Abstract
Depolarizing sodium (Na+) leak currents carried by the NALCN channel regulate the resting membrane potential of many neurons to modulate respiration, circadian rhythm, locomotion and pain sensitivity1,2,3,4,5,6,7,8. NALCN requires FAM155A, UNC79 and UNC80 to function, but the role of these auxiliary subunits is not understood3,7,9,10,11,12. NALCN, UNC79 and UNC80 are essential in rodents2,9,13, and mutations in human NALCN and UNC80 cause severe developmental and neurological disease14,15. Here we determined the structure of the NALCN channelosome, an approximately 1-MDa complex, as fundamental aspects about the composition, assembly and gating of this channelosome remain obscure. UNC79 and UNC80 are massive HEAT-repeat proteins that form an intertwined anti-parallel superhelical assembly, which docks intracellularly onto the NALCN–FAM155A pore-forming subcomplex. Calmodulin copurifies bound to the carboxy-terminal domain of NALCN, identifying this region as a putative modulatory hub. Single-channel analyses uncovered a low open probability for the wild-type complex, highlighting the tightly closed S6 gate in the structure, and providing a basis to interpret the altered gating properties of disease-causing variants. Key constraints between the UNC79–UNC80 subcomplex and the NALCN DI–DII and DII–DIII linkers were identified, leading to a model of channelosome gating. Our results provide a structural blueprint to understand the physiology of the NALCN channelosome and a template for drug discovery to modulate the resting membrane potential.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The NALCN–FAM155A–UNC79–UNC80–CaM coordinates are deposited in the PDB (7SX3 and 7SX4) and cryo-EM data are deposited in the Electron Microscopy Data Bank (EMD-25492 and EMD-25493) for conformation 1 and conformation 2, respectively. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE54 partner repository with the dataset identifier PXD027213 and details are included in the Supplementary Information. Source data are provided with this paper.
References
Lear, B. C. et al. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron 48, 965–976 (2005).
Lu, B. et al. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell 129, 371–383 (2007).
Jospin, M. et al. UNC-80 and the NCA ion channels contribute to endocytosis defects in synaptojanin mutants. Curr. Biol. 17, 1595–1600 (2007).
Xie, L. et al. NLF-1 delivers a sodium leak channel to regulate neuronal excitability and modulate rhythmic locomotion. Neuron 77, 1069–1082 (2013).
Flourakis, M. et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848 (2015).
Lutas, A., Lahmann, C., Soumillon, M. & Yellen, G. The leak channel NALCN controls tonic firing and glycolytic sensitivity of substantia nigra pars reticulata neurons. eLife 5, e15271 (2016).
Shi, Y. et al. Nalcn is a “leak” sodium channel that regulates excitability of brainstem chemosensory neurons and breathing. J. Neurosci. 36, 8174–8187 (2016).
Philippart, F. & Khaliq, Z. M. Gi/o protein-coupled receptors in dopamine neurons inhibit the sodium leak channel NALCN. eLife 7, e40984 (2018).
Lu, B. et al. Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron 68, 488–499 (2010).
Chua, C. H., Wulf, M., Weidling, C., Rasmussen, L. P. & Pless, S. A. The NALCN channel complex is voltage sensitive and directly modulated by extracellular calcium. Sci. Adv. 6, eaaz3154 (2020).
Bouasse, M., Impheng, H., Servant, Z., Lory, P. & Monteil, A. Functional expression of CLIFAHDD and IHPRF pathogenic variants of the NALCN channel in neuronal cells reveals both gain- and loss-of-function properties. Sci. Rep. 9, 11791 (2019).
Wie, J. et al. Intellectual disability-associated UNC80 mutations reveal inter-subunit interaction and dendritic function of the NALCN channel complex. Nat. Commun. 11, 3351 (2020).
Nakayama, M., Iida, M., Koseki, H. & Ohara, O. A gene-targeting approach for functional characterization of KIAA genes encoding extremely large proteins. FASEB J. 20, 1718–1720 (2006).
