLA-A*0201, HLA-A*0301, HLA-DRB1*0701, HLA-DRB1*1501, and HLA-DRB1*0301) to predict the cytotoxic T lymphocyte (CTL) epitopes and helper T lymphocyte (HTL) epitopes.T-cell epitopes must be processed by antigen-presenting cells and recognized by T-cell receptors. When antigen-presenting cells (APC) present antigen peptides to T cells, they are assisted by both MHC I and MHC II molecules. Cytotoxic T lymphocytes (CTL) recognize MHC I, while helper T lymphocytes (HTL) recognize MHC II. In this study, we predicted 8 B cell epitopes using IEDB and ABCpred. In order to optimize immunogenicity, 8 B cell epitopes and 15 T cell epitope were combined with flexible lingers, to produce a novel vaccine. The B-cell epitopes and T-cell epitopes added in vaccine help to enhance the humoral and cellular immune response36.
In our study, several unique elements are included in mRNA vaccines for improving mRNA stability and translation efficiency. The 5' and 3' UTR elements on both sides of the coding sequence can originate from viruses or eukaryotic genes37. This significantly impacts the stability and translation of mRNA, leading to a substantial increase in mRNA half-life and expression38. A 5' cap structure is necessary for the effective translation of mRNA into protein39. The poly(A) tail plays an important role in mRNA translation and stability, and can be directly added to mRNA using poly(A) polymerase40. In ribosome translation, protein synthesis will stop when it encounters the conservative stop codons: UAA, UAG, or UGA41. Kozak sequence is recognized by ribosomes as translation initiation site42. TPA43 is a sequence located before the coding region in a structural gene, which can be transcribed but not translated. The open reading frame is the translation area of mRNA, and replacing rare codons with common codons is beneficial to the efficiency of subsequent translation44. We added a 6xHis tag to the C-terminal of the fused amino acid sequence to facilitate protein purification through affinity chromatography45.
An adjuvant is a substance that can nonspecifically enhance or alter the specific immune response of the body to the corresponding antigen, increase the antigen's immunogenicity, or modify the type of immune response, but does not have antigenicity itself46. Adjuvants in mRNA vaccines are typically categorized into three groups: (1) RNA with inherent adjuvant activity from its nucleotide sequence or encoded protein; (2) components of the delivery system, particularly ionizable cationic lipids in lipid nanoparticles47; and (3) exogenous immunostimulants48. Lipid nanoparticles (LNPs) have become one of the most popular and widely used tools for delivering mRNA49. LNP typically consists of four components: ionizable cationic lipids, which facilitate self-assembly into virus-sized particles (approximately 100 nm) and enable the release of mRNA endosomes into the cytoplasm; Lipid-linked polyethylene glycol (PEG), which prolongs the preparation's half-life; Cholesterol as a stabilizer; and naturally occurring phospholipids that maintain the lipid bilayer structure32. In this study, the adjuvant is not directly connected to the vaccine, because mRNA vaccines often are supplemented with lipid nanoparticles, serving as both a carrier vector and an adjuvant, boosting the immune response.
One of the key issues in vaccine is the capacity to elicit the desired immune response and generate immune factors following injection into the body. In our study, the Online website C-ImmSim was employed to perform Immune Simulation Response.After simulating the three-needle injection reaction, B cells can differentiate into plasma cells when stimulated by antigens, and plasma cells can produce and release antibodies. After the simulated injection, there is an increase and peak in the amount of B cells, along with elevated levels of antibody IgM and IgG. This demonstrates the production of an effective humoral immunity. The rise in the quantity of natural killer (NK) cells and dendritic cells (DC) suggests that vaccination typically generates innate immunity. Additionally, it can trigger alterations such as the presence of IFN and IL-2 cytokines.
Conclusion
The physical and chemical characteristics of the LptD-BTuB mRNA immunization formulated in this study fulfill the fundamental criteria for a vaccine. After simulating immunity with the C-ImmSim server, it has been theoretically proven that the vaccine can result in optimal cellular and humoral immunity post-injection. However, to ascertain the practical utility of the vaccine in brucellosis prevention, further experiments are crucial, despite challenges that would be expected during the experimental process.
Materials and methods
Obtaining protein sequences
Epitopes refer to specific regions on the surface of antigens that are identified by the immune system50. In this study, the conserved region of the chosen protein was retrieved from the National Center for Biotechnology Information (NCBI)51 (https:// www.ncbi.nlm.nih.gov/accessed) and were (1) LPS assembly protein LptD (accession number WP_011068938.1) lptD, (2)TonB-dependent receptor Outer membrane BTuB(accession number WP_004691650.1 )1), which were identified as potential protective antigens19.
