Abstract
Genotype by environment interactions (G × E) are frequently observed in herbage production. Understanding the underlying biological mechanisms is important for achieving stable and predictive outputs across production environments. The microbiome is gaining increasing attention as a significant contributing factor to G × E. Here, we focused on the soil microbiome of perennial ryegrass (Lolium perenne L.) grown under field conditions and investigated the soil microbiome variation across different ryegrass varieties to assess whether environmental factors, such as seasonality and nitrogen levels, affect the microbial community. We identified bacteria, archaea, and fungi operational taxonomic units (OTUs) and showed that seasonality and ryegrass variety were the two factors explaining the largest fraction of the soil microbiome diversity. The strong and significant variety-by-treatment-by-seasonal cut interaction for ryegrass dry matter was associated with the number of unique OTUs within each sample. We identified seven OTUs associated with ryegrass dry matter variation. An OTU belonging to the Solirubrobacterales (Thermoleophilales) order was associated with increased plant biomass, supporting the possibility of developing engineered microbiomes for increased plant yield. Our results indicate the importance of incorporating different layers of biological data, such as genomic and soil microbiome data to improve the prediction accuracy of plant phenotypes grown across heterogeneous environments.
Similar content being viewed by others
Introduction
Perennial ryegrass (Lolium perenne L.) is a forage species widely cultivated in temperate grasslands1. Grasslands make a significant contribution to human food security by providing fodder for ruminants used for meat and milk production. However, frequent occurrence of genotype by environment interactions (G × E, reflected in the varying response of different genotypes to changing environmental factors) in herbage yield production results in heterogeneous herbage production across years and seasons, which complicates the prediction of yield and quality2,3,4,5,6. Understanding the contributing mechanisms that drive inconsistent output for perennial ryegrass yield is essential for optimizing the breeding programs to achieve stable and predictive output across different and variable environments.
The plant microbiome, in general, has gained increasing attention through the past years. It has become evident that the soil microbiome interplays with the plants, affecting both yield and plant performance in a specific environment7,8,9,10. Often, the host plant relies on the soil microbiome to provide essential nutrients, whereas the plant cultivates its microbiome, for example by releasing specific metabolites in the soil, adjusting the soil pH, and reducing competition among beneficial microbes7. Because of this close relationship between plants and the surrounding soil microbiome, it is not surprising that the composition of microbial community varies among plant species8. It has been shown that the genotype of the plant is involved in structuring the microbiome of maize10,11,12, Arabidopsis thaliana13,14,15, barley16, rice17,18,19, wheat20. Likewise, certain components of the microbiome have been shown to contribute with enhanced plant resistance towards biotic and abiotic stressors, for example wilt resistance in tomato21, or drought resistance in Capsicum annuum22. The soil microbiome was shown to interfere with physiological processes in the plant, for example altering flowering time through influencing the metabolic network of phytohormones23. Moreover, the soil microbiome was suggested to mediate positive relationships between plant diversity and plant productivity24. The soil microbiome functions not only as a plant health indicator, but also as an indicator of soil health25.
Despite years of research attempting to uncover the association between host plant and the soil microbiome composition26,27,28, little is known about the associated soil microbial community of perennial ryegrass. Chen et al. investigated in a controlled experiment the root-associated bacterial microbes of ryegrass across different compartments of the roots and soil and found differences in bacterial composition across the compartments, as well as across soil types29. Understanding the interactions between the host plant and the soil microbiome and eventually identifying the genetic basis controlling such interactions, will be of great significance for plant breeding30. The aim of this study was to determine to what extent the soil microbiome varied among 20 ryegrass varieties grown under field conditions, whether the soil microbial community was modified by nitrogen level and how the season affected the soil microbiome community.
Results
Composition of the soil microbial community
Across the 240 samples (20 varieties, 2 nitrogen treatments, 3 samplings at seasonal cuts and 2 replicates) a total of 10,010 bacterial OTUs were initially detected, with the number of sequences per sample ranging between 29,710—54,850 (mean 41,850). We detected 6,130 fungal OTUs, with the number of sequences per sample ranging between 37,959—287,558 (mean 131,026). A total of 6,092 bacterial and 2,907 fungal OTUs remained after removing OTUs with frequency < 0.1% (Supplementary Tables S4, S5).
The most abundant bacteria phyla were Actinobacteria, Chloroflexi, Proteobacteria, and Firmicutes (Fig. 1A), and the most abundant fungi phylum was Ascomycota, followed by Basidiomycota and Zygomycota (Fig. 1B). A considerable proportion of the fungal OTUs had no BLAST hits (Fig. 1B).
Representation of the (A) bacteria and (B) fungi composition at phylum level. Each bar represents the relative abundance (%) averaged across samples within collection points (A: 24.05.2017, B: 07.07.2017, C: 17.08.2017). Only taxonomic groups with relative abundance > 1% are shown (remaining is agglomerated in the category “Others”).
The OTU richness is the number of unique OTUs within each sample, whereas Shannon’s Index also accounts for both OTU abundance and evenness. Phylogenetic diversity is a measure of biodiversity that incorporates the differences in phylogenetic diversity between OTUs, such that related individuals increase the measure of phylogenetic biodiversity less than unrelated individuals do. The OTU richness and Shannon’s Index were lower for fungi than for bacteria (Fig. 2), indicating that the within-sample diversity of bacteria was larger compared to the fungal community (Supplementary Table S6). Contrary, the phylogenetic diversity was two-fold larger for the fungal community than for bacteria (Fig. 2). These trends are generalized across all samples and comparing the α-diversity measures between bacteria and fungi showed no correlation within samples (Fig. 2), but strong correlation was observed among diversity metrics within bacteria and within fungi (Supplementary Fig. S1).