Cochet-Bissuel, M., Lory, P. & Monteil, A. The sodium leak channel, NALCN, in health and disease. Front. Cell. Neurosci. 8, 132 (2014).
Bramswig, N. C. et al. Genetic variants in components of the NALCN-UNC80-UNC79 ion channel complex cause a broad clinical phenotype (NALCN channelopathies). Hum. Genet. 137, 753–768 (2018).
Lee, J. H., Cribbs, L. L. & Perez-Reyes, E. Cloning of a novel four repeat protein related to voltage-gated sodium and calcium channels. FEBS Lett. 445, 231–236 (1999).
Kang, Y., Wu, J. X. & Chen, L. Structure of voltage-modulated sodium-selective NALCN-FAM155A channel complex. Nat. Commun. 11, 6199 (2020).
Kschonsak, M. et al. Structure of the human sodium leak channel NALCN. Nature 587, 313–318 (2020).
Xie, J. et al. Structure of the human sodium leak channel NALCN in complex with FAM155A. Nat. Commun. 11, 5831 (2020).
Lu, B. et al. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature 457, 741–744 (2009).
Yeh, E. et al. A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 6, e55 (2008).
Lear, B. C. et al. UNC79 and UNC80, putative auxiliary subunits of the NARROW ABDOMEN ion channel, are indispensable for robust circadian locomotor rhythms in Drosophila. PLoS ONE 8, e78147 (2013).
Speca, D. J. et al. Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 6, e1001057 (2010).
Stray-Pedersen, A. et al. Biallelic mutations in UNC80 cause persistent hypotonia, encephalopathy, growth retardation, and severe intellectual disability. Am. J. Hum. Genet. 98, 202–209 (2016).
Perez, Y. et al. UNC80 mutation causes a syndrome of hypotonia, severe intellectual disability, dyskinesia and dysmorphism, similar to that caused by mutations in its interacting cation channel NALCN. J. Med. Genet. 53, 397–402 (2016).
Shamseldin, H. E. et al. Mutations in UNC80, encoding part of the UNC79-UNC80-NALCN channel complex, cause autosomal-recessive severe infantile encephalopathy. Am. J. Hum. Genet. 98, 210–215 (2016).
Humphrey, J. A. et al. A putative cation channel and its novel regulator: cross-species conservation of effects on general anesthesia. Curr. Biol. 17, 624–629 (2007).
Holm, L. Benchmarking fold detection by DaliLite v.5. Bioinformatics 35, 5326–5327 (2019).
Wang, H. & Ren, D. UNC80 functions as a scaffold for Src kinases in NALCN channel function. Channels (Austin) 3, 161–163 (2009).
Topalidou, I. et al. The NCA-1 and NCA-2 ion channels function downstream of Gq and Rho to regulate locomotion in Caenorhabditis elegans. Genetics 206, 265–282 (2017).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Cardone, G., Heymann, J. B. & Steven, A. C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 184, 226–236 (2013).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D 74, 814–840 (2018).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Banerjee, R. et al. Bilobal architecture is a requirement for calmodulin signaling to CaV1.3 channels. Proc. Natl Acad. Sci. USA 115, E3026–E3035 (2018).
Kang, P. W. et al. Elementary mechanisms of calmodulin regulation of NaV1.5 producing divergent arrhythmogenic phenotypes. Proc. Natl Acad. Sci. USA 118, e2025085118 (2021).
Leitner, A., Walzthoeni, T. & Aebersold, R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat. Protoc. 9, 120–137 (2014).
Mohammadi, A., Tschanz, A. & Leitner, A. Expanding the cross-link coverage of a carboxyl-group specific chemical cross-linking strategy for structural proteomics applications. Anal. Chem. 93, 1944–1950 (2021).
Walzthoeni, T. et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods 9, 901–903 (2012).