Immunological characterization
As antigenicity is a crucial factor in vaccine development, we initially predicted antigenicity. As antigenicity is a crucial factor in vaccine development, we initially predicted the antigenicity52. The ANTIGENpro (https://scratch.proteomics.ics.uci.edu/) website was conducted to confirm the antigenicity of the selected protein,which is a part of Scratch protein predicter. The physicochemical properties of the protein were analyzed using the online software ProtParam (http://web.expasy.org/protparam/), including molecules the number of amino acids, the molecular formula, molecular weight, the instability index, and the overall mean of the water solubility (GRAVY). In order to conduct a more comprehensive analysis of the selected proteins.
AlgPred (https://webs.iiitd.edu.in/raghava/algpred/submission.html) predicts the allergenicity of bacterial protein sequences which choose prediction a MEME/MAST motif aproach. Using the online website Gneg-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/Gneg-multi/ )to predict the subcellular localization of two quantitative proteins again.
Prediction of signal peptides
When analyzing the structure of a protein and predicting the epitope, the signal peptide sequence should be removed first53.To predict the presence of a signal peptide in the selected protein, we uesd SignalP 5.054 and LiPOP 1.0 software (https://services.healthtech.dtu.dk/service.php?LipoP-1.0) to analyze the protein sequence for potential signal peptides. SignalP 5.0 can predict signal peptides (SPs) and their associated cleavage sites in proteins from Gram-negative bacteria. Meanwhile, LipoP 1.0 functions as a signal peptide (SP) prediction tool that differentiates between lipoprotein SPs, other SPs, and N-terminal membrane helices in Gram-negative bacteria55. We will combine the two results to remove the signal peptide.
T cell epitopes prediction
T cells can only recognize antigen-presenting cells (APCs) through their interaction with major histocompatibility complex (MHC) molecules, rather than directly identifying epitopes56. MHC molecules are primarily classified into two categories: class I (MHC I) and class II (MHC II)57. Cytotoxic T lymphocyte (CTL) cells recognize endogenous antigenic peptides presented by MHC I molecules, while helper T lymphocyte (HTL) cells recognize exogenous antigenic peptides presented by MHC II molecules58. Therefore, we found it necessary to analyze the CD8 + and CD4 + T cell epitopes separately. For this study, we chose to use the high-frequency alleles of Xinjiang, which consist of HLA-A*1101 (13.46%), HLA-A*0201 (12.50%), HLA-A*0301 (10.10%), HLA-DRB1*0701 (16.35%), HLA-DRB1*1501 (8.65%), and HLA-DRB1*0301 (7.69%), to predict the CTL and HTL epitopes59.
The CTL epitopes were predicted using IEDB (http://tools.immuneepitope.org/) and the NetCTLpan1 server (https://services.healthtech.dtu.dk/service.php?NetCTLpan-1.1). Amino acids with a length of 10 mers were selected based on the previously mentioned alleles, with no modification to the original parameters. The top ten epitopes with the highest scores from the software were chosen for further investigation. HTL epitopes of target proteins will be predicted using the IEDB and NetMHC-IIpan-4.0 (https://services.healthtech.dtu.dk/service.php?NetMHCIIpan-4.0), with an amino acid length of 15 mer. The default thresholds for NetMHC-IIpan-4.0 will remain unchanged. After running the predictions on both servers, the top ten epitopes will be selected for further analysis.
B cell epitopes Prediction
Antigens use B cell epitopes to recognize B cells and stimulate the B cell immune response60. We decided to use Ellipro from IEDB (http://tools.iedb.org/ellipro/) and ABCpred (https://webs.iiitd.edu.in/raghava/abcpred/ABC_submission.html) to identify B-cell epitopes. The length of the epitopes was set at 16 mer. We will select the top ten epitopes from the epitope prediction results provided by the online servers, and screen for cross-reactive epitopes for subsequent verification.
Homology of predicted peptides with humans
Non-homologous proteins can reduce the likelihood of stimulating the body to produce an immune response61. By using the NCBI BlastP database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins), a comparison was made between Homo sapiens (TaxID: 9606) and predicted peptides to mitigate the risk of autoimmunity. Peptides with an E value greater than 0.05 were considered non-homologous and suitable for use in vaccine development62.