The within-sample bacteria and fungi diversity varied across season (Supplementary Tables S7, S8) with the highest species richness latest in the season for most perennial ryegrass varieties (Fig. 3). For bacteria, only samples collected in July (cut-B) showed that species richness was influenced by different levels of nitrogen treatment, where the samples with lower nitrogen supplement had higher species richness (P = 0.012, Supplementary Table S6). Separation of soil samples by seasonal cut, replicate, horizontal position, treatment, and ryegrass variety using bacterial and fungal OTUs is presented in Supplementary Fig. S2 and S3, respectively. Within-sample complexity did not differ significantly between ryegrass varieties or their ploidy when tested within collection time points or nitrogen treatment level (Supplementary Table S6).
Species richness for bacteria and fungi. Comparison of bacterial species richness (S16S) for all ryegrass varieties as function of nitrogen treatment (A). The bacterial species richness (S16S) for normal (B) and low nitrogen treatment (C) across the three seasonal cuts. The fungi species richness (SITS2) for normal (D) and low nitrogen treatment (E) across the three seasonal cuts. The P-values are from ANOVA testing the effect of treatment (A), and seasonal cuts for normal (B,D) and low nitrogen supplement (C,E).
Across all samples, within the three seasonal collection time points, we observed vast interactions between the relative abundances among bacterial and fungal OTUs, where the evolution through the season was also evident (Supplementary Fig. S4). Correlation network analyses revealed positive and negative connections not only within the bacterial and within the fungal taxa, but also between bacteria and fungi (Fig. S4).
Diversity within the soil microbiome
We computed distance matrices adjusting for OTU abundance and phylogenetic distance among OTUs to estimate the across sample diversity. Principal coordinate analysis showed a clear separation of samples obtained from the different collection time points (Fig. 4). Next, we partitioned the variation in the soil microbiome diversity (Table 1) and found that 30.5% of the total variation in the bacterial microbiome could be explained by the experimental setup, with the largest fraction of variation explained by collection time point, i.e. seasonal cut (13.7%, P = 0.001), followed by ryegrass variety (8.15%, P = 0.001). Similarly, 27.70% of the variation in fungal microbiome diversity could be explained by the experimental variables, where 11.92% (P = 0.001) could be explained by the three collection time points and 7.87% by ryegrass variety (P = 0.001). Comparison of the distance matrices based on 16S OTUs and ITS2 OTUs for seasonal cut A, B and C are presented in Supplementary Fig. S5.
Principal coordinates 1 and 2 from PCoA of bacterial (A) and fungal (B) OTU communities. The color coding indicates which seasonal cut each sample belongs to. The centroid of each group is shown as larger dot in its respective color (see legend). The percent variation explained by the first two components is indicated on the axes.
Variation in soil microbiome and plant dry matter yield
Based on dry matter yield of a total of 64 ryegrass varieties, we estimated the proportion of dry matter variation attributable to the different ryegrass varieties (i.e. broad-sense heritability, \({\widehat{H}}^{2}\)) to \({\widehat{H}}^{2}=0.37\), with strong evidence of variety-by-treatment-by-seasonal cut interaction (P = 1.87 × 10−6; Supplementary Table S9 for fixed effect analysis). For the subset of 20 ryegrass varieties used for the soil microbiome analysis, the heritability estimate was \({\widehat{H}}^{2}=0.37\).
We found that bacteria and fungi species richness was negatively associated with ryegrass dry matter yield (Fig. 5). Similar effects were found for Shannon’s Index (not for fungi) and Faith’s phylogenetic diversity (Supplementary Fig. S6). Contrast plot of ryegrass dry matter (DM kg/plot, points) as function of replicate, horizontal groups, seasonal group and by low (n) or normal (N) nitrogen treatment are presented in Supplementary Fig. S7.
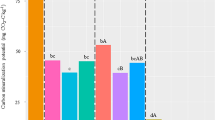
Because species richness was strongly associated with ryegrass dry matter, we then investigated if any OTUs were associated with ryegrass dry matter across season and nitrogen treatment. We identified four bacteria and three fungi OTUs that showed statistical association with ryegrass dry matter (Table 2, detailed in Supplementary Tables S10, S11). The bacteria OTUs belong to the phylum Actinobacteria, Proteobacteria and Acidobacteria, one fungi OTUs belongs the phylum Basidiomycota, whereas the other two had no BLAST hit. OTU richness was associated with decreased plant mass, except for bacteria from the order Solirubrobacterales (OTU_4991), which was associated with increased plant biomass (Supplementary Figs. S8, S9).
Discussion
Despite the increasing number of soil microbial community studies, still limited information is available related to how the microbial composition variation contributes to herbage production across seasons and production years. In the present study, we characterized the soil microbial community of perennial ryegrass under field conditions in order to investigate to what extent the soil microbiome was influenced by plant genotype, whether the microbial community was stable across the season and if the microbiome diversity was altered by a 20% difference in applied nitrogen levels. We focused on the top 15 cm, the soil layer with the largest plant biomass31,32, with previous reports observing no significant effect of seasonality on perennial ryegrass root counts in soil layers deeper than 7 cm33. We identified several bacteria and fungi OTUs and showed that seasonality and ryegrass variety were the two most important factors in explaining the microbial diversity. We observed strong variety-by-environment interaction for ryegrass dry matter yield and this was associated with bacterial and fungal OTU richness. Finally, we identified seven OTUs associated with variation in ryegrass dry matter yield.