Rinner, O. et al. Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 (2008).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
Acknowledgements
We thank members of the Pless and Ben-Johny laboratories, P. Picotti, and Genentech colleagues in the BioMolecular Resources and Structural Biology departments for their support, and appreciate the encouragement of A. Rohou, C. Koth, S. Hymowitz, V. Dixit and A. Chan. Members of the Ben-Johny group acknowledge support from the NIH National Institute of Neurological Disorders and Stroke (R01 NS110672). Members of the Pless group acknowledge the Carlsberg Foundation (CF16-0504), the Independent Research Fund Denmark (7025-00097A and 9124-00002B) and the Lundbeck Foundation (R252-2017-1671) for financial support. Reagents are available under a material transfer agreement with Genentech or the appropriate party.
Author information
Authors and Affiliations
Contributions
M.K. established protein purification and reconstitution methods with input from C.L.N. T.C., C.T. and N.P. generated key protein expression reagents. M.K and C.P.A. optimized cryo-EM sample preparation and data collection. M.K. determined the structure, with guidance from C.C. H.C.C. established methods to record the function of the NALCN complex. H.C.C., C.W. and K.S. performed the molecular biology and two-electrode voltage clamp electrophysiology experiments. A.L. performed and analysed the crosslinking mass spectrometry experiments. N.C. and M.B.-J. performed the single-channel experiments and analyses. M.K., H.C.C., C.W., N.C., K.S., A.L., M.B.-J., C.C., S.A.P. and J.P. analysed the data. H.C.C. performed and analysed the linker competition experiments with input from M.K., S.A.P. and J.P. M.K., H.C.C., C.C., S.A.P. and J.P. wrote the manuscript with input from all authors. A.L., M.B.-J., C.C., S.A.P. and J.P. supervised the project and are co-senior authors.
Corresponding authors
Ethics declarations
Competing interests
M.K., C.L.N., T.C., C.T., N.P., C.P.A., C.C. and J.P. are or were employees of Genentech/Roche; all remaining authors declare no competing interests.
Peer review
Peer review information
Nature thanks Paul DeCaen, Vera Moiseenkova-Bell and David Spafford for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
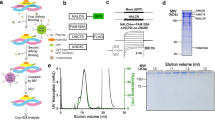
Extended Data Fig. 1 Purification and structure determination of the NALCN channelosome.
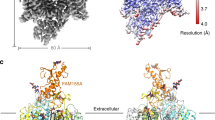
a, NALCN channelosome protein expression and purification scheme. b, Example size exclusion chromatogram and SDS-PAGE of nanodisc-reconstituted NALCN channelosome sample. c, Example cryo-EM micrograph image of the NALCN channelosome-MSP1E3D1 complex. d, Representative 2D-class averages of selected particles from 200 classes and approximately 130,000 particles. e, Data collection and processing workflow. f, Conformation 1, FSC between two half datasets yields a global resolution estimate of approximately 3.1 Å resolution from the refinement using an overall mask of the NALCN channelosome, 3.2 Å resolution from the focused NALCN-CaM-FAM, 2.9 Å from the focused UNC crossover, 3.7 Å from the focused UNC N-C and 3.8 Å from the focused UNC C-N refinements. g, Conformation 1, heat map representation of the distribution of assigned particle orientations. h, Conformation 2, FSC between two half datasets yields a global resolution estimate of approximately 3.5 Å resolution. i, Conformation 2, heat map representation of the distribution of assigned particle orientations.
Extended Data Fig. 2 Select cryo-EM map regions of the NALCN channelosome.
a–g, Example 3D map overlay for indicated regions.
Extended Data Fig. 3 NALCN and FAM155A structures in the NALCN channelosome and NALCN-FAM155A subcomplex.
a, Comparison of the NALCN subunit in the NALCN-FAM155A subcomplex and NALCN channelosome (with UNC79 and UNC80 removed for clarity). b, Superposition of the FAM155A subunit in the NALCN-FAM155A subcomplex and NALCN channelosome. c, Pore radius of the ion conduction pathway in the NALCN channelosome or NALCN-FAM155A subcomplex structures (PDB 6XIW) calculated by HOLE55.