Antigenicity, allergenicity and toxicity evaluation of epitopes
All selected epitopes need to be evaluated for their antigenicity, allergenicity, and toxicity63. To assess antigenicity, the VaxiJen online web server (http://www.ddg-pharmfac.net/Vaxijen/VaxiJen/VaxiJen.html ) was performed with the bacteria parameter and a threshold of 0.4 were selected. Using the website of AllerTop V.2.0 (http://www.ddg-pharmfac.net/AllerTOP) to analyze the allergenicity of epitopes, which predicts using a Tanimoto coefficient to predict64. The ToxinPred server was utilized to evaluate the toxicity of the selected epitopes. Ultimately, only epitopes that were antigenic, non-toxic, and non-allergenic were chosen for further analysis.
Molecular docking of T-cell epitopes to HLA alleles
The human leukocyte antigen (HLA) system is an integral component of the human immune system. The trait is controlled by genes located on chromosome 6 and encodes cell surface molecules. These molecules present antigen peptides to T cell receptors (TCR) on T cells65. In order to maximize vaccine coverage within the population, it is important to consider the frequency of HLA alleles66. The HLA DRB*0101 allele is associated with susceptibility and is present in 95% of the population67. HLA class I (HLA-A*02:01) is one of the most common HLA in the world68. The HDOCK server69 was utilized to molecular docking, following selection of HLA class I (HLA-A*02:01) and HLA class II (HLA-DRB1*01:01) alleles were chosen for molecular docking with T cell epitopes. The resulting interactions between the alleles and T cell epitopes were subsequently identified.
Construction of mRNA vaccine
The construction structure of mRNA vaccine from N-terminus to C-terminus includes the following components:5’m7GCap-5’UTR-Kozak sequence-tPA (Signal peptide)-GPGPG (linker)-HTL Epitopes-KK (linker)-B Cell Epitopes-AAY (Linker)-CTL Epitopes-GGGS (linker)-HHHHHH-Stop codon-3’UTR-Poly (A) tail. We have given this vaccine the name LptD-BTuB mRNA vaccine.
GPGPG70, KK, AAY71 and GGGS72 linkers were used to connect all of the predicted epitopes. The linkers' purpose is to separate epitopes, ensuring that their functions—flexibility, cuttability, and solidity—are not interfered with. The KK linker can maintain the independent immune activity of B-cell epitopes in vaccines73. AAY linker can improve the immunogenicity of an epitope vaccine74. The GPGPG linker will facilitate the immune processing and presentation processes75.To detect and purify recombinant proteins later on, we plan to add a group of 6xHis tag sequences to the C-terminal of the amino acid sequences mentioned above 76. MRNA vaccine17 must be have a Kozak sequence that contains a start codon AUG, at the beginning of ORF and a stop codon TGA ended translation. Moreover, This construction contains three structures. Firstly, the leader sequence of tissue plasminogen activator (tPA) (UniProt ID: P00750) is located in the 5 regions of the construct, which ensures correct translation of the target gene fragment. Secondly, the 5' and 3' Untranslated regions (UTRs) will consist of human β globin (NCBI Gene ID: 3043) and α globin (NCBI Gene ID: 3039), respectively.
Physicochemical properties, antigenicity, allergenicity, and toxicity of vaccine
Using the VaxiJen77 and ANTIGENpro(http://scratch.Proteomics.ics.uci.edu/) servers to perform the assessment of antigenicity of the constructed mRNA vaccines, which only the amino acid sequence was included, without the tPA or MITD sequences. The allergy potential of the construct was assessed using the AllerTOP 2.0 server, while the ToxinPred server was utilized to evaluate its toxicity78. Physicochemical parameters of the construct, such as amino acid concentration, theoretical isoelectric point (PI), molecular weight, Aliphatic Index (AI), Instability Index (II), and Grand Average of Hydropathicity (GRAVY), were estimated using the program website ExPASy-ProtParam(https://web.expasy.org/protparam/)79. And the SOLpro (http://scratch.proteomics.ics.uci.edu/) was applied to predict the solubility80.
Immune simulation response
The Online website C-ImmSim simulation (http://150.146.2.1/C-IMMSIM/index.php) was used to evaluate the efficacy of immunological simulation with the vaccine. Its mechanism is as follows, It can simulate the immune response of epitope interacting with T cell receptor. We have chosen to analyze high frequency alleles in Xinjiang, specifically HLA-A*1101, HLA-A*0201, HLA-B*5101, HLA-B*3501, HLA-DRB1*0701, and HLA-DRB1*1501.The server is capable of simulating three components of mammalian immune system: bone marrow, thymus, and lymph nodes. The intervals between the three vaccine injections were 1, 84 and 168 time-steps respectively. 1 time-steps equivalence of 8 h of real-life, the three vaccine injections was administered at intervals of 0, 28, and 56 days67. Each injection contains a dose of 1000 units75.Ultimately, the simulation parameters were set as follows: random seed 12,345, simulation volume of 50, and simulation step of 1050.