Based on a total of 240 soil samples, we identified over 6000 bacterial and close to 3000 fungal OTUs. Over 80% of the bacterial OTUs were found within four phyla, where Actinobacteria clearly dominated through the whole season, in all three collection time points. This is similar to what was observed in other studies34,35. The soil samples were likewise dominated by the fungal phyla Ascomycota, as expected based on previous reports36. Notable is however, that 30–40% of all fungal OTUs could not be annotated due to lack of BLAST hits. While this limits the interpretation of results, it also highlights the ample amount of research and work needed in this research field.
The major source of soil microbiome variation was attributable to the three sample collection time points (Table 1, Supplementary Tables S4, S5), which is most likely due to abiotic environmental changes caused by natural seasonality. Although both bacterial and fungal communities were influenced by the collection time points, their evolution through the season showed differences related to abundance, positive and negative associations, but also with respect to the evolution of the community across the season (Fig. 4, Supplementary Fig. S4). While bacteria showed a gradual change through the collection time points (Fig. 4A), the notable change in the fungal community was observed late in the season, at the third cut (Fig. 4B). Even with collection time point accounting for the largest amount of variation in the soil microbiome of perennial ryegrass, the individual varieties had a strong influence on this effect (Fig. 3).
In contrast to other studies where increased nitrogen supplements led to reduced bacterial richness37,38,39, we did not observe this trend (Supplementary Fig. S2, S5, and Supplementary Table S7). Although we did identify a significant effect of nitrogen treatment on species richness for cut-B, in July (Fig. 3, Supplementary Table S6), we noticed rather strong variety-specific effects of the nitrogen treatment (Fig. 3, Fig. S5), in accordance with a previous report for barley39. Wang et al. observed no significant impact of nitrogen treatment on perennial ryegrass soil microbiome structure and composition across different farm niches40, in an experiment comparing three nitrogen application rates of 0, 150 and 300 kg ha−1 year−1. The soil samples were collected from similar depth level, but the differences between nitrogen treatments are significantly larger compared to our study. The reduction in the amount of applied N was only around 20% in our study. Wang et al. noticed however a significant reduction of the microbiome network connectivity in soil. Variety specific effects could explain the differences observed between these studies, as our results indicate that plant genotype affects both the bacterial and fungal soil community. We observed that the soil microbiome of different varieties responded differently to the two nitrogen levels across the three cuts. For example, as seen in Fig. 3 for cut-B, the soil microbiome of the variety Kerry showed no difference between the two nitrogen levels, while a clear response to nitrogen treatment was observed for the variety Hopi. This suggests a different potential for the genetic background of different varieties, which could be further utilized in breeding. Escudero-Martinez et al. identified genomic loci having a major effect on the barley rhizosphere microbiome composition, and with no association to the investigated root phenotypic traits41. One of these loci is located on chromosome 3H, with three candidate genes suggested to control the heritable component of the barley rhizosphere microbiome. Furthermore, individual genes were observed to have different effects on fungi and bacteria. The recent advances in elucidating the perennial ryegrass genome42 will facilitate the identification of genomic regions involved in shaping its microbiome.
Different patterns were observed in case of both bacteria and fungi through the season depicted by the three cuts, depending on variety (Fig. 3, Supplementary Fig. S2). We were not able to directly identify within-variety differences in microbiome community composition (Supplementary Table S7) and this is most probably due to the limited per-variety samples with only two biological replicates. However, when comparing the entire soil microbiome community across the different ryegrass varieties, it was evident that the different varieties captured a significant proportion of the total microbiome variation (Table 1). Previous studies have also reported correlations between soil microbiome and plant genotype in other species39,43,44,45,46, while a cultivar effect was reported for the perennial ryegrass seed microbiome47. Future studies based on perennial ryegrass should be focused on elucidating the specific genetic elements involved in shaping its soil microbiome, to develop genetic markers to be employed in breeding.
Additional support for the existence of host genotype–microbiome interactions was obtained by incorporating plant phenotypic measurements of plant biomass. We estimated the proportion of phenotypic variation that could be explained by the different ryegrass varieties to 0.37 (independent on whether we used the entire dataset of 64 varieties or the subset of 20 varieties) and found a negative correlation between bacterial and fungal richness and ryegrass dry matter yield (Fig. 5). These results suggest that the local soil microbiome was associated with plant genotype and with plant phenotype; here, plant biomass.
Seven OTUs were statistically associated with ryegrass dry matter yield (Table 2), where six of them showed a negative association. Two of these OTUs belong to the phylum Acidobacteria, one of the most dominant soil bacteria detected in a wide range of different soil habitats48,49,50,51,52. In particular subgroup 6 has been shown to respond to high content of soil Ca, Mg, Mn, and B53. Our results suggest their presence and/or activity in the soil to be less beneficial for the plants. Another OTU belongs to the Enterobacteriaceae family, which is a class of bacteria known for its many pathogens, but also for harmless symbionts. The identified OTU belongs to the Buchnera genus, which is not well described. But given the observed negative association, it is less likely that these bacteria are neutral to the plant. The fungal OTU_99 belongs to Tetragoniomyces uliginosus and has previously been described to have a mycoparasitic lifestyle54, which probably explains the negative correlation observed in this study. Although the two other fungi OTUs with significant negative correlation to dry matter yield are unknown at this point, our results suggest a negative effect of these on plant performance. The only OTU found to be positively associated with increased ryegrass dry matter (Supplementary Fig. S8) belongs to the Actinobacteria order Solirubrobacterales. This group of bacteria has previously been described as cosmopolitan and widely present in the soil55, more abundant in the soil during the dry season56 and more abundant in conventionally managed soil compared to organic agroecosystem57. Also, it was previously associated with transportation of chemical contamination58 and with the ability of decomposing lignin59. Some endophytic members of this group were also described60. Solirubrobacteraceae family showed different correlation patterns to specific metabolites in Arabidopsis accessions differing in resistance to Fusarium oxysporum15 and was found to supress common scab disease of potato61. Although limited information is available related to the Solirubrobacterales members, a recent review summarizes the direct and indirect plant growth promoting role of Actinobacteria, highlighting the potential of some of the better characterized strains as natural fertilizer and pesticides62. Our results suggest that the presence of OTU_4991 in the soil microbiome has a positive effect on perennial ryegrass yield.