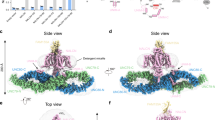
Extended Data Fig. 4 UNC79 and UNC80 are HEAT-repeat proteins.
a, Side- and top-views of UNC79. Position of disordered loops >50 residues in length are indicated. b, Side- and top-views of UNC80. Position of disordered loops >50 residues in length are indicated. c, Open-book view of UNC79-UNC80 interface showing contact surface. d, Same as part c, showing electrostatic surface. e, Same as part c, showing surface conservation across selected vertebrate species calculated by ConSurf56.
Extended Data Fig. 5 Structure-function analysis of UNC79 and UNC80.
a, Schematic of UNC79 and UNC80 with corresponding amino acid residues of fragments indicated. Example current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79 and UNC80, where various isolated ~500 residue fragments of either UNC79 or UNC80 are co-expressed in trans, with steps from +80 to −80 mV, 40 mV increments, in ND96 recording solution. Right shows summary of mean current amplitudes elicited at +80 mV (top bar graph) or −80 mV (bottom bar graph) from a holding potential of 0 mV for indicated construct combinations. Numbers of biological replicates (n) are indicated. Recordings were performed on four to five oocytes from two different batches. Data are shown as mean±SD. ****, * and ns represent p value < 0.0001, < 0.05 and >0.05, respectively; one-way analysis of variance (ANOVA), Dunnett’s test (against +H2O). P values are adjusted to account for multiple comparisons and are presented in Supplementary Table 5. For electrophysiological measurements, see Source Data. b, Schematic of UNC80 with corresponding truncated constructs indicated. Example current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79 and UNC80, where wild-type UNC80 or truncated constructs are expressed, with steps from +80 to −80 mV, 20 mV increments, in ND96 recording solution. Star (#) indicates constructs that contain fusion of a C-terminal GFP-Flag tag. Right shows summary of mean current amplitudes elicited at +80 mV (top bar graph) or −80 mV (bottom bar graph) from a holding potential of 0 mV for indicated construct combinations. Numbers of biological replicates (n) are indicated. Recordings were performed on four to six oocytes from two different batches. Data are shown as mean±SD. ****, **, and * represent p value < 0.0001, < 0.01 and < 0.05, respectively; one-way analysis of variance (ANOVA), Dunnett’s test (against UNC80#). P values are adjusted to account for multiple comparisons and are presented in Supplementary Table 5. For electrophysiological measurements, see Source Data. c, Schematic of UNC80 with corresponding nonsense mutation constructs indicated (note, R1265X, L2586X, and Y2796X mutations have been previously reported by us10 and representative data are shown here for comparison). Example current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79 and UNC80, where wild-type UNC80 or nonsense mutation constructs are expressed, with steps from +80 to −80 mV, 20 mV increments, in ND96 recording solution. Right shows summary of mean current amplitudes elicited at +80 mV (top bar graph) or −80 mV (bottom bar graph) from a holding potential of 0 mV for indicated construct combinations. Numbers of biological replicates (n) are indicated. Recordings were performed on three to six oocytes from two to three different batches. Data are shown as mean±SD. **** represents p value < 0.0001; one-way analysis of variance (ANOVA), Dunnett’s test (against UNC80). P values are adjusted to account for multiple comparisons and are presented in Supplementary Table 5. For electrophysiological measurements, see Source Data. d, Example current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79 and UNC80, where wild-type UNC80 or missense mutation constructs are expressed, with steps from +80 to −80 mV, 20 mV increments, in ND96 recording solution. Right shows summary of mean current amplitudes elicited at +80 mV (top bar graph) or −80 mV (bottom bar graph) from a holding potential of 0 mV for indicated construct combinations. Numbers of biological replicates (n) are indicated. Recordings were performed on two to eight oocytes from two to three different batches. Data are shown as mean±SD. **** and ns represent p value < 0.0001 and >0.05, respectively; one-way analysis of variance (ANOVA), Dunnett’s test (against UNC80). P values are adjusted to account for multiple comparisons and are presented in Supplementary Table 5. For electrophysiological measurements, see Source Data. e, Close-in views of select missense mutations previously identified in UNC80 mapped onto the UNC79-UNC80 subcomplex structure. Note, exact p values are presented in Supplementary Table 5.