Optimization of Codon of mRNA Vaccine and in silico cloning
Firstly, step one was transform amino acid sequence into a nucleotide sequence ,which would use the online server EMBOSS Backtranseq(https://www.ebi.ac.uk/Tools/st/emboss_backtranseq/) to complete. The criteria for optimization were a Codon Adaptation Index (CAI) greater than 0.8 and a GC content between 40–70%. In the second step of the process, we utilized the JCat tool (http://www.jcat.de/Literature.jsp) to optimize the codon usage for the mRNA vaccine. We also eliminated the XHOI and BamHI restriction endonuclease sites. These efforts resulted in the acquisition of the LptD-BTuB mRNA vaccine DNA sequence according to the generated output. The next step in the in silico cloning process was to select pVAX1 as the vector. SnapGene software (https://www.snapgene.com/) was then utilized to design primers and conduct a polymerase chain reaction (PCR) with primer lengths ranging from 15 to 30 base pairs, Tm values set at 60°C, an annealing temperature of 1°C, and GC content between 40 and 60%. Finally, the LptD-BTuB mRNA vaccine DNA fragments will be inserted into the cloned plasmid using the two restriction sites that were selected.
Structure prediction for mRNA vaccine and peptides
The ViennaRNA Package 2.0's RNAfold tool (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) was utilized to predict the secondary structure of the mRNA vaccine we created. The McCaskill method was used to calculate the minimum free energy (MFE) of the expected secondary structure.
In order to determine the secondary and tertiary structures structure of the peptides, we utilized both the SOPMA tool (http://npsa-pbil.ibcp.fr/cgibin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) and the Robetta service81 (https://robetta.bakerlab.org/) were performed separately. The SOPMA will analyzes and evaluates the target protein structure for alpha helix, beta-turn, random coil, and extended strand formations. The Robetta website is capable of predicting five distinct 3D structures, which are then assessed to optimize the tertiary structure of the protein through the GalaxyRefine server (http://galaxy.seoklab.org/), thus improving the quality of the local structure. The GalaxyRefine modeling method involves ab initio calculation, refining the loop or terminus region in the first-order protein 3D model82. However, the rationality of the optimized three-level structure still needs to be verified later.
Quality evaluation of tertiary structure model
The SWISS-MODEL online service structural assessment (https://swissmodel.expasy.org/assess), is a homology modeling server that predicts predict the quality of the tertiary structural model. First, the protein that is homologous to the LptD-BTuB mRNA vaccine was located in the ExPDB database to serve as a modeling template83. ExPDB is derived from the Protein Data Bank (PDB) (https://www.rcsb.org/) and offers a distinct file for each protein chain84. Then, the two sequences were compared and replaced, and the resulting structural model was predicted, evaluated, and refined through repeated steps until achieving a satisfactory model. The final outcomes will be shown in a Ramachandran plot.
In order to enhance the reliability of the results, we utilize verification tools such as Verify 3D, ERRAT to assess the quality of the three-dimensional structure85. Meanwhile using ProSA-web server(https://prosa.services.came.sbg.ac.at/prosa.php) to evaluate and explain the potential errors in predicting the tertiary structure of proteins based on the predicted value of Z-score value86.
Molecular docking
The Toll-like receptor (TLR) family, including Toll-like receptor 4 (TLR-4) (PDB ID: 3FXI), is involved in innate immune response, and can recognize bacterial endotoxin or lipopolysaccharide-induced inflammatory response87. To ensure proper binding of the TRL4 immune cell receptor to the LptD-BTuB mRNA vaccine and its subsequent effective immune response, we will simulate the docking of immune molecules88. The HDOCK server (http://hdock.phys.hust.edu.cn/) was used to dock the LptD-BTuB mRNA vaccine peptides with TLR-4 (PDB ID: 3FXI). The resulting three-dimensional structure was visualized with PyMOL.
Molecular dynamic simulation
The iMOD server was utilized to conduct dynamic simulation analysis of the TLR-4-vacine complexes (http://imods.Chaconlab.org/), which verified the stability and motion of atoms and molecules in the vaccines. These calculations provided us with a better understanding of the atomic-level interactions and conformational changes that take place inside the vaccine when activating the TLR-4 receptor. These insights have set a foundation for the creation of more effective vaccine candidates through rational design. We submitted the PDB file format of LptD-BTuB mRNA, including atomic coordinates. Choose a coarse-grained (CG) model from three options. The Cα atom accounts for the entire residue mass, which can be explained. Then, we selected JAVA in the JSmol plug-in to conserve memory. Finally, the other parameters remain unchanged, and Normal Mode Analysis (NMA) was chosen89.
