Abstract
Voltage-gated sodium channels (NaV) are pivotal proteins responsible for initiating and transmitting action potentials. Emerging evidence suggests that proteolytic cleavage of sodium channels by calpains is pivotal in diverse physiological scenarios, including ischemia, brain injury, and neuropathic pain associated with diabetes. Despite this significance, the precise mechanism by which calpains recognize sodium channels, especially given the multiple calpain isoforms expressed in neurons, remains elusive. In this work, we show the interaction of Calpain-10 with NaV's C-terminus through a yeast 2-hybrid assay screening of a mouse brain cDNA library and in vitro by GST-pulldown. Later, we also obtained a structural and dynamic hypothesis of this interaction by modeling, docking, and molecular dynamics simulation. These results indicate that Calpain-10 interacts differentially with the C-terminus of NaV1.2 and NaV1.6. Calpain-10 interacts with NaV1.2 through domains III and T in a stable manner. In contrast, its interaction with NaV1.6 involves domains II and III, which could promote proteolysis through the Cys-catalytic site and C2 motifs.
Similar content being viewed by others
Introduction
Voltage-gated sodium channels (NaV's) are vital in initiating action potentials within excitable cells. Composing a central alpha subunit responsible for pore formation, these channels can also associate with one or two ß-subunits (ß1–4), which concurrently function as cell adhesion proteins1. The alpha subunits consist of approximately 2000 amino acids organized into four domains (I-IV) interconnected by intracellular loops. Both the N- and C-termini are situated intracellularly. Each domain encompasses six transmembrane segments (SI–S6) alongside a pivotal loop between S5 and S6 that forms the pore and constructs the selectivity filter. Within each domain, the S4 segments contribute to the voltage sensor and activation gate. The linkage between domains III and IV (linker III–IV) actively participates in the function of the inactivation gate. The linkers I–II, II–III, and the C- and N-termini seem more important for protein interactions. Nine isoforms of alpha subunits of sodium channels named NaV1.1 to NaV1.9 have been identified2. Since the discovery of the association of alpha subunits with auxiliary ß- subunits, our knowledge of the interactions of sodium channels with other proteins has grown to the point of considering that sodium channels are part of signaling macromolecular complexes3; however, the impact of these interactions on the channel function, or the function of its interacting partners is only starting to unveil. Such interactions may help to understand spatial and temporal fine-tuning of sodium channel expression. For example, NaV1.2 and NaV1.6 alpha subunits contain 86% amino acid similarity in the transmembrane domains4 and rather subtle differences in electrophysiological properties5. Also, they have a different subcellular distribution in neurons6,7, and controlling its expression during development and survival after birth seems essential. For example, in SNC8A null mice (medtg/medtg), the absence of NaV1.6 channels results in ataxia and early death of mice8, even though NaV1.1 and NaV1.2 fill the spots usually occupied for NaV1.6 and compensate for the lack of NaV1.69,10.
Here, we show evidence that Calpain-10 binds the C-terminals of NaV1.2 and NaV1.6 in vitro and in silico. The Calpain family is a set of 14 non-lysosomal calcium-dependent proteases that perform limited proteolysis to modulate the function of its targets11. Calpains exhibit four domains: domain I in the N-terminus can be proteolyzed when the calpains are active, domain II possesses the Cys-catalytic site, and domain III exhibits C2 motifs. The C-terminus containing domain IV is very divergent. Depending on the version of this domain, calpains are classified as typical or atypical. Typical calpains exhibit a penta-EF-hand domain, while atypical calpains have different versions of the C-terminal domain12. Although typical calpains, such as Calpain-1 and -2, have been reported before to proteolyse sodium channels in isolated membranes13, it is unknown if other calpains can target sodium channels. Calpain-10 is considered an atypical calpain; its C-terminus was first known as domain T because of its homology to the nematode calpain TRA-314. However, because both domain III and domain T present a beta-sandwich structure related to the calcium-binding C2 domain of phospholipase C, domain T has received several names: domain III’, C2L or C2-like, and CBSW domain (calpain-beta sandwich). In this work, after demonstrating the physical interaction between Calpain-10 with NaV1.2 and NaV1.6, we offer a structural and dynamical hypothesis to explain the interactions in atomistical terms. The docking analysis and molecular dynamics simulation showed that the C-terminus might be relevant for Calpain-10 to dock at the C-terminus of sodium channels, suggesting this process as a regulatory mechanism of neural activity.
Results
Experimental evidence of binding between Calpain-10 and NaV’s C-terminals
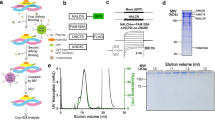
Interactions of sodium channels with proteins have proven capable of regulating channel function. We have reported before that over 50 proteins are able to bind the C-terminus of NaV1.2 in the yeast-two hybrid assay15. We screened a cDNA library from mouse brain, from which we recovered binding partners previously reported, like FHF4, Calmodulin, and mSin3B. Here we report another binding partner found in this screening. We found two clones containing 215 aa of Calpain-10 (amino acids 452 to 666), including the last 36 aa of domain III and the whole of domain T. In Fig. 1A, we show that cells expressing NaV1.2 or NaV1.6, but not negative controls (LaminA, CoREST, and Pincher), exhibit ß-galactosidase activity as a result of the interaction between the C-terminus of these channels and domains III and T of Calpain 10 encoded in clone T22–322. Another clone (T22–82) from the same YTH screening expressing calmodulin, a well-known interacting protein of sodium channels, is also shown as a positive control. To further verify this interaction, the plasmid recovered from yeast was subcloned to produce a His6-tagged version of Calpain 10 with a calculated molecular weight of 28.9 kDa (His6-Calpain10) to be used in a GST-pull down assay. Once again, GST-tagged versions of NaV1.2 and NaV1.6 could retain the His6-Calpain10, while GST alone did not (Fig. 1B). Calpain 10 scheme shown in Fig. IC depicts clone T22-322, including part of DIII and DT.