The limitations of the present study need to be considered when interpreting the presented results. First, the study was performed at a single location in Denmark, while a replicate in another location with a similar environmental profile would be advantageous. Second, the experimental test field had a vertical gradient which could have influenced the soil humidity and thereby the plants’ access to water. We have accounted for the exact location in the field in the statistical analysis, however, micro-environmental variability in water access could still play a role in both plant dry matter and microbial composition. Third, sequencing was performed for two biological replicates, which limits the statistical power and the accuracy with which we can estimate parameters. Despite the practical limitations posed especially by the only two biological replicates for each nitrogen treatment, our results are promising and lay foundation for future large studies to uncover the specific components of the microbiome positively affecting plant traits of interest. Biostimulants based on nitrifying bacteria have already been shown to exhibit positive effects on perennial ryegrass performance, even under nutritional stress conditions63. Large scale studies, with sufficient biological replication and comprising a large number of varieties ideally to be tested in multiple locations should be designed to confirm the effects of specific soil microbiome components on plant performance.
There was a clear difference in the development and evolution of fungal and bacterial community through the season, with positive and negative interactions between components of perennial ryegrass soil microbiome, within and across phyla. Our results indicate that the plant variety has a strong influence on the soil microbiome, while the microbiome has a strong influence on plant variety performance. This opens for possibilities towards developing new varieties harnessing the potential and attributes of the microbiome. Moreover, the potential for continuation of the perennial ryegrass bacterial microbiome from seed, through plant maturation and to seed was suggested, highlighting its hereditary potential64. It is envisioned that a specially tailored microbiome could be used as an enhancer for traits of interest, repressor or natural pest control, natural fertilizer, promoting a more sustainable and efficient agriculture and opening for the next Green Revolution. There is a clear need for further studies in these directions. The cooperative modulating role of the host and environmental variables emphasizes the importance of these variables for future development and application of synthetic microbiomes.
Material and methods
Field trial
This study was based on a larger field experiment which included a total of 64 ryegrass varieties. The soil samples were collected from plots established at Tystofte Foundation, located in the south-western region of Zealand, Denmark. The soil in this region is characterized as fine sandy clay65,66,67,68,69, with the composition of 10–15% clay, 0–30% silt, 40–90% fine sand, 55–90% sand and less than 10% humus. The seeds from 64 perennial ryegrass varieties were sown in August 2016, alternating a normal nitrogen treatment and an approximately 20% lower nitrogen treatment, using inorganic N fertilizer (Table 3), with two replicates for each treatment. Each variety was sown in a plot of size 1.5 m × 10 m. In the following year, after the plants were established, a total of twenty (five diploid and fifteen tetraploid) varieties were subjected to the soil microbiome study. The field layout and location of the sampled varieties within the field are illustrated in Supplementary Table S1.
Soil sample collection
This study considered 20 of the 64 ryegrass varieties (Supplementary Table S1). Three collection time points were selected as the day after the first, second, and third cut of the plants (cut-A: 24.05.2017, cut-B: 07.07.2017, cut-C: 17.08.2017). This study included thus 20 varieties × 2 treatments × 2 biological replicates × 3 sample collection time points; in total 240 soil samples. The soil samples were collected using a sampling tube, to a depth of 15 cm, from five points for each plot: the middle and 40 cm distance on the diagonals from each corner; these were afterwards mixed to represent the plot, as a sample for one variety. The soil samples were stored at − 20 °C, then freeze-dried. Roots were removed through agitation and sifting.
Perennial ryegrass dry matter yield
Dry matter content was measured using near infrared reflectance spectroscopy, and DM kg/plot was computed from dry matter content and total green mass yield harvested, as previously described70. The phenotyping data was kindly provided by the breeding company DLF, who performed the phenotyping according to their custom protocol as part of the GreenSelect project, funded by the Green Development and Demonstration Research Programme.
DNA extraction and sequencing
Total DNA was extracted from 0.25 g of soil using the DNeasy PowerLyzer PowerSoil kit (Qiagen) according to the manufacturer’s instructions. DNA concentration was assessed with Quant-iT PicoGreen dsDNA assay kit (Invitrogen, Oregon, USA). The sequencing was performed as custom paid service by Novogene (Novogene Corporation) using their TruSeq® DNA PCR-Free Library Prep workflow and the Illumina HiSeq platform, resulting in 250 bp paired-end reads. Region V3-V4 with a fragment length of 466 bp was targeted for the bacterial 16S amplicon sequencing and the ITS2 region with a fragment length of 350 bp was used to target fungi.
Sequence analysis and OTU identification
Paired-end reads were merged using FLASH (V1.2.7)71 and chimera sequences were detected with the UCHIME algorithm72,73. 30.2% of the 16S and 18% of the ITS2 sequences were chimera or low quality (Phred score < 20) and were removed (Supplementary Tables S2, S3). Sequences with 97% nucleotide similarity were clustered into operational taxonomic units (OTUs) using Uparse software74. For the 16S data, sequences were screened for annotation against the SSUrRNA database of the SILVA Database75 using Mothur76 with a threshold of 0.8–1 for taxonomic ranks. For the ITS2, sequence annotation was obtained by BLAST with Qiime77 and the Unit database78.