Data availability
The data was derived from public domain information: Uniprot database (https://www.uniprot.org/) and PDB library (https://www.rcsb.org/). The data that support the findings of this study are available in the methods and/or supplementary material of this article. The data that support the findings of this study are available from the corresponding author upon request. There are no restrictions on data availability. If you have any questions, please contact me.
References
Heidary, M. et al. Evaluation of brucellosis vaccines: A comprehensive review. Front. Vet. Sci. 9, 925773 (2022).
Casabuono, A. C. et al. New features in the lipid a structure of Brucella suis and Brucella abortus Lipopolysaccharide. J. Am. Soc. Mass Spectrom. 28, 2716–2723 (2017).
Liu, Z., Gao, L., Wang, M., Yuan, M. & Li, Z. Long ignored but making a comeback: A worldwide epidemiological evolution of human brucellosis. Emerg. Microbes. Infect. https://doi.org/10.1080/22221751.2023.2290839 (2023).
Hernández-Mora, G. et al. Brucellosis in mammals of Costa Rica: An epidemiological survey. PLoS ONE 12, e0182644 (2017).
Rossetti, C. A., Arenas-Gamboa, A. M. & Maurizio, E. Caprine brucellosis: A historically neglected disease with significant impact on public health. PLoS Negl. Trop. Dis. 11, e0005692 (2017).
Jiang, H., O’Callaghan, D. & Ding, J.-B. Brucellosis in China: History, progress and challenge. Infect. Dis. Poverty 9, 55 (2020).
Nagalingam, M. et al. Comparative evaluation of the immunodominant proteins of Brucella abortus for the diagnosis of cattle brucellosis. Vet. World 14, 803–812 (2021).
Di Bonaventura, G., Angeletti, S., Ianni, A., Petitti, T. & Gherardi, G. Microbiological laboratory diagnosis of human brucellosis: An overview. Pathogens 10, 1623 (2021).
Głowacka, P., Żakowska, D., Naylor, K., Niemcewicz, M. & Bielawska-Drózd, A. Brucella—virulence factors, pathogenesis and treatment. Pol. J. Microbiol. 67, 151–161 (2018).
Amjadi, O., Rafiei, A., Mardani, M., Zafari, P. & Zarifian, A. A review of the immunopathogenesis of Brucellosis. Infect. Dis. 51, 321–333 (2019).
Xu, J. et al. Sustained and differential antibody responses to virulence proteins of Brucella melitensis during acute and chronic infections in human brucellosis. Eur. J. Clin. Microbiol. Infect. Dis. 32, 437–447 (2013).
Shevtsov, A. et al. Antimicrobial susceptibility of Brucella melitensis in Kazakhstan. Antimicrob. Resist. Infect. Control. 6, 130 (2017).
Rezaei Shahrabi, A., Moradkasani, S., Goodarzi, F., Beig, M. & Sholeh, M. Prevalence of Brucella melitensis and Brucella abortus tetracyclines resistance: A systematic review and meta-analysis. Microb. Pathog. 183, 106321 (2023).
Hull, N. C. & Schumaker, B. A. Comparisons of brucellosis between human and veterinary medicine. Infect. Ecol. Epidemiol. 8, 1500846 (2018).
Elbehiry, A. et al. The development of diagnostic and vaccine strategies for early detection and control of human brucellosis particularly endemic areas. Vaccines 11, 654 (2023).
Khan, T. et al. Evaluation of the whole proteome of Achromobacter xylosoxidans to identify vaccine targets for mRNA and peptides-based vaccine designing against the emerging respiratory and lung cancer-causing bacteria. Front. Med. 8, 825876 (2021).
Naveed, M. et al. Designing mRNA- and peptide-based vaccine construct against emerging multidrug-resistant Citrobacter freundii: A computational-based subtractive proteomics approach. Medicina 58, 1356 (2022).
Litvinova, V. R., Rudometov, A. P., Karpenko, L. I. & Ilyichev, A. A. mRNA vaccine platform: mRNA production and delivery. Russ. J. Bioorg. Chem. 49, 220–235 (2023).
Hisham, Y. & Ashhab, Y. Identification of cross-protective potential antigens against pathogenic Brucella spp. through combining pan-genome analysis with reverse vaccinology. J. Immunol. Res. 2018, 1474517 (2018).