Calpain-10 binds to the C-termini of sodium channels NaV1.2 and NaV1.6 through domains III and T in Y2H assay. (A) L40 yeast expressing Calpain-10 were mated with AMR70 cells previously transfected with plasmids encoding NaV1.2 CT, NaV1.6 or a series control proteins (Lamin A, CoREST and Pincher). The interaction between transfected proteins induced expression of ß-galactosidase reporter gene. The presentation of ß-galactosidase produces blue color in presence of the hydrolyzable substrate X-gal. Only those cells expressing Calpain-10 and NaV1.2CT or NaV1.6CT could hydrolyze X-gal. As a positive control, we show the same experiment with cells expressing calmodulin. (B) In vitro binding of His6Calpain-10 to the C-terminus of NaV1.2 and NaV1.6. Recombinant proteins GST, GST-NaV1.2CT or NaV1.6CT, bound to glutathione-beads, were incubated with recombinant His6-Calp10. After extensive washing, the proteins bound to glutathione-beads were analyzed by western blot with an anti-poly-Histidinde antibody. A single band of ~ 29 kDa, matching the expected molecular weight of the His6-Calpain10, was identified, indicating that it is able to bind to GST-NaV1.2CT and GST-NaV1.6CT but does not bind to GST alone. Full length blots are shown in Supplementary Fig. S3. (C) Scheme shows the fragment of Calpain-10 identified initially as a protein capable of binding dependent sodium channels voltage corresponding mainly to the domain T. Numbers under scheme 2D of NaV's corresponding at alpha helix number.
Modeling and docking analysis
Explaining molecular mechanisms based on a peptide interaction model is valuable as it can offer an atomistic description of cellular processes, such as the role of peptides as toxins16, conjugated peptides with anticancer activity17, or antimicrobial peptides18, to name a few. In our group, we have jointly used homology modeling, docking, and molecular dynamics to model the interaction of an immunogenic peptide with the Major Histocompatibility Complex II (MHC II)19 or even RNA–Protein interaction20.
Here, we first generated 3D models for the C-terminus of NaV1.2 and NaV.16 using automatically selected templates by the comparative modeling method implemented in the Robetta server35. Then, the models were analyzed using PROCHECK, which revealed that 88.9% of residues in NaV1.2 and 90.2% of NaV1.6 were in the most favored regions of the Ramachandran plot, and 10.1% and 8.8%, respectively, of residues were in the additional allowed regions. It is worth mentioning that the most distal part of both sequences are de novo predictions since the available templates cover roughly the first half or less of the sequence. It is also important to remember that the C-terminus of sodium channels becomes more variable among isoforms towards the second half of the sequence (see Supplementary Fig. S4).
Another important novelty of this work is the use of the whole Calpain-10 model to study protein–protein interaction since previous studies on Calpain-10 molecular dynamics and docking have focused on the catalytic domain and its interaction with inhibitors, such as calpastatin21,22. Instead, the roles of domains III and T have been overlooked despite their potential for binding targets. Here we used a template from Alphafold and refined it with Robbeta. The resulting model has a 91.7% of amino acids within the most favored region and 7.9% in the allowed region.
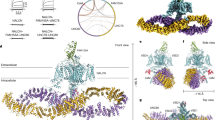
The HDock and Cluspro 2.0 programs provided the complexes of Calpain-10 with the NaV1.2CT and NaV1.6CT, using a cutoff of 5 Å. Later, the interactions in each complex were found by PDBsum. We analyzed the 20 more energetically favorable models, ten from each docking program. Then, we determined the most frequent interacting amino acids to obtain a more representative interaction model. Noteworthy, all of the docking poses of NaV1.6 were different from those of NaV1.2. This result was unexpected, given the similarity of a good part of the amino acid sequences (see Supplementary Fig. S4). Figure 2A,B presents a representative model that summarizes these interactions.
Molecular recognition of C-terminus of NaV1.2 and NaV1.6 with Calpain-10. Docking of Calpain-10 to NaV1.2CT (A) and NaV1.6CT (B) is shown as a 3D model. The domains of Calpain-10 were in different colors: Domain II in Cyan color, domain III in green color, and domain T in red color. NaV1.2 and NaV1.6 structure is shown in yellow (non interacting surfaces) and dark blue, while in blue color are the interactions. Amino acids relevant to docking on both proteins are listed right into the 3D models. Hydrogen bonds (blue solid lines), salt bridges (red solid lines), and non-bonded contacts (orange dashed lines) are shown. Amino acids color code: positive (blue), negative (red), neutral (green), aliphatic (gray), aromatic (purple), Pro and Gly (orange), and Cys (yellow). Amino acids of NaV1.2 and NaV1.6 correspond to amino acids 1778-2006 of NaV1.2 (Uniprot: B1AWN6; NCBI ID: BAC27748.1) and 1765-1978 of NaV1.6 (Uniprot: Q9WTU3; NCBI ID: NP_035453.2). (C) Secondary structure schematic highlights the regions more relevant to the interaction between the C-terminal ends of the NaV1.2 and NaV1.6 channels and the characteristic domains of Calpain-10.
Docking of NaV1.2CT with Calpain-10
We docked the C-terminus of NaV1.2 with amino acids 452 to 666 of mouse Calpain-10 and analyzed the amino acids that participate in the interaction between these two proteins. The PDBsum interaction analysis revealed that eight salt bridges, 23 hydrogen bonds, and 318 nonbonded contacts were formed between NaV1.2CT with Calpain-10. In these interactions, 36 residues of NaV1.2CT and 47 of Calpain-10 are involved, for which four double hydrogen bonds were formed between Thr1784–Val598, Glu1785–Lys469, Arg1915-Glu657 and Asp2000-Lys665. The eight salt bridges are formed between residues Glu1785-Lys469, Glu1869-Arg552, Arg1915-Glu649, Arg1915-Glu657, Arg1918-Glu657, Asp2000-Lys665, Lys2006-Glu649, and Lys2006-Glu657 (Fig. 2A). Of the 318 nonbonded contacts, 128 occurred on the atoms of the first nine amino acids of the C-terminus of NaV1.2; at these same residues, two salt bridges occurred. The eight salt bridges form hydrogen bonds and nonbonded contacts, suggesting an important role in promoting this interaction. The docking score for the NaV1.2CT/Calpain-10 complex was − 227.56 and a ΔG of -20 kcal/mol with a confidence score of 0.8251 (Table 1).