The raw OTU tables were scaled to the minimum number of reads across all samples (16S = 29,710 and ITS2 = 37,959) prior to statistical analyses, using the “rarefy” approach. OTUs with presence less than 0.1% sequences across all N samples were filtered out resulting in 6092 bacterial OTUs (based on 16S rRNA) and 2907 fungi OTUs (based on ITS2). The relative abundance within sample was computed for each OTU. To get the phylogenetic relationship among the OTUs (16S and ITS2 separate) we used MUSCLE79 for multiple sequence alignment and to construct a UPGMA (Unweighted Pair Group Method with Arithmetic Mean) phylogenetic tree.
Within sample complexity
Different measures of within sample OTU diversity (α-diversity) were computed from the bacteria and fungi OTU tables using the R packages RAM80 and picante81. We computed the OTU richness (S), Shannon’s Index (H), and Faith’s phylogenetic diversity (PD).
We compared the within sample diversity metrics (\({y}_{\alpha }\)) across ryegrass varieties (\({y}_{\alpha }=rep+horz+variety+e\)), ryegrass ploidy level (\({y}_{\alpha }=rep+horz+ploidy+e\)), nitrogen treatment level (\({y}_{\alpha }=rep+horz+variety+treatment+e\)) and seasonal cuts (\({y}_{\alpha }=rep+horz+variety+cut+e\)) using linear models, and comparing the full model to a reduced model neglecting the term of interest. For all comparisons we included a replicate effect (rep) and the horizontal position effect (horz) corresponding to the position in the field.
We conducted a combined correlation network analysis by computing Spearman’s correlation of the relative abundance among all bacteria and fungi OTUs. We only considered correlations that were significant (Bonferroni adjusted P value < 0.001) and had correlation coefficients below − 0.5 or above 0.5. The correlation networks were visualized with iGraph82.
Across sample complexity
While the α-diversity measures the within-sample diversity, the β-diversity is the OTU diversity across samples. First, we computed a distance metric over the OTUs using the generalized UniFrac distance83 using the R package GUniFrac84, which is a distance measure that incorporates the genetic distance of the OTUs in each sample to the OTUs in all the other samples. We then performed a permutational multivariate analysis of variance—PERMANOVA, implemented in the R vegan package85 to partition the variation within the distance matrix to seasonal cuts, replicate, in field location/horizontal placement, treatment and ryegrass variety. The PERMANOVA tests for differences in the centroid and variances among the groups (thus, seasonal cuts, replicate, horizontal placement, treatment and ryegrass variety) and also provides estimate of proportion of total variance explained by each factor. The results were visualized by plotting the first two principal coordinate axes from the principal coordinate analyses (PCoA).
Association between microbiome diversity and ryegrass dry matter
We estimated the proportion of total phenotypic variation (of dry matter yield) explained by ryegrass varieties (\({\widehat{H}}^{2}\)) by fitting a linear mixed model with the R package lme486 assuming independence among the ryegrass varieties. We obtained estimates of \({\widehat{H}}^{2}\) using the entire data set of 64 ryegrass varieties and for the subset of 20 varieties used in the soil microbiome study by fitting \(y=hrz+rep+cut+tr+cut:tr+id+id:tr:cut\), where \(y\) was a vector of dry matter content, the horizontal groups (\(hrz\)), replicate (\(rep\)), seasonal cut (\(cut\)), treatment (\(tr\)), and the interaction between cut and treatment (\(cut:tr\)) were considered as fixed effects. The ryegrass varieties (\(id\)) and the interaction between ryegrass varieties and treatment and seasonal cut (\(id:tr:cut\)) were considered as random effects. The broad sense heritability was estimated as \({\widehat{H}}^{2}=\frac{{\widehat{\sigma }}_{id}^{2}+{\widehat{\sigma }}_{id:tr:cut}^{2}}{{\widehat{\sigma }}_{id}^{2}+{\widehat{\sigma }}_{id:tr:cut}^{2}+{\widehat{\sigma }}_{e}^{2}}\), where \({\widehat{\sigma }}_{id}^{2}\) is the variance among ryegrass varieties, \({\widehat{\sigma }}_{id:tr:cut}^{2}\) is the variety-treatment-cut interaction variance, and \({\widehat{\sigma }}_{e}^{2}\) is the residual variance.
Finally, we investigated if any bacteria or fungi OTUs were associated with variation in ryegrass dry matter. We fitted one OTU at a time (\({otu}_{i}\)) specified as a fixed effect \(y={otu}_{i}+hrz+rep+cut+tr+cut:tr+id+id:tr:cut\), and compared the model to one neglecting the OTU using a likelihood ratio test. Following, all P values were adjusted for multiple testing using a false discovery rate (FDR) < 0.05 as significance threshold.
Data availability
Sequencing data is deposited in the EMBL-EBI/MGnify and will be made publicly available upon publication of the study.
Abbreviations
- OTU:
-
Operational taxonomic unit
- G × E:
-
Genotype by environment interactions
References
Humphreys, M. O., Feuerstein, U. & Vandewalle, M. Ryegrasses. In Fodder Crops and Amenity Grasses (eds Boller, B. et al.) 211–260 (Springer, 2010).
Talbot, M. Yield variability of crop varieties in the U.K. J. Agric. Sci. 102, 315–321 (1984).
Ravel, C. & Charmet, G. A comprehensive multisite recurrent selection strategy in perennial ryegrass. Euphytica 88, 215–226 (1996).