Extended Data Fig. 6 Characterization of NALCN-NaV1.4 chimeras.
a, Schematic of human NALCN and human NALCN-rat NaV1.4 chimeric constructs. Example current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79 and UNC80, where wild-type or chimeric truncated NALCN constructs are expressed, with steps from +80 to −80 mV, 20 mV increments, in ND96 recording solution. Numbers of biological replicates (n) are indicated in b. Recordings were performed on two to five oocytes from two to five different batches. For electrophysiological measurements, see Source Data. b, Summary of mean current amplitudes elicited at +80 mV (top bar graph) or −80 mV (bottom bar graph) from a holding potential of 0 mV for indicated constructs. Data are shown as mean±SD. **** and ns represent p value < 0.0001 and >0.05, respectively; one-way analysis of variance (ANOVA), Dunnett’s test (against NALCN). P values are adjusted to account for multiple comparisons and are presented in Supplementary Table 5.
Extended Data Fig. 7 Structure-function of the NALCN DI-DII and DII-DIII linkers.
a, Current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79 and UNC80, where the NALCN subunit is wild-type or has indicated mutations, insertions or deletions in the DI-DII linker. Steps from +80 to −100 mV, 20 mV increments, in ND96 recording solution. Right shows summary of mean current amplitudes elicited at +80 mV (top bar graph) or −80 mV (bottom bar graph) from a holding potential of 0 mV for indicated constructs. b, Representative Western blot of total lysate and surface fraction proteins extracted from Xenopus oocytes expressing the indicated constructs where wild-type or mutant NALCN was co-expressed with wild-type UNC80, UNC79 and FAM155A. For gel source data, see Supplementary Fig. 3. c, Current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79 and UNC80, where the NALCN subunit is wild-type or has indicated insertions and deletions in the DII-DIII linker. Steps from +80 to −100 mV, 20 mV increments, in ND96 recording solution. Right shows summary of mean current amplitudes elicited at +80 mV (top bar graph) or −80 mV (bottom bar graph) from a holding potential of 0 mV for indicated constructs. a, c, Numbers of biological replicates (n) are indicated. Recordings were performed on four to five oocytes from two different batches. Data are shown as mean±SD. ****, ***, **, * and ns represent p value < 0.0001, < 0.001, < 0.01, < 0.05 and >0.05, respectively; one-way analysis of variance (ANOVA), Dunnett’s test (against WT). P values are adjusted to account for multiple comparisons and are presented in Supplementary Table 5.
Extended Data Fig. 8 Structure-function analysis of the NALCN C-terminal domain.