Docking of NaV1.6CT with Calpain-10
Following the same strategy depicted above, we analyzed the interaction between Calpain-10 and NaV1.6. The interaction analysis revealed that the NaV1.6CT/Calpain-10 interaction is constituted of 11 salt bridges, 36 hydrogen bonds, and 407 nonbonded contacts, in turn, formed by 46 residues from the C-terminus of NaV1.6 and 57 residues of Calpain-10 (Fig. 2B). The hydrogen bonds formed by the NaV1.6CT/Calpain-10 complex include five double bonds between the residues Glu1784-Ser400, Arg1905-Glu507, Glu1802-Arg601, Arg1910-Leu651 and Arg1916-Gln-656. On the other hand, 9 of the 11 salt bridges also form hydrogen bonds and non-bonded contacts. These are comprised between Glu1869-Arg386, Thr1798-Arg601, Ala1873-Arg386, Glu1869-Arg388, Arg1905-Glu507, Glu1802-Arg601, Arg1907-Glu507, Lys1917-Glu649 and Lys1932-Glu657, while Glu1869-Arg388 and Lys1917-Glu649 form only salt bridges and non-bonded contacts. The docking score for the NaV1.6CT/Calpain-10 complex was − 308.57 and a ΔG of − 21.1 kcal/mol with a confidence score of 0.9597 (Table 1).
Notably, a key distinction arises in the interaction mechanisms of NaV1.2CT/Calpain-10 and NaV1.6CT/Calpain-10. While the former utilizes its initial nine N-terminal residues for interaction, the latter does not involve these residues. Furthermore, although in both cases the interaction domain encompasses the alpha helix 6, NaV1.6CT/Calpain-10 also involves five extra amino acids (GFICRK) unique to the NaV1.6 sequence, further demarcating the divergent characteristics between these channels. A schematic representation of these differences is shown in Fig. 2C.
Geometrical and dynamical parameters of Calpain-10, NaV1.2CT/Calpain-10, and NaV1.6CT/Calpain-10
To investigate the protein stability, residue fluctuations, and dynamic behavior, 50 ns of MD simulations of the Calpain-10, NaV1.2CT/Calpain-10, and NaV1.6CT/Calpain-10 were performed (Fig. 3). The RMSD of the backbones, as a parameter of structural stability during the simulations, was calculated and compared with the reference structure at t = 0. As shown in Fig. 3, Calpain-10 alone seems stable (RMSD = 0.677 ± 0.087 nm), and NaV’s gain stability through its interaction with Calpain-10. RMSDs for NaV1.2CT/Calpain-10 complex and NaV1.2CT are shown in Fig. 3B. After an initial increase of RMSD during the first 12 ns, both simulations reach a fluctuant stability around 0.842 ± 0.168 nm and 0.892 ± 0.286 nm, respectively. This result indicates minor conformational changes in NaV1.2CT when bound to Calpain-10. It also indicates a stable binding over 40 ns of NaV1.2/Calpain-10; actually, the complex seems more stable than the NaV1.2CT alone beyond this point. NaV1.6 presented higher RMSD values (2.44 ± 0.445 nm) than NaV1.2 and also higher values than NaV1.6CT/Calpain-10 complex (1.242 ± 0.248 nm) and Calpain-10 alone (Fig. 3B). However, its binding with Calpain-10 resulted in lesser RMSD values for the complex NaV1.6CT/Calpain-10 compared to NaV1.6CT alone, roughly by 50%. This result indicates that the interaction of the C-terminus with Calpain-10 provides them with greater stability. Furthermore, the NaV1.2CT/Calpain-10 complex starts from a more stable conformation than the NaV1.6CT/Calpain-10 complex.
RMSD of Calpain-10 and bound to NaV1.2CT or Nav1.6CT. (A) Calpain-10 RMSD value remains stable through the simulation. (B) NaV1.2CT RMSD values are more stable when in complex with Calpain-10 than for NaV1.2 alone; (C) RMSD values for NaV1.6CT are higher than those observed for NaV1.2 suggesting a more flexible structure. Again a reduction of the RMSD for the NaV1.6/Calpain 10 is noticeable, indicating a more stable structure.
The differences in RMSD behavior are consistent with the evolution in the number of hydrogen bonds and the number of electrostatic interactions at the initial structure and the final of the NaVs-Calpain-10 complexes. For NaV1.2/Calpain-10, although there were fluctuations in quantity, the number of these remained relatively constant when the initial and final structures were compared, approximately 620 HBonds and 8 to 6 salt bridges. However, for NaV1.6/Calpain-10, the HBonds were initially 650 and were reduced to 620 in the final structure, and the number of salt bridges almost doubled from 7 to 13 during the simulation.
The RMSF of the Cα atoms was calculated to analyze the residues' fluctuations. To clarify how the results are shown, we must mention that Fig. 4B,C show a number greater than 800, representing the sum of residues in each C-terminus of NaV’s with Calpain-10. For Calpain-10, as observed in the RMSF, its structure remains stable throughout the MD; we found three peaks that exceed 0.4 nm. The first is in domain III (Val379-Asp410), while the second and the third are in the T domain between Ile551-Leu595 (Fig. 4A). In Fig. 4B, the RMSF of the complex NaV1.2/Calpain-10 is shown. The RMSF values for NaV1.2CT indicate more flexibility than the observed for Calpain-10 since most structures show values above 0.4 nm. However, these values decrease when it is in complex with Calpain-10, except for the α6 helix and the zone of most significant divergence between sodium channels (Ser153-Leu179), which can be considered a zone of important movements for the binding with Calpain-10 (Fig. 4c).