Jafari, A., Connolly, V. & Walsh, E. Genetic analysis of yield and quality in full-sib families of perennial ryegrass (Lolium perenne L.) under two cutting managements. Irish J. Agric. Food Res. 42, 275–292 (2003).
Conaghan, P., Casler, M., McGilloway, D., O’Kiely, P. & Dowley, L. Genotype x environment interactions for herbage yield of perennial ryegrass sward plots in Ireland. Grass Forage Sci. 63, 107–120. https://doi.org/10.1111/j.1365-2494.2007.00618.x (2008).
Fè, D. et al. Genomic dissection and prediction of heading date in perennial ryegrass. BMC Genom. 16, 921. https://doi.org/10.1186/s12864-015-2163-3 (2015).
Bais, H., Weir, T., Perry, L., Gilroy, S. & Vivanco, J. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. https://doi.org/10.1146/annurev.arplant.57.032905.105159 (2006).
Kuske, C. et al. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68, 1854–1863. https://doi.org/10.1128/AEM.68.4.1854-1863.2002 (2002).
Giovannetti, M., Salvioli di Fossalunga, A., Stringlis, I. A., Proietti, S. & Fiorilli, V. Unearthing soil-plant-microbiota crosstalk: Looking back to move forward. Front. Plant Sci. 13, 1082752. https://doi.org/10.3389/fpls.2022.1082752 (2023).
Aira, M., Gomez-Brandon, M., Lazcano, C., Baath, E. & Dominguez, J. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 42, 2276–2281. https://doi.org/10.1016/j.soilbio.2010.08.029 (2010).
Bouffaud, M. et al. Is diversification history of maize influencing selection of soil bacteria by roots?. Mol. Ecol. 21, 195–206. https://doi.org/10.1111/j.1365-294X.2011.05359.x (2012).
Peiffer, J. et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 110, 6548–6553. https://doi.org/10.1073/pnas.1302837110 (2013).
Gomes, E. et al. Root-associated microbiome of maize genotypes with contrasting phosphorus use efficiency. Phytobiomes J. 2, 129–137. https://doi.org/10.1094/PBIOMES-03-18-0012-R (2018).
Bulgarelli, D. et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. https://doi.org/10.1038/nature11336 (2012).
Kudjordjie, E. N. et al. Fusarium oxysporum disrupts microbiome-metabolome networks in Arabidopsis thaliana roots. Microbiol. Spectr. 10, e0122622. https://doi.org/10.1128/spectrum.01226-22 (2022).
Bulgarelli, D. et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403. https://doi.org/10.1016/j.chom.2015.01.011 (2015).
Edwards, J. et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 112, E911–E920. https://doi.org/10.1073/pnas.1414592112 (2015).
Edwards, J. A. et al. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 16, e2003862. https://doi.org/10.1371/journal.pbio.2003862 (2018).
Edwards, J. et al. Soil domestication by rice cultivation results in plant-soil feedback through shifts in soil microbiota. Genome Biol. 20, 221. https://doi.org/10.1186/s13059-019-1825-x (2019).
Kavamura, V. et al. Wheat dwarfing influences selection of the rhizosphere microbiome. Sci. Rep. 10, 1–11. https://doi.org/10.1038/s41598-020-58402-y (2020).
Kwak, M. J. et al. Author Correction: Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1117. https://doi.org/10.1038/nbt1118-1117 (2018).
Marasco, R. et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One 7, e48479. https://doi.org/10.1371/journal.pone.0048479 (2012).
Lu, T. et al. Investigation of rhizospheric microbial communities in wheat, barley, and two rice varieties at the seedling stage. J. Agric. Food Chem. 66, 2645–2653. https://doi.org/10.1021/acs.jafc.7b06155 (2018).
Wang, G. et al. Soil microbiome mediates positive plant diversity-productivity relationships in late successional grassland species. Ecol. Lett. 22, 1221–1232. https://doi.org/10.1111/ele.13273 (2019).
Dubey, A. et al. Soil microbiome: A key player for conservation of soil health under changing climate. Biodivers. Conserv. 28, 2405–2429. https://doi.org/10.1007/s10531-019-01760-5 (2019).
Berendsen, R., Pieterse, C. & Bakker, P. The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. https://doi.org/10.1016/j.tplants.2012.04.001 (2012).
Mendes, R., Garbeva, P. & Raaijmakers, J. M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. https://doi.org/10.1111/1574-6976.12028 (2013).
Bakker, P. et al. The soil-borne identity and microbiome-assisted agriculture: Looking back to the future. Mol. Plant 13, 1394–1401. https://doi.org/10.1016/j.molp.2020.09.017 (2020).
Chen, L. et al. Structural and functional differentiation of the root-associated bacterial microbiomes of perennial ryegrass. Soil Biol. Biochem. 98, 1–10. https://doi.org/10.1016/j.soilbio.2016.04.004 (2016).
Trivedi, P., Batista, B., Bazany, K. & Singh, B. Plant-microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 234, 1951–1959. https://doi.org/10.1111/nph.18016 (2022).
Cougnon, M. et al. In situ quantification of forage grass root biomass, distribution and diameter classes under two N fertilisation rates. Plant Soil 411, 409–422. https://doi.org/10.1007/s11104-016-3034-7 (2017).
Humphreys, M. et al. Root imaging showing comparisons in root distribution and ontogeny in novel Festulolium populations and closely related perennial ryegrass varieties. Food Energy Secur. https://doi.org/10.1002/fes3.145 (2018).
Wedderburn, M., Crush, J., Pengelly, W. & Walcroft, J. Root growth patterns of perennial ryegrasses under well-watered and drought conditions. N. Z. J. Agric. Res. 53, 377–388. https://doi.org/10.1080/00288233.2010.514927 (2010).
