Abstract
Anthropogenic activities have been shown to significantly affect marine life. Water pollution and oil spills are particularly deleterious to the fish population, especially during their larval stage. In this study, Sobaity-sea bream Sparidentex hasta (Valenciennes, 1830) larvae were exposed to serial dilutions of water-accommodated fraction of Kuwait crude oil (KCO-WAF) for varying durations (3, 6, 24, 48, 72 or 96 h) in acute exposure regime. Gene expression was assessed using RNA sequencing and validated through RT-qPCR. The RNA sequencing data were aligned to the sequenced genome, and differentially expressed genes were identified in response to treatment with or without KCO-WAF at various exposure times. The highest number of differentially expressed genes was observed at the early time point of 6 h of post-exposure to KCO-WAF. The lowest number of differentially expressed genes were noticed at 96 h of treatment indicating early response of the larvae to KCO-WAF contaminant. The acquired information on the differentially expressed genes was then used for functional and pathway analysis. More than 90% of the differentially expressed genes had a significant BLAST match, with the two most common matching species being Acanthopagrus latus and Sparus aurata. Approximately 65% of the differentially expressed genes had Gene Ontology annotations, whereas > 35% of the genes had KEGG pathway annotations. The differentially expressed genes were found to be enriched for various signaling pathways (e.g., MAPK, cAMP, PI3K-Akt) and nervous system-related pathways (e.g., neurodegeneration, axon guidance, glutamatergic synapse, GABAergic synapse). Early exposure modulated the signaling pathways, while KCO-WAF exposure of larvae for a longer duration affected the neurodegenerative/nervous system-related pathways. RT-qPCR analysis confirmed the differential expression of genes at each time point. These findings provide insights into the underlying molecular mechanisms of the deleterious effects of acute exposure to oil pollution—on marine fish populations, particularly at the early larval stage of Sparidentex hasta.
Similar content being viewed by others
Introduction
Anthropogenic activities are widely acknowledged to have a significant adverse impact on marine ecosystems and biodiversity globally1,2. Water pollution from various sources, such as agricultural, municipal, and industrial sources has become a major concern, which affects marine life. Further, the effects are profoundly evident in fish populations3. Multiple studies have reported a decline in the marine fauna as a result of anthropogenic activities owing to urbanization, and industrialization4,5.
Crude oil is one of the major pollutants released into the water and drastically affects the marine ecosystem. The Deepwater Horizon oil spill, also known as the largest oil spill in history, occurred in 2010 in the Gulf of Mexico. Approximately 4.4 million barrels of oil were released into the sea, resulting in a catastrophic impact on the marine ecosystem and coastal communities6. Despite the passing of a decade since this catastrophic event, researchers are still uncovering a vast amount of information regarding its effect on the marine environment6,7. For example, in the aftermath of the Deepwater Horizon disaster, it was discovered that the offshore fish population was in a lymphopenic or immunocompromised state due to the exposure to polycyclic aromatic hydrocarbons (PAHs)8. A study conducted on the short-term impact of the 2019 oil spill in Brazil on fisheries, indicated the prevalence of low molecular weight PAHs, mainly naphthalenes in the edible tissues of 34 finfish and shellfish species within the first 3 months time9. Oil spills not only affect the fish species but also other marine fauna. Oil spills are responsible for the mortalities of four different species of sea snakes in the Gulf of Oman10. The study showed that the majority (~ 85%) of sea snakes had oil covering on 75–100% of their bodies. Furthermore, snouts and eyes of ~ 91.4% of sea snakes were observed with oil covering. In addition, a large proportion (26–41%) of sea snakes had oil content in their mouth, esophagus and stomach. Furthermore, climate change and crude oil spills have a combined effect on fish species11. Polar cod embryos and larvae were exposed to low crude oil levels and a 2.3 °C increase in water temperature. The synergistic effects of increased temperature and crude oil exposure on early life stages were most prevalent in the first feeding larval stages, highlighting potential long-term consequences for survival, growth, and reproduction11. In addition, produced water in oil drilling, contains high levels of aromatic hydrocarbon and heavy metals. Its release can particularly impact the vulnerable larval and juvenile stages of marine organisms, leading to long-term ecological consequences12. These findings highlight the urgent need for effective measures to prevent oil spills and mitigate their long-lasting effects on marine fauna.
Kuwait is located in the North of the Arabian Gulf with an extensive coastline of 290 km long, taking into account the inclusion of its associated islands13. The effects of anthropogenic activities on marine life, particularly fish population has also been investigated in Kuwaiti waters. Alqattan and Gray discussed the nature of pollution in Kuwaiti waters, its causes, and measures that could be taken to reduce it14. The article also investigates whether pollution is the primary reason for the decline in fish stocks in Kuwaiti waters. Although the study indicates the existence of pollution in Kuwaiti waters, it fails to establish a direct link between oil pollution and the decline in fish stocks. In another study, Edmonds et al. assessed the state of Kuwait's marine biodiversity by reviewing data on the occurrence, distribution, and threats to key marine habitats and associated indicator organisms15.
The effects of oil spills on increased mortality of fish has been demonstrated in few other studies too16,17. However, eggs and larvae of fish species exhibited more vulnerability to the toxicity of crude oil due to their smaller size, poorly developed membrane and detoxification mechanisms17. Multiple studies have shown that PAHs in the crude oil even at low concentrations can cause lethal or sub-lethal damage to fish eggs and larvae18,19,20,21. This could result in morphological deformities, reduced feeding and growth rates resulting in starvation, and further increases the vulnerability to predators. A few studies have also indicated increased mortality of eggs and larvae at the spill sites22,23.