RMSF of Calpain-10 and bound to NaV1.2CT or NaV1.6CT. Calpain-10 RMSF is presented in (A). Notice a more pronounced variation towards the N- and C-termini. Consistent with the RMSD, NaV1.2CT(B) individual residue fluctuations are smaller compared with that of NaV16CT(C). In both cases these fluctuations are reduced by interaction with Calpain-10 (olive green). Notice that Calpain-10 RMSF profile remains basically unchanged by the interaction with either NaV1.2CT or NaV1.6CT.
The RMSF for NaV1.6CT shows a profile similar to that observed for NaV1.2CT up to Leu145, consistent with the 93% similarity in amino acid sequence between them; however, the sequences are divergent from this residue. The residues with the highest flexibility in the NaV1.6 complex are located from Ile150 and up to Cys214. Interestingly, this region is involved in the interaction with Calpain-10 (Fig. 4C) and exhibits residues forming hydrogenous bridges, unconnected contacts, and salt bridges (Lys168-Glu657; Arg152-Glu657; Lys168-Lys665) (Fig. 2B). Thus, the interaction with Calpain-10 stabilizes the sodium channel structure mainly within the highly conserved region, but it does not reduce the peaks observed in the divergent region. It seems to increase the mobility of the last α-helix in NaV1.6CT (Pro188-Cys214). Sodium channels are reported to bind calmodulin through an IQ motif in the α6 helix of NaV1.2 C-terminus. Although the binding surface for Calpain-10 does not overlap with the IQ motif, the surface involves part of the alpha helix needed to bind calmodulin. Therefore, some competition might be expected. In summary, at the far left of the graph of 4B and C, we observed a comparison of the NaV1.2CT and NaV1.6CT free or bound to Calpain-10. Thus, we conclude that this interaction stabilizes the C-terminal ends, significantly affecting NaV1.6CT.
Secondary structure evolution
The evolution of the secondary structure as a function of time and the number of residues for Calpain-10, NaV1.2CT, and NaV1.2CT/Calpain-10 are shown in Supplementary Fig. S1. The secondary structure of Calpain-10 is well preserved during the MD simulation (Supplementary Fig. S1A) except for the amino and carboxyl-terminal ends. NaV1.2CT loses part of the α6 helix, which contains the IQ domain important for calmodulin binding (Supplementary Fig. S1B). Conversely, in the NaV1.2CT/Calpain-10 complex, the N- and C-terminus of α6-helix seem to be structurally more robust; the same is more evident in the α-helices 4, 5, suggesting more structural stability (Supplementary Fig. S1C). Also, complex NaV1.2CT /Calpain-10 induces partial unfolding of Calpain-10 in its α-helices 4,5, 7 (DII), and in the β-sheets 18 and 19 (DIII), while aid to stabilize both the N-and C-termini. In contrast to NaV1.2CT, the MD of NaV1.6CT alone (Supplementary Fig. S2B) shows that its α-helices are more preserved along the simulation. However, the interaction with the CT’s seems to destabilize the structure of Calpain-10, specifically α-helices 9, 16 y 17 (Supplementary Fig. S2). In contrast, the N- and C-terminus of Calpain-10 tend to be more stable when in the complex with NaV1.6, although domain II α-helices and ß-strands remain the same. The loop joining PC1 and PC2 (catalytic subdomains) seem to be less rigid.
These results indicate that the secondary structure of Calpain-10 undergoes more changes when it is in complex with NaV1.2 than with NaV1.6, suggesting a different behavior with its targets. Based on the results of the RMSD, RMSF, and secondary structure evolution and describing the effect on Calpain-10 due to its binding to NaVCTs, we can observe that the interaction with NaV1.6 does not alter its secondary structure, even though Calpain-10 has a significant stabilizing effect on this C-terminus. Interestingly, consolidating Calpain-10's interaction with NaV1.2CT costs the protein a portion of its secondary structure.
Discussion
Here, we show evidence that Calpain-10 binds to the carboxyl termini of NaV1.2 and NaV1.6. First, the yeast two-hybrid assay found that a region comprising 36 amino acids of domain III and the whole of domain T of Calpain-10 could interact with the C-termini of sodium channels NaV1.2 and NaV1.6. The ability of this region to bind sodium channels was then confirmed in a pull-down assay. Given these experimental findings, we use 3D homology models to predict macromolecular docking and molecular dynamics of the interaction. Thus, we found that a surface spanning the sixth alpha helix of NaV1.2 and the seventh helix in NaV1.6 may provide several interactions with domains III and T of Calpain-10.
A novelty of this work is the use of the whole Calpain-10 model to study protein–protein interaction since previous studies on Calpain-10 molecular dynamics and docking have focused on the catalytic domain and its interaction with inhibitors, such as calpastatin21,22. Instead, the roles of domains III and T have been overlooked despite their potential for binding targets. We found that Calpain-10 binding of the carboxy terminus of NaV1.2 and NaV1.6 has a favorable free energy consistent with the results of the GST-pulldown assay. Likewise, the RMSD analysis indicated that the carboxy-terminus of both NaV1.2 and NaV1.6 become more stable upon interaction with Calpain-10, which explains why the complex lasts long enough to recruit the transcription machinery to the reporter genes HIS3 and beta-galactosidase in the YTH assay.
Recent crystallographic studies of heterologously expressed channels often resort to the removal of linkers I-II, II-III, and C-terminus to improve expression, thermostability, and sample homogeneity compared with full-length wild type channels23,24. Although it results in a channel with biophysical properties similar to full-length channels, the need to remove intracellular amino acid chains that account roughly 50% of the protein remarks the importance of these regions to downregulate expression of channels in non-excitable cells and the need to study their interactions with other proteins. Establishing the interactions of the C-terminus with multiple proteins may help refine the search for an allosteric modulator specific to the interacting partner, as already assayed for FGF1425. We selected NaV1.2 and NaV1.6 to search for new binding partners because these two channels are the most abundant voltage-gated channels in the adult brain, and despite their overall similarity, there is a clear segregation of their distribution at subcellular level in neurons6,7. We selected the C-terminus region as bait in the Y2H assay because of its divergence among sodium channels, hoping to find binding partners that showed a preference for a particular type of sodium channels. However, we have found that almost every binding partner for NaV1.2 can also bind NaV1.6. Nevertheless, our analysis indicates the interaction between Calpain-10 and NaV1.6 differs from that with NaV1.2, possibly due to differences in the c-terminal ends, despite the high identity between the NaV’s (Supplementary Fig. S4).