Kaiser, K. et al. Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci. Rep. 6, 33696. https://doi.org/10.1038/srep33696 (2016).
Cao, C. et al. Land-use changes influence soil bacterial communities in a meadow grassland in Northeast China. Solid Earth 8, 1119–1129. https://doi.org/10.5194/se-8-1119-2017 (2017).
Zifcakova, L., Vetrovsky, T., Howe, A. & Baldrian, P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ. Microbiol. 18, 288–301. https://doi.org/10.1111/1462-2920.13026 (2016).
Zhou, J. et al. Consistent effects of nitrogen fertilization on soil bacterial communities in black soils for two crop seasons in China. Sci. Rep. 7, 3267. https://doi.org/10.1038/s41598-017-03539-6 (2017).
Wang, H. et al. Nitrogen addition reduces soil bacterial richness, while phosphorus addition alters community composition in an old-growth N-rich tropical forest in southern China. Soil Biol. Biochem. 127, 22–30. https://doi.org/10.1016/j.soilbio.2018.08.022 (2018).
Terrazas, R. A. et al. Nitrogen availability modulates the host control of the barley rhizosphere microbiota. bioRxiv 605204 (2020).
Wang, X. et al. Soil nitrogen treatment alters microbiome networks across farm niches. Front. Microbiol. 12, 786156. https://doi.org/10.3389/fmicb.2021.786156 (2022).
Escudero-Martinez, C. et al. Identifying plant genes shaping microbiota composition in the barley rhizosphere. Nat. Commun. 13(1), 3443. https://doi.org/10.1038/s41467-022-31022-y (2022).
Nagy, I. et al. Chromosome-scale assembly and annotation of the perennial ryegrass genome. BMC Genom. 23, 505. https://doi.org/10.1186/s12864-022-08697-0 (2022).
Wagner, M. et al. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7, 12151. https://doi.org/10.1038/ncomms12151 (2016).
Li, Y. et al. Plant phenotypic traits eventually shape its microbiota: A common garden test. Front. Microbiol. 9, 2479. https://doi.org/10.3389/fmicb.2018.02479 (2018).
Liu, F. et al. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 19, 201. https://doi.org/10.1186/s12866-019-1572-x (2019).
Alegria Terrazas, R. et al. A footprint of plant eco-geographic adaptation on the composition of the barley rhizosphere bacterial microbiota. Sci. Rep. 10, 12916. https://doi.org/10.1038/s41598-020-69672-x (2020).
Tannenbaum, I., Rodoni, B., Spangenberg, G., Mann, R. & Sawbridge, T. An assessment of the Lolium perenne (perennial ryegrass) seedborne microbiome across cultivars, time, and biogeography: Implications for microbiome breeding. Microorganisms https://doi.org/10.3390/microorganisms9061205 (2021).
Chow, M., Radomski, C., McDermott, J., Davies, J. & Axelrood, P. Molecular characterization of bacterial diversity in Lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol. Ecol. 42, 347–357. https://doi.org/10.1016/S0168-6496(02)00392-6 (2002).
Quaiser, A. et al. Acidobacteria form a coherent but highly diverse group within the bacterial domain: Evidence from environmental genomics. Mol. Microbiol. 50, 563–575. https://doi.org/10.1046/j.1365-2958.2003.03707.x (2003).
Janssen, P. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728. https://doi.org/10.1128/AEM.72.3.1719-1728.2006 (2006).
Singh, B., Munro, S., Potts, J. & Millard, P. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecol. 36, 147–155. https://doi.org/10.1016/j.apsoil.2007.01.004 (2007).
Singh, B. et al. Investigating microbial community structure in soils by physiological, biochemical and molecular fingerprinting methods. Eur. J. Soil Sci. 57, 72–82. https://doi.org/10.1111/j.1365-2389.2005.00781.x (2006).
Navarrete, A. et al. Acidobacterial community responses to agricultural management of soybean in Amazon forest soils. FEMS Microbiol. Ecol. 83, 607–621. https://doi.org/10.1111/1574-6941.12018 (2013).
Bauer, R. & Oberwinkler, F. Direct cytoplasm-cytoplasm connection: An unusual host-parasite interaction of the tremelloid mycoparasite Tetragoniomyces uliginosus. Protoplasma 154, 157–160 (1990).
Seki, T., Matsumoto, A., Omura, S. & Takahashi, Y. Distribution and isolation of strains belonging to the order Solirubrobacterales. J. Antibiot. 68, 763–766. https://doi.org/10.1038/ja.2015.67 (2015).
Postma, A., Slabbert, E., Postma, F. & Jacobs, K. Soil bacterial communities associated with natural and commercial Cyclopia spp. FEMS Microbiol. Ecol. 92(3), fiw016. https://doi.org/10.1093/femsec/fiw016 (2016).
Schmidt, J., Kent, A., Brisson, V. & Gaudin, A. Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 7, 146. https://doi.org/10.1186/s40168-019-0756-9 (2019).
Jiao, S., Chen, W. & Wei, G. Resilience and assemblage of soil microbiome in response to chemical contamination combined with plant growth. Appl. Environ. Microbiol. 85, e02523-18. https://doi.org/10.1128/AEM.02523-18 (2019).
Wilhelm, R., Singh, R., Eltis, L. & Mohn, W. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 13, 413–429. https://doi.org/10.1038/s41396-018-0279-6 (2019).
Wei, L., Ouyang, S., Wang, Y., Shen, X. & Zhang, L. Solirubrobacter phytolaccae sp. nova, an endophytic bacterium isolated from roots of Phytolacca acinosa Roxb. Int. J. Syst. Evol. Microbiol. 64, 858–862. https://doi.org/10.1099/ijs.0.057554-0 (2014).