Lim, B. L. TonB-dependent receptors in nitrogen-fixing nodulating bacteria. Microbes. Environ. 25, 67–74 (2010).
Gomez, G. et al. Immunogenic and invasive properties of Brucella melitensis 16M outer membrane protein vaccine candidates identified via a reverse vaccinology approach. PLoS ONE 8, e59751 (2013).
Simpson, B. W., May, J. M., Sherman, D. J., Kahne, D. & Ruiz, N. Lipopolysaccharide transport to the cell surface: Biosynthesis and extraction from the inner membrane. Philos. Trans. R. Soc. Lond B Biol. Sci. 370, 20150029 (2015).
Gong, W. et al. Peptide-based vaccines for tuberculosis. Front. Immunol. 13, 830497 (2022).
Zhu, L. et al. Brucella suis strain 2 vaccine is safe and protective against heterologous Brucella spp. infections. Vaccine 34, 395–400 (2016).
Liu, L. et al. Research progress on live attenuated vaccine against African swine fever virus. Microb. Pathog. 158, 105024 (2021).
Perkins, S. D., Smither, S. J. & Atkins, H. S. Towards a Brucella vaccine for humans. FEMS Microbiol. Rev. 34, 379–394 (2010).
Sun, S. et al. Safety and Transcriptome analysis of live attenuated Brucella vaccine strain S2 on non-pregnant Cynomolgus monkeys without abortive effect on pregnant Cynomolgus monkeys. Front. Vet. Sci. 8, 641022 (2021).
Gheibi, A., Khanahmad, H., Kashfi, K., Sarmadi, M. & Khorramizadeh, M. R. Development of new generation of vaccines for Brucella abortus. Heliyon 4, e01079 (2018).
Mehla, K. & Ramana, J. Identification of epitope-based peptide vaccine candidates against enterotoxigenic Escherichia coli: A comparative genomics and immunoinformatics approach. Mol. Biosyst. 12, 890–901 (2016).
Yang, N. et al. Subunit vaccines for Acinetobacter baumannii. Front. Immunol. 13, 1088130 (2022).
Yang, X. et al. Progress in Brucella vaccine development. Front. Biol. 8, 60–77 (2013).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
Zhang, L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell. Mol. Immunol. 15, 182–184 (2018).
Lu, R. et al. Accurately cleavable goat β-lactoglobulin signal peptide efficiently guided translation of a recombinant human plasminogen activator in transgenic rabbit mammary gland. Biosci. Rep. https://doi.org/10.1042/BSR20190596 (2019).
Zheng, Y. et al. Predictive analysis of the number of human brucellosis cases in Xinjiang China. Sci. Rep. 11, 11513 (2021).
Ejazi, S. A. et al. Immunoproteomic identification and characterization of Leishmania membrane proteins as non-invasive diagnostic candidates for clinical visceral Leishmaniasis. Sci. Rep. 8, 12110 (2018).
Kirshina, A. et al. Effects of combinations of untranslated-region sequences on translation of mRNA. Biomolecules 13, 1677 (2023).
Ross, J. & Sullivan, T. D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood 66, 1149–1154 (1985).
Gallie, D. R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes. Dev. 5, 2108–2116 (1991).
Holtkamp, S. et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108, 4009–4017 (2006).
Brown, A., Shao, S., Murray, J., Hegde, R. S. & Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496 (2015).
Jaan, S., Zaman, A., Ahmed, S., Shah, M. & Ojha, S. C. mRNA vaccine designing using Chikungunya virus E Glycoprotein through Immunoinformatics-guided approaches. Vaccines 10, 1476 (2022).
Wang, X. et al. Strong immune responses and protection of PcrV and OprF-I mRNA vaccine candidates against Pseudomonas aeruginosa. NPJ Vaccines 8, 76 (2023).
Gustafsson, C., Govindarajan, S. & Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 22, 346–353 (2004).
Deng, A. & Boxer, S. G. Structural insight into the photochemistry of split green fluorescent proteins: A unique role for a His-Tag. J. Am. Chem. Soc. 140, 375–381 (2018).
Coffman, R. L., Sher, A. & Seder, R. A. Vaccine adjuvants: Putting innate immunity to work. Immunity 33, 492–503 (2010).
Gebre, M. S. et al. Novel approaches for vaccine development. Cell 184, 1589–1603 (2021).
Xie, C., Yao, R. & Xia, X. The advances of adjuvants in mRNA vaccines. NPJ Vaccines 8, 162 (2023).