A most interesting observation is that two amino acids (Arg 1870 and Leu 1863) of NaV1.6 relevant to the interaction with calpain also have naturally occurring variants associated with epilepsy26,27,28. Mainly, Arg1872 of human NaV1.6, Arg1870 in mice (see Supplementary Fig. S5) has been reported to be a hotspot for missense mutations26,27,28 related to several forms of epilepsy, including Arg1872Trp, Arg1872Gln and a mutant that produces a truncated C-terminus. The impact of these mutations on binding not only Calpain-10 but other known interactors, such as calmodulin and FGF14 (Fibroblast Growth Factor 14), may help better understand the changes in excitability and its relationship to the survival of these cells. The binding of FGF14 to the C-terminus region of NaV1.6 has also been studied to an atomistic level29. Notably, charged residues such as Arginine and Glutamate, and an uncharged polar residue such as Serine participated significantly in the interaction, as reported by Ali et al.29, for the FGF14/NaV1 complex. 6. We found three residues relevant to Calpain-10 binding also interact with FGF14 (Ser1836, Arg1890 and Ile1884). Therefore, some degree of competition to bind this region may be expected, as it happens with calmodulin, which has also been found in association with the C-terminus of all types of sodium channels through an IQ motif that, in NaV1.6, becomes an LQ motif. In fact, in our YTH assay, calmodulin was the most frequently binding partner found (over 400 clones!), followed by mSin3B and FGF14 (See Supplementary Table S1).
Early experiments have shown that calpains do not affect the activity of the kinetics of sodium channel NaV1.5 as other proteases do30, while in chromaffin cells (expressing mainly NaV1.7), the calpain inhibitor calpastatin prevented a Ca-induced reduction of [3H]STX binding sites31. These early findings suggest that not all sodium channel alpha subunits interact with calpains. The docking analysis herein indicates that the first nine residues of NaV1.2 (ENFSVATEES) are relevant for the interaction with Calpain-10. Surprisingly, although this sequence is also present in NaV1.6, our modeling predicts that this set of residues is not involved in the interaction with Calpain-10, instead a cluster of residues from the most variable region 1912–1918 (GFICRK) becomes highlighted (see Supplementary Fig. S4), especially Arg1916 establishes four hydrogen bonds to residues 655, 656 and 659 in Calpain-10 domain T. This observation also highlights the relevance of the divergent sequences found in the carboxy-terminus of the sodium channel, where the sequence of amino acids is highly conserved through the first 180 amino acids, but the rest of the amino acid chain is highly divergent both in length and sequence among sodium channels. Further analysis of every NaV would be necessary to determine if Calpain-10 can regulate the members of the sodium channel family differentially.
On the other hand, little is known about the physiological effects of partial proteolysis of sodium channels by calpains. In vitro studies on calpain-mediated proteolysis have led to conclude that calpains targets brain sodium channels, or more specifically NaV1.2 and NaV1.1, in models of injury, such as brain injury, stretch injury, and ischemia13. However, these models do not allow the detail of the particular contribution of each calpain expressed in the neurons, which would be essential to define since there are at least five calpains expressed in the mouse brain, calpains 1, 2, 5, 7, and 1032. Other efforts employ purified Calpain-1 to assess its ability to proteolyse sodium channels, but more information should be obtained regarding other calpains. Additionally, these studies use calcium concentrations that are hard to reach in physiological conditions13,33,34. In a model of spinal cord injury (SCI), an increase in sodium persistent current could be associated with partial cleavage of sodium channels by calpains and generation of a ~ 120 kDa fragment34. Similarly, NaV1.6 exposed to Calpain-1 degradation, in HEK293 cells, increased persistent current, indicating a gain of function after proteolysis. Also, some authors have suggested that the 120 and 160 kDa fragments of NaV1.2 produced by calpains remain associated for some time before being withdrawn from the membrane13, but its effect on current is unknown. However, as we now realize, Calpain-10 might interact differently with either NaV1.2 or NaV1.6 and thus differ from the interaction with Calpain-1. Therefore, more studies should be performed before drawing any conclusions.
The interaction between Calpain-10 and sodium channels may be especially interesting in the context of diabetes and the development of neuropathic pain, a common complication of diabetes. In most ß-cell types studied to date, NaV1.3 and NaV1.7 appear to play different roles, and in human ß-cells alpha-subunits NaV1.7 and NaV1.6 are present35. Also, it has been reported that sodium channels inhibitor carbamazepine positively affects β-cell survival and insulin production36. On the other hand, some gene variants of Calpain-10 have been implicated as risk factors for the development of type 2 diabetes37, and diabetic patients exhibit an increase in Calpain-10 expression in B-cells38. A down-regulation of sodium channels activity, mediated by Calpain-10 proteolysis, may be part of the standard mechanisms of regulation of ß-cells excitability, thus indirectly regulating calcium entry through voltage-gated calcium channels and insulin release. Interestingly, Calpain-10 expression is also elevated in glial cells in patients with Alzheimer's disease39, where sodium channel NaV1.6 is the predominant isoform40. Docking analysis indicates that the binding between Calpain-10 and NaV1.2 or NaV1.6 is different, making a contact surface rather unique that may be helpful later on to design inhibitors targeting specific protein–protein interactions. Docking analysis with other sodium channels may help to refine drug design. Likewise, docking analysis of other intracellular chains may further our understanding of the interaction between sodium channels and calpains.