Sarikhani, E., Sagova-Mareckova, M., Omelka, M. & Kopecky, J. The effect of peat and iron supplements on the severity of potato common scab and bacterial community in tuberosphere soil. FEMS Microbiol. Ecol. 93(1), fiw206. https://doi.org/10.1093/femsec/fiw206 (2017).
Boukhatem, Z. F., Merabet, C. & Tsaki, H. Plant growth promoting Actinobacteria, the most promising candidates as bioinoculants?. Front. Agric. https://doi.org/10.3389/fagro.2022.849911 (2022).
De Luca, V., de Barreda, D., Lidon, A. & Lull, C. Effect of nitrogen-fixing microorganisms and amino acid-based biostimulants on perennial ryegrass. HortTechnology 30, 280–291. https://doi.org/10.21273/HORTTECH04236-19 (2020).
Tannenbaum, I. et al. Profiling the Lolium perenne microbiome: From seed to seed. Phytobiomes J. 4, 281–289. https://doi.org/10.1094/PBIOMES-03-20-0026-R (2020).
Madsen, H. B. A pedological soil classification system for Danish soils. Pedologie 33, 171–197 (1983).
Madsen, H. B. & Jensen, N. H. The establishment of pedological soil databases in Denmark. Geografisk Tidsskrift-Danish Journal of Geography 85, 1–8 (1985).
Breuning-Madsen, H. & Jensen, N. H. Pedological regional variations in well-drained soils. Geografisk Tidsskrift-Danish Journal of Geography 92, 61–69 (1992).
Adhikari, K. et al. High-resolution 3-D mapping of soil texture in Denmark. Soil Sci. Soc. Am. J. 77, 860–876. https://doi.org/10.2136/sssaj2012.0275 (2013).
Adhikari, K., Minasny, B., Greve, M. & Greve, M. Constructing a soil class map of Denmark based on the FAO legend using digital techniques. Geoderma 214, 101–113. https://doi.org/10.1016/j.geoderma.2013.09.023 (2014).
Fè, D., Pedersen, M. G., Jensen, C. S. & Jensen, J. Genetic and environmental variation in a commercial breeding program of perennial ryegrass. Crop Sci. Soc. Am. Spec. Publ. 55(2), 631–640 (2015).
Magoc, T. & Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. https://doi.org/10.1093/bioinformatics/btr507 (2011).
Edgar, R., Haas, B., Clemente, J., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. https://doi.org/10.1093/bioinformatics/btr381 (2011).
Haas, B. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. https://doi.org/10.1101/gr.112730.110 (2011).
Edgar, R. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. https://doi.org/10.1038/NMETH.2604 (2013).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. https://doi.org/10.1093/nar/gks1219 (2013).
Schloss, P. et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. https://doi.org/10.1128/AEM.01541-09 (2009).
Caporaso, J. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. https://doi.org/10.1038/nmeth.f.303 (2010).
Koljalg, U. et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. https://doi.org/10.1111/mec.12481 (2013).
Edgar, R. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. https://doi.org/10.1093/nar/gkh340 (2004).
Chen, W., Simpson, J. & Levesque, C. A. RAM: R for amplicon-sequencing-based microbial-ecology. R package version 1.2. 1.7. http://cran.r-project.org/package=RAM (2018).
Kembel, S. et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. https://doi.org/10.1093/bioinformatics/btq166 (2010).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst., 1695. https://igraph.org (2006).
Chen, J. et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28, 2106–2113. https://doi.org/10.1093/bioinformatics/bts342 (2012).
Chen, J., Zhang, X. & Yang, L. GUniFrac: Generalized UniFrac distances, distance-based multivariate methods and feature-based univariate methods for microbiome data analysis. https://cran.r-project.org/web/packages/GUniFrac/index.html (2008).
Oksanen, J. et al. vegan: Community ecology package. R package version 2.6-2. https://cran.r-project.org/package=vegan
Bates, D., Machler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. https://doi.org/10.18637/jss.v067.i01 (2015).
Acknowledgements
We are very grateful to Stephan Hentrup, Istvan Nagy (Aarhus University), Maria de la O Leyva Perez (formerly Aarhus University) for their help with the collection of soil samples. Many thanks to Rumakanta Sapkota and Enoch Nahr Kudjordjie (Aarhus University) for their kind advice and inspiring discussions related to the experimental setup, practical things, as well as data analyses. We kindly acknowledge TystofteFonden for performing the field trial, with many thanks especially to Anders Søndergaard Larsen and Jon R. Hedegaard. The perennial ryegrass cultivars were kindly provided by DLF Seeds. This project was funded by the Center for Genomic Selection in Animals and Plants (GenSAP), funded by the Danish Council for Strategic Research under grant number 12-132452. The field trials were funded by the GreenSelect project, funded by the Green Development and Demonstration Programme, under grant number 34009-15-0952.
Author information
Authors and Affiliations
Contributions
Conceptualization: JJ, TA. Methodology: JJ, TA, PDR, CP. Soil sample processing and laboratory work: CP, MF. Data analyses: PDR. Interpretation of results: PDR, CP. Visualization: PDR. Writing original draft: PDR, CP, MF. Review and editing: PDR, CP, TA, JJ, MF, PBH.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paina, C., Fois, M., Asp, T. et al. The soil microbiome of Lolium perenne L. depends on host genotype, is modified by nitrogen level and varies across season. Sci Rep 14, 5767 (2024). https://doi.org/10.1038/s41598-024-56353-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-56353-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.