a, Current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79 and UNC80, where the NALCN subunit is wild-type or has indicated CTD-deletions or mutations. Steps from +80 to −100 mV, 20 mV increments, in ND96 recording solution. Right shows summary of mean current amplitudes elicited at +80 mV (top bar graph) or −80 mV (bottom bar graph) from a holding potential of 0 mV for indicated constructs. Numbers of biological replicates (n) are indicated. Recordings were performed on three to six oocytes from two different batches. Data are shown as mean±SD. * and ns represent p value < 0.05 and >0.05, respectively; one-way analysis of variance (ANOVA), Dunnett’s test (against WT). P values are adjusted to account for multiple comparisons and are presented in Supplementary Table 5. b, Current traces from Xenopus oocytes expressing NALCN, FAM155A, UNC79, UNC80 and control (H2O) or an isolated NALCN-CTD construct in trans. Steps from +80 to −100 mV, 20 mV increments, in ND96 recording solution. Right shows summary of mean current amplitudes elicited at +80 mV from a holding potential of 0 mV for indicated conditions. Numbers of biological replicates (n) are indicated. Recordings were performed on four to five oocytes from two different batches. Data are shown as mean±SD. Two-sided unpaired t-test, p = 0.1286 (+80 mV), p = 0.0037 (-80 mV). c, Current traces from Xenopus oocytes expressing wild-type NALCN (1-1738) or indicated C-terminal truncation constructs recorded in ND96 (top) and divalent cation (X2+)-free buffer (bottom). Steps from +80 to −100 mV, 20 mV increments. Right shows fold-increase in inward current elicited at −100 mV for wild-type NALCN and indicated truncation mutants in response to removal of divalent cations. Numbers of biological replicates (n) are indicated. Recordings were performed on three to four oocytes from two different batches. Data are shown as mean±SD. ns represents p value >0.05; one-way analysis of variance (ANOVA), Dunnett’s test (against WT). P values are adjusted to account for multiple comparisons and are presented in Supplementary Table 5. d, Current traces from Xenopus oocytes expressing wild-type NALCN, FAM155A, UNC79 and UNC80, where a “CaM-chelator” construct (which is composed of four tandem IQ motifs (IQ3-6: residues L766–K920) of mouse myosin Va (Uniprot Q99104) that diminishes free concentration of apo-CaM48,49) is co-expressed. Numbers of biological replicates (n) are indicated. Recordings were performed on four to five oocytes from three different batches. Data are shown as mean±SD. Two-sided unpaired t-test, p = 0.0002 (+80 mV), p = 0.0421 (−80 mV).
Extended Data Fig. 9 Cell-attached single channel recordings of the NALCN channelosome.
a, Exemplar traces from an individual patch show absence of channel openings when HEK293 cells are transfected with only UNC79, UNC80, and FAM155A. Gray slanted line, unitary current relationship for wild-type NALCN. b, Ensemble average current obtained from 154 sweeps for the individual patch shown in panel a. No detectible channel activity was observed in a total of n = 8 cells (852 sweeps). c, Exemplar traces show additional examples of single-channel openings of wild-type NALCN channelosome from the same patch shown in Fig. 4b. Gray slanted line, unitary current relationship. d, Ensemble average current obtained from 90 sweeps for the single-channel patch of NALCN channelosome as shown in panel c. e, Diary plot of single-trial PO values from the one-channel patch of wild-type NALCN, as shown in panel c, reveal distinct active (A; traces with brief and flickery openings) and quite (Q; traces with no discernible activity) gating modes. f, Exemplar traces show additional examples of single-channel openings of Y578S mutant channels from the individual patch shown in Fig. 4b. Gray slanted line, unitary current relationship. g, Ensemble average current obtained from 40 sweeps for the single-channel patch of Y578S mutant channel shown in panel f. h, Diary plot of single-trial PO for the one-channel patch for Y578S mutant displayed in panel f. Y578S mutant channel also show switching between active and quite gating modes.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1–3 and Supplementary Tables 1 and 3–5.
Supplementary Table 2
Complete cross-linking mass spectrometry of the NALCN channelosome.
Rights and permissions
About this article
Cite this article
Kschonsak, M., Chua, H.C., Weidling, C. et al. Structural architecture of the human NALCN channelosome. Nature 603, 180–186 (2022). https://doi.org/10.1038/s41586-021-04313-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04313-5
This article is cited by
-
Two rare autosomal recessive neurological disorders identified by combined genetic approaches in a single consanguineous family with multiple offspring
Neurological Sciences (2024)
-
Architecture of the human NALCN channelosome
Cell Discovery (2022)
-
Structure and mechanism of NALCN-FAM155A-UNC79-UNC80 channel complex
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.