Methods
Yeast two-hybrid assay (Y2H)
The procedure followed for Y2H screens was described before by Vega et al.15. For the initial screen, yeast cells of strain L40 were co-transfected with the bait plasmid pSN12CT together with a commercial plasmid library derived from adult mouse brain cDNA in fusion with GAL4 activation domain (Clontech, catalog #ML4008AH). Transfected cells were plated on -his -leu -trp medium, on which L40 yeast cells are able to grow only if they contain both bait and prey plasmids and if the LexA/GAL4 activator is formed by interaction of bait and prey proteins. After 3 days at 30 °C, white colonies > 2 mm in diameter were picked and replated on the triple-deficient medium. Colonies were tested for LexA-driven β-galactosidase activity, after 3 rounds of replating and testing for ß-galactosidase activity, positive colonies were released of trophic pressure to reproduce the bait plasmid, by plating them in -Leu medium, resulting prey-only cells. To determine specificity of prey interaction with sodium channels, these cells were then crossed with mating-proficient AMR70 yeast cells transfected with negative control bait-plasmids, or with the pSN12CT or pSN16CT plasmid to reconfirm the original interaction. Colonies were then tested for β-galactosidase activity, as an index of bait-prey interaction. Afterwards, prey plasmids were recovered and sequenced.
Plasmids
Plasmids pSN12CT and pSN16CT15 contain bases 5512–6198 (NM_001099298.3) and 5293–5934 (AF049617) of NaV1.2 and NaV1.6 coding regions, corresponding to amino acids 1778-2006 and 1765-1978 of the respective α-subunits. They express the C-terminus of NaV1.2 or NaV1.6 in frame with the DNA-binding domain of LexA in two-hybrid bait plasmid pSTT91. Bait plasmids were confirmed not to have endogenous GAL4 activity when expressed in yeast. Negative-control bait plasmids encoding LexA-LaminA, LexA-CoREST and LexA-Pincher. C-terminal cDNAs were also subcloned into pGEX to produce GST-tagged versions of NaV1.2CT and NaV1.6CT (plasmids pGN12CT and pGN16CT). Two prey plasmids recovered from yeast matched bases 1819–2877 of mouse Calpain 10 (NM_011796.2), encoding amino acids 452–666 of sequence NP_035926.2. This partial calpain-10 sequence was subcloned into pET-28 plasmid to produce a 6His-tagged version (pEcalp10 plasmid). Full-length mouse Calpain-10 cDNA was amplified by PCR from a plasmid obtained from the Mammalian Gene collection, MGC-13731, IMAGE: 4159512, EcoRI and BamHI sites were introduced into the amplification primers to clone into pEGFP-C1(Clontech).
Glutathione S-transferase (GST) pull-down assay
Detailed explanations about the GST-pull down assay have been published before15. GST, GST-NaV1.2CT, and GST-NaV1.6CT proteins were obtained in E. coli BL21 cells transformed with pGEX, pGN12CT, and pGN16CT, respectively. Cells were induced with 1 mM IPTG for 4 h at 37 °C. Then lysed by snap freezing and digestion with lysozyme, DNA was degraded by sonication. Glutathione sepharose beads were incubated with the lysate for 1 h at 4 °C and then washed extensively with ice-cold binding buffer (50 mM Tris–HCl, pH 7.5, 120 mM NaCl, 2 mM EGTA, 0.1% triton X-100, 2 mM DTT). BL21 cells transformed with pECalp10 were used to produce a His6-tagged recombinant Calp10 (His6-Calp10) expressing a, and then lysed as described above. Beads preloaded with GST, GST-NaV1.2CT, or GST-NaV1.6CT were incubated with the Calpain lysate for 1 h at 37 °C, in presence or absence of calcium. Beads were washed extensively with binding buffer and resuspended in 2X Laemmli reducing sample buffer (RSB). Pulled-down proteins were resolved by standard SDS-PAGE, electro blotted, and then subjected to western blot with mouse anti-polyhistidine antibody (1:6000, Sigma Aldrich H1029) and HRP-conjugated anti-mouse IgG secondary antibody (Santa Cruz Biotechnology). Blocking and incubation with antibodies were carried out in blotto (20 mM Tris–HCl, 150 mM NaCl, 4% non-fat milk, pH8.0). Immunoreactive bands were visualized by chemiluminescence and captured in film. After exposure, the film was hand-developed and digitized with a Canon PowerShot G5. Digitized images of three different experiments, including the blot shown in Fig. 1, are presented in full length in Supplementary Fig. S3.
Molecular modeling
3D models of the mouse Calpain-10 (with NCBI Ref. Seq NP_035926.2), NaV1.2, and NaV1.6 (BAC27748.1, NP_035453.2 respectively) were constructed by homology in the Robetta server41. The mouse Calpain-10 model in full (666 amino acids), based on a mouse Calpain-10 structure (ID: AF-Q9ESK3-F1) predicted in Alphafold42, while NaV1.2 and NaV1.6 were modeled only by the cytosolic C-terminals, NaV1.2 model includes 229 amino acids (1778-2006), and NaV1.6 includes 214 amino acids (1765-1978), corresponding to the same amino acids expressed in the Y2H and the GST-pulldown assays. The structures of C-termini of NaV1.2 and 1.6 were constructed using partial crystallographic models of the human NaV1.2, 1.4, and 1.5 C-termini as templates (PDB: 4JPZ, 6MBA, and 2KBI), selected automatically by Robetta. To assess the geometry and restriction violations of these protein structures we use PROCHECK free software43, thus we calculate the degree of agreement of the model structures with the experimental data, and the quality of its geometric properties.
Protein–protein coupling of NaV1.2CT and NaV1.6CT with Calpain-10 were performed with ClusPro 2.0 and the HDOCK server (http://hdock.phys.hust.edu.cn/), which is based on a search ensemble through homology, template-based modeling, structure prediction, macromolecular docking, biological information incorporation, and data-management works for a robust and rapid protein–protein coupling. With input information on the receptor and ligand molecules (either amino acid sequences or structures from the protein databank), the server automatically predicts their interaction through a hybrid template-based and templateless-matching algorithm44. The HDOCK server globally samples all possible binding modes between the two proteins through an algorithm based on the fast Fourier transformation45. Then, all sampled binding modes were evaluated using the iterative knowledge-based scoring function ITScorePP46. Finally, the binding energy assessed the macromolecules' binding mode and classified them according to their coupling energies.