Yu, X. et al. Validation of an HPLC-CAD method for determination of lipid content in LNP-encapsulated COVID-19 mRNA vaccines. Vaccines 11, 937 (2023).
Lopez-Martinez, E., Manteca, A., Ferruz, N. & Cortajarena, A. L. Statistical analysis and tokenization of epitopes to construct artificial Neoepitope libraries. ACS Synth. Biol. 12, 2812–2818 (2023).
Rangwala, S. H. et al. Accessing NCBI data using the NCBI sequence viewer and genome data viewer (GDV). Genome Res. 31, 159–169 (2021).
Alam, R. et al. In silico formulation of a next-generation multiepitope vaccine for use as a prophylactic candidate against Crimean-Congo hemorrhagic fever. BMC Med. 21, 36 (2023).
Zhu, Y. et al. Contriving a novel of CHB therapeutic vaccine based on IgV_CTLA-4 and L protein via immunoinformatics approach. J. Biomol. Struct. Dyn. https://doi.org/10.1080/07391102.2023.2234043 (2023).
Narang, P. K. et al. Functional annotation and sequence-structure characterization of a hypothetical protein putatively involved in carotenoid biosynthesis in microalgae. South Afr. J. Bot. 141, 219–226 (2021).
Singhal, N. et al. Efficacy of signal peptide predictors in identifying signal peptides in the experimental secretome of Picrophilous torridus, a thermoacidophilic archaeon. PLoS ONE 16, e0255826 (2021).
Nakayama, M. Antigen presentation by MHC-dressed cells. Front. Immunol. 5, 672 (2014).
Rock, K. L., Reits, E. & Neefjes, J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. 37, 724–737 (2016).
Sha, T. et al. Bioinformatics analysis of candidate proteins Omp2b, P39 and BLS for Brucella multivalent epitope vaccines. Microb. Pathog. 147, 104318 (2020).
Shen, C. et al. Allele polymorphism and haplotype diversity of HLA-A, -B and -DRB1 loci in sequence-based typing for Chinese Uyghur ethnic group. PLoS ONE 5, e13458 (2010).
Liu, J. & Zhang, W. Databases for B-cell epitopes. Methods Mol. Biol. 1184, 135–148 (2014).
Galperin, M. Y., Kristensen, D. M., Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Microbial genome analysis: The COG approach. Brief. Bioinform. 20, 1063–1070 (2019).
Naveed, M. et al. Evaluation of the whole proteome to design a novel mRNA-based vaccine against multidrug-resistant Serratia marcescens. Front. Microbiol. 13, 960285 (2022).
Asadollahi, P. et al. Immunoinformatics Insights into the Internalin A and B Proteins to Design a Multi-Epitope Subunit Vaccine for L. monocytogenes. Int. J. Pept. Res. Ther. 28, 47 (2022).
Akhtar, N., Joshi, A., Kaushik, V., Kumar, M. & Mannan, M.A.-U. In-silico design of a multivalent epitope-based vaccine against Candida auris. Microb. Pathog. 155, 104879 (2021).
Ahmad, B., Ashfaq, U. A., Rahman, M.-U., Masoud, M. S. & Yousaf, M. Z. Conserved B and T cell epitopes prediction of ebola virus glycoprotein for vaccine development: An immuno-informatics approach. Microb. Pathog. 132, 243–253 (2019).
Kim, Y., Sette, A. & Peters, B. Applications for T-cell epitope queries and tools in the immune epitope database and analysis resource. J Immunol Methods 374, 62–69 (2011).
Ud-Din, M. et al. Vaccinomics to design a multi-epitopes vaccine for Acinetobacter baumannii. Int. J. Environ. Res. Public Health 19, 5568 (2022).
Szeto, C. et al. Molecular basis of a dominant SARS-CoV-2 spike-derived epitope presented by HLA-A*02:01 recognised by a public TCR. Cells 10, 2646 (2021).
Yan, Y., Tao, H., He, J. & Huang, S.-Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 15, 1829–1852 (2020).
Saadi, M., Karkhah, A. & Nouri, H. R. Development of a multi-epitope peptide vaccine inducing robust T cell responses against brucellosis using immunoinformatics based approaches. Infect. Genet. Evol. 51, 227–234 (2017).
Li, M. et al. Design of a multi-epitope vaccine candidate against Brucella melitensis. Sci. Rep. 12, 10146 (2022).
Chen, H. et al. Influences of various peptide linkers on the Thermotoga maritima MSB8 Nitrilase displayed on the spore surface of Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 27, 64–71 (2017).