The docking model with the lowest energy score and the highest ligand root mean square deviation (RMSD) was selected for analysis of binding energy (Kd) scores using the PRODIGY server47. PRODIGY is a robust predictive system that uses the structural properties of protein–protein interactions, the number of interfacial contacts, and non-interacting surfaces to calculate the binding affinity of proteins48. In addition, the residual interactions of the three-dimensional model of the protein complexes were analyzed via the PDBsum server (http://www.ebi.ac.uk/pdbsum). Bound and unbound interacting residues between protein–protein interactions were examined.
Molecular dynamics simulation
MD simulations were performed through the GROMACS 5.1.4 package49 using OPLS-aa force fields and the SP3 water model50. A dodecahedral box with 2 Å of freedom on the sides was utilized. The system was electrically neutralized with sodium ions. The Particle Mesh Ewald (PME) algorithm calculated the long-range electrostatic interaction. Subsequently, the energy minimization of all systems was computed using the steepest descent algorithm. The systems were then equilibrated using NVT and NPT with 400 ps steps, respectively, using a V-rescaled Berendsen thermostat and a Parrinello-Rahman NPT array progressively directed the equilibration of each system and the Nose–Hoover algorithm maintained temperature51,52. During NPT equilibrium, the Parrinello-Rahman barostat maintained pressures at 1 atm while the systems' heating gradually increased from 0 to 310 K. The MD simulation was conducted for the complexes in 25 ns. As a part of the analysis, we calculated root mean square deviation (RMSD), root mean square fluctuation (RMSF) of the Cα backbone, the secondary structure and the H-bonds evolution of the proteins using Gromacs tools. To estimate the electrostatic contacts, we calculate the number of interactions in the initial and final structure, using PDBsum.
References
Hull, J. M. & Isom, L. L. Voltage-gated sodium channel β subunits: The power outside the pore in brain development and disease. Neuropharmacology 132, 43–57 (2018).
Catterall, W. A., Goldin, A. L. & Waxman, S. G. Voltage-gated sodium channels (NaV) in GtoPdb v.2023.1. IUPHARBPS Guide Pharmacol. CITE 2023, (2023).
Catterall, W. A. Signaling complexes of voltage-gated sodium and calcium channels. Neurosci. Lett. 486, 107–116 (2010).
Schaller, K., Krzemien, D., Yarowsky, P., Krueger, B. & Caldwell, J. A novel, abundant sodium channel expressed in neurons and glia. J. Neurosci. 15, 3231–3242 (1995).
Safo, P. et al. Distinction among neuronal subtypes of voltage-activated sodium channels by μ-conotoxin PIIIA. J. Neurosci. 20, 76–80 (2000).
Boiko, T. et al. Functional specialization of the axon initial segment by isoform-specific sodium channel targeting. J. Neurosci. 23, 2306–2313 (2003).
Trimmer, J. S. & Rhodes, K. J. Localization of voltage-gated ion channels IN mammalian brain. Annu. Rev. Physiol. 66, 477–519 (2004).
Burgess, D. et al. Mutation of a new sodium channel gene, SenSa, in the mouse mutant ‘motor endplate disease’. 10, (1995).
Van Wart, A. & Matthews, G. Impaired firing and cell-specific compensation in neurons lacking Nav 1.6 sodium channels. J. Neurosci. 26, 7172–7180 (2006).
Vega, A. V., Henry, D. L. & Matthews, G. Reduced expression of Nav1.6 sodium channels and compensation by Nav1.2 channels in mice heterozygous for a null mutation in Scn8a. Neurosci. Lett. 442, 69–73 (2008).
Spinozzi, S., Albini, S., Best, H. & Richard, I. Calpains for dummies: What you need to know about the calpain family. Biochim. Biophys. Acta BBA Proteins Proteomics 1869, 140616 (2021).
Ono, Y., Saido, T. C. & Sorimachi, H. Calpain research for drug discovery: challenges and potential. Nat. Rev. Drug Discov. 15, 854–876 (2016).
von Reyn, C. R. et al. Calpain mediates proteolysis of the voltage-gated sodium channel α-subunit. J. Neurosci. 29, 10350–10356 (2009).
Horikawa, Y. et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 26, 163–175 (2000).
Vega, A. V., Avila, G. & Matthews, G. Interaction between the transcriptional corepressor Sin3B and voltage-gated sodium channels modulates functional channel expression. Sci. Rep. 3, 2809 (2013).
Sanches, K., Wai, D. C. C. & Norton, R. S. Conformational dynamics in peptide toxins: Implications for receptor interactions and molecular design. Toxicon 201, 127–140 (2021).
Rico, M. I., Lebedenko, C. G., Mitchell, S. M. & Banerjee, I. A. Molecular dynamics simulations, docking and MMGBSA studies of newly designed peptide-conjugated glucosyloxy stilbene derivatives with tumor cell receptors. Mol. Divers. 26, 2717–2743 (2022).
Peptides, A. Methods and protocols Vol. 1548 (Springer, 2017).
Carrillo-Vazquez, J. P. et al. A continuous peptide epitope reacting with pandemic influenza AH1N1 predicted by bioinformatic approaches: Immunogenic Peptide against Pandemic Influenza AH1N1. J. Mol. Recognit. 28, 553–564 (2015).
Ospina-Villa, J. D. et al. Importance of amino acids Leu135 and Tyr236 for the interaction between EhCFIm25 and RNA: a molecular dynamics simulation study. J. Mol. Model. 24, 202 (2018).
Romo-Mancillas, A., Lemus, R., Pérez-Estrada, R., Kuribreña-Romero de Terreros, F. & Domínguez-Ramírez, L. Molecular dynamic simulations of the catalytic subunit of calpains 1, 2, 5, and 10: Structural analysis with an aim toward drug design. Chem. Biol. Drug Des. 93, 38–49 (2019).
da Silva, R. C., de Alencar, N. A. N., Alves, C. N. & Lameira, J. Analysis of the structure of calpain-10 and its interaction with the protease inhibitor SNJ-1715. Comput. Biol. Med. 43, 1334–1340 (2013).
Jiang, D. et al. Structure of the cardiac sodium channel. Cell 180, 122-134.e10 (2020).
Li, Y. et al. Structure of human NaV1.6 channel reveals Na+ selectivity and pore blockade by 4,9-anhydro-tetrodotoxin. Nat. Commun. 14, 1030 (2023).
Dvorak, N. M., Wadsworth, P. A., Wang, P., Zhou, J. & Laezza, F. Development of allosteric modulators of voltage-gated Na+ channels: A novel approach for an old target. Curr. Top. Med. Chem. 21, 841–848 (2021).
Trump, N. et al. Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. J. Med. Genet. 53, 310–317 (2016).
Larsen, J. et al. The phenotypic spectrum of SCN8A encephalopathy. Neurology 84, 480–489 (2015).
Wagnon, J. L. et al. Pathogenic mechanism of recurrent mutations of SCN8A in epileptic encephalopathy. Ann. Clin. Transl. Neurol. 3, 114–123 (2016).
Ali, S. R., Singh, A. K. & Laezza, F. Identification of amino acid residues in fibroblast growth factor 14 (FGF14) required for structure-function interactions with voltage-gated sodium channel Nav1.6. J. Biol. Chem. 291, 11268–11284 (2016).
Benz, I. et al. Two types of modified cardiac Na + channels after cytosolic interventions at the α-subunit capable of removing Na + inactivation. Eur. Biophys. J. 25, 189–200 (1997).
Kobayashi, H. et al. Regulation of voltage-dependent sodium channel expression in adrenal chromaffin cells. Ann. N. Y. Acad. Sci. 971, 127–134 (2002).
Singh, R. et al. Calpain 5 is highly expressed in the central nervous system (CNS), carries dual nuclear localization signals, and is associated with nuclear promyelocytic leukemia protein bodies*. J. Biol. Chem. 289, 19383–19394 (2014).
Von Reyn, C. R., Mott, R. E., Siman, R., Smith, D. H. & Meaney, D. F. Mechanisms of calpain mediated proteolysis of voltage gated sodium channel α-subunits following in vitro dynamic stretch injury: Mechanisms of calpain mediated NaCh proteolysis. J. Neurochem. 121, 793–805 (2012).
Brocard, C. et al. Cleavage of Na+ channels by calpain increases persistent Na+ current and promotes spasticity after spinal cord injury. Nat. Med. 22, 404–411 (2016).
Zhang, Q. et al. Na+ current properties in islet α- and β-cells reflect cell-specific Scn3a and Scn9a expression. J. Physiol. 592, 4677–4696 (2014).
Szabat, M. et al. High-content screening identifies a role for Na+ channels in insulin production. R. Soc. Open Sci. 2, 150306 (2015).
Horikawa, Y. Calpain-10 (NIDDM1) as a susceptibility gene for common type 2 diabetes. Endocr. J. 53, 567–576 (2006).
Ling, C., Groop, L., Guerra, S. D. & Lupi, R. Calpain-10 expression is elevated in pancreatic islets from patients with type 2 diabetes. PLoS ONE 4, e6558 (2009).
Garwood, C. et al. Calcium dysregulation in relation to Alzheimer-type pathology in the ageing brain. Neuropathol. Appl. Neurobiol. 39, 788–799 (2013).
Pappalardo, L. W., Black, J. A. & Waxman, S. G. Sodium channels in astroglia and microglia. Glia 64, 1628–1645 (2016).
Kim, D. E., Chivian, D. & Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 (2004).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R. & Thornton, J. M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996).
Yan, Y., Tao, H., He, J. & Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 15, 1829–1852 (2020).
Yan, Y. & Huang, S.-Y. Pushing the accuracy limit of shape complementarity for protein-protein docking. BMC Bioinformatics 20, 696 (2019).
Huang, S.-Y. & Zou, X. An iterative knowledge-based scoring function for protein–protein recognition. Proteins Struct. Funct. Bioinforma. 72, 557–579 (2008).
Xue, L. C., Rodrigues, J. P., Kastritis, P. L., Bonvin, A. M. & Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 32, 3676–3678 (2016).
Vangone, A. & Bonvin, A. M. Contacts-based prediction of binding affinity in protein–protein complexes. eLife 4, e07454 (2015).
Tan, C. et al. Implementation of residue-level coarse-grained models in GENESIS for large-scale molecular dynamics simulations. PLOS Comput. Biol. 18, e1009578 (2022).
Schmid, N. et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 40, 843–856 (2011).
Evans, D. J. & Holian, B. L. The Nose-Hoover thermostat. J. Chem. Phys. 83, 4069–4074 (1985).
Van Der Spoel, D. et al. GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005).
Acknowledgements
LexA-LaminA, LexA-CoREST and LexA-Pincher were generous gifts from Drs. Rolf Sternglanz, Gail Mandel, and Simon Halegoua, respectively.
Funding
This work was supported by CONAHCYT grant A1-S-36984 and PAPIIT-UNAM grant IN220223 to Dr. Ana V Vega, CONAHCYT grant 287002 and SIP-IPN (20220704/20230875) to Dr. Absalom Zamorano, and CONAHCYT scholarship 704048 to Luis M Arratia.
Author information
Authors and Affiliations
Contributions
AVV, AZC and LMAC contributed to the conception and design of the study. EARM, JCLR, MLH and JAJM collaborated in either experiments and data collection and/or analysis. LMAC and JDBC wrote the first draft of the manuscript. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Arratia, L.M., Bermudes-Contreras, J.D., Juarez-Monroy, J.A. et al. Experimental and computational evidence that Calpain-10 binds to the carboxy terminus of NaV1.2 and NaV1.6. Sci Rep 14, 6761 (2024). https://doi.org/10.1038/s41598-024-57117-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-57117-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.