Sarkar, B., Ullah, M. A., Araf, Y., Islam, N. N. & Zohora, U. S. Immunoinformatics-guided designing and in silico analysis of epitope-based polyvalent vaccines against multiple strains of human coronavirus (HCoV). Expert. Rev. Vaccines 21, 1851–1871 (2022).
Yang, Y. et al. In silico design of a DNA-based HIV-1 multi-epitope vaccine for Chinese populations. Hum. Vaccin. Immunother. 11, 795–805 (2015).
Dey, J. et al. Designing of multi-epitope peptide vaccine against Acinetobacter baumannii through combined immunoinformatics and protein interaction-based approaches. Immunol. Res. 71, 639–662 (2023).
Booth, W. T. et al. Impact of an N-terminal Polyhistidine tag on protein thermal stability. ACS Omega 3, 760–768 (2018).
Dey, J. et al. Exploring Klebsiella pneumoniae capsule polysaccharide proteins to design multiepitope subunit vaccine to fight against pneumonia. Expert Rev. Vaccines 21, 569–587 (2022).
Sharma, N., Naorem, L. D., Jain, S. & Raghava, G. P. S. ToxinPred2: an improved method for predicting toxicity of proteins. Brief Bioinform. https://doi.org/10.1093/bib/bbac174 (2022).
Mahapatra, S. R. et al. Immunoinformatics-guided designing of epitope-based subunit vaccine from Pilus assembly protein of Acinetobacter baumannii bacteria. J. Immunol. Methods 508, 113325 (2022).
Mahapatra, S. R., Dey, J., Raj, T. K., Misra, N. & Suar, M. Designing a next-generation multiepitope-based vaccine against staphylococcus aureus using reverse Vaccinology approaches. Pathogens 12, 376 (2023).
Dey, J. et al. Designing a novel multi-epitope vaccine to evoke a robust immune response against pathogenic multidrug-resistant enterococcus faecium bacterium. Gut Pathog. 14, 21 (2022).
Ko, J., Park, H., Heo, L. & Seok, C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 40, W294-297 (2012).
Waterhouse, A. et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Fischer, M., Thai, Q. K., Grieb, M. & Pleiss, J. DWARF–a data warehouse system for analyzing protein families. BMC Bioinform. 7, 495 (2006).
Narang, P. K. et al. Genome-based identification and comparative analysis of enzymes for carotenoid biosynthesis in microalgae. World J. Microbiol. Biotechnol. 38, 8 (2021).
Ahmad, I. et al. Development of multi-epitope subunit vaccine for protection against the norovirus’ infections based on computational vaccinology. J. Biomol. Struct. Dyn. 40, 3098–3109 (2022).
Zhang, Y., Liang, X., Bao, X., Xiao, W. & Chen, G. Toll-like receptor 4 (TLR4) inhibitors: Current research and prospective. Eur. J. Med. Chem. 235, 114291 (2022).
Ismail, S. et al. Design of a multi-epitopes vaccine against hantaviruses: An immunoinformatics and molecular modelling approach. Vaccines 10, 378 (2022).
López-Blanco, J. R., Aliaga, J. I., Quintana-Ortí, E. S. & Chacón, P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 42, W271-276 (2014).
Acknowledgements
This work was supported by Youth Science and Technology Top Talent Program (2022TSYCCX0112); Xinjiang Uygur Autonomous Region Science and Technology Support Project (2022E02061); State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia Fund (SKL-HIDCA2021-JH11); Science and Technology Department of Xinjiang Uygur Autonomous Region, Outstanding Youth Project(2022D01E69); National Natural Science Foundation of China and regional science foundation projects(32160182)
Author information
Authors and Affiliations
Contributions
Juan Shi is responsible for the writing of the introduction, materials, methods and results, Yuejie Zhu is responsible for the writing of the abstract, discussion,and revising of full-text grammar,Zhengwei Yin is responsible for the analysis of the results, Yueyue He is responsible for the review of the whole article format, Yujiao Li is responsible for secondary and tertiary structure analysis of vaccine, Gulishati Haimiti is responsible for analysis silicon clone immunity, Xinru Xie is responsible for analysis mRNA secondary structure,Ce Niu is responsible for analysis molecular dynamics simulation, and Wenhong Guo modify the full text, and Fengbo Zhang is a correspondent,responsible for the review and revision of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, J., Zhu, Y., Yin, Z. et al. In silico designed novel multi-epitope mRNA vaccines against Brucella by targeting extracellular protein BtuB and LptD. Sci Rep 14, 7278 (2024). https://doi.org/10.1038/s41598-024-57793-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-57793-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.