High-throughput technologies, such as microarray and RNA sequencing are powerful tools to understand the effect of oil pollution on the biology of fish species. RNA sequencing was performed to study the effect of water-accommodated fraction (WAF) of crude oil exposure in the gills of Japanese flounder, Paralichthys olivaceus, and the study provided insights into the mechanisms of WAF-induced toxicity in P. olivaceus24. Crude oil exposure weakens the immune function and increases the susceptibility to Vibrio anguillarum, a causative agent of vibriosis in southern flounder25. The authors demonstrated decreased expression of immunoglobulin M, the major systemic fish antibody, and downregulation of the genes related to immune function, response to stimulus and hemostasis in the fish specimens exposed the crude oil. RNA sequencing of liver and gill tissues of lined sole fish was performed to assess the transcriptomic changes in response to WAF of light crude oil26. WAF treatment resulted in the induction of hypoxia-regulated genes and the regulation of multiple signaling pathways in lined sole fish. Jantzen et al.,27 demonstrated the effect of three different polyfluorinated compounds on morphometric, behavior, and gene expression in both yolk sac fry and larval zebrafish. Administration of crude oil to Atlantic haddock in short term exposures at two developmental stages resulted in the impairment of calcium homeostasis by affecting calcium-regulated developmental pathways, including cardiogenesis2.
The Sobaity seabream, Sparidentex hasta (Valenciennes, 1830), is an economically important species and is considered as one of the most promising species for aquaculture due to its good adaptation to captivity, rapid growth, and high market value. The toxicity of WAF and chemically enhanced water-accommodated fraction (CEWAF) of Kuwait crude oil with three dispersants (Corexit® 9500, Corexit® 9527, and Slickgone® NS) on the larvae of S. hasta was investigated28. The authors demonstrated that the dispersed and undispersed KCO resulted in toxicity manifestations in different magnitudes. WAF of the Kuwait crude oil and CEWAF with of Corexit® 9527 had higher toxicity, whereas, CEWAFs with Corexit® 9500 and Slickgone® NS had lower toxicity on fish larvae28. Nevertheless, there remains a research gap in investigating the effects of crude oil on S. hasta larvae at the molecular-level.
In the current study, we used RNA sequencing to investigate the effects of WAF of Kuwait crude oil (KCO-WAF) on S. hasta larvae at various exposure times. The larvae were incubated in seawater with or without KCO-WAF for 3, 6, 24, 48, 72 or 96 h and differentially expressed genes were identified using RNA sequencing. The functional and pathway analyses of the genes were performed to determine the effects of KCO-WAF on fish larvae at the molecular level. Additionally, we analyzed the effects of early, intermediate, and longer exposures of KCO-WAF by studying the various sets of differentially expressed genes. Furthermore, we have also validated a selected set of genes differentially expressed at each time point through RT-qPCR.
Materials and methods
Fish larvae exposure assay
Wild broodstock fish caught from the Kuwait waters were domesticated in tanks for 2 years at the Mariculture and Fisheries Department, Kuwait Institute for Scientific Research (KISR), Salmiya, Kuwait. All experiments on fish and larvae were performed as per the approved institutional guidelines of KISR. The cultured broodstock spawned naturally in tanks without any hormonal induction. Twenty-four hours post-hatch larvae of Sobaity-sea bream weighing around 0.1–0.75 mg were procured from the Aquaculture facility of KISR and acclimated to the laboratory conditions at the Ecotoxicology Laboratory at KISR. Kuwait Crude Oil (KCO) of API gravity 30 was procured from Petroleum Research Center (PRC) of KISR. The acute toxicity test was done according to the guidelines of the OCED (Organization of Economic Co-Operation and Development) for the Fish Embryo Toxicity (FET) Test29,30. KCO WAF prepared according to Singer et al.31, and previously standardized method within the research team's laboratory32,33. A known amount of KCO (0.25, 0.5, or 1 g) was placed in aspirator bottles containing 1 L filtered seawater, stirred for 24 h and then allowed to stay for the separation of the aqueous layer. WAF fraction was drained and collected in amber bottles, characterized for TPH and used in WAF exposure experiments. Based on the results of the preliminary experiments, the WAF prepared by mixing 0.5 g KCO/L concentration was used for the assay at 50% dilution of WAF (1:1) with clean sea water containing larvae (for each replicate n = 40–50). The larvae were then exposed to KCO-WAF in triplicate for 3, 6, 24, 48, 72, or 96 h in a static non-renewal test regime according to ASTM: E 729-9634. The control treatment consisted of larvae samples exposed to filtered seawater without KCO-WAF for the same duration at each time point. Each treatment was conducted in individual 100 ml beakers containing 40–50 larvae in filtered sea water with our without WAF.
RNA isolation
The total RNA was isolated from the control and KCO-WAF-exposed larvae using TRIzol LS reagent (Invitrogen, USA). Each experimental replicate beaker containing 40–50 larvae was sieved for collecting the larvae and immediately frozen in liquid nitrogen. The samples were homogenized with 0.75 mL TRIzol LS reagent and stored at − 80 ℃ until further use. RNA isolation was carried out following the instructions provided by the manufacturer. The total yield of RNA was determined using a Nanodrop spectrophotometer and Qubit fluorometer, and the purity of RNA was assessed by measuring the ratio of absorbance at 260 nm and 280 nm and by agarose gel electrophoresis.
Library preparation and sequencing
The mRNA molecules were purified from total RNA using oligo(dT)-attached magnetic beads and fragmented using fragmentation reagent. First-strand cDNA was generated using random hexamer-primed reverse transcription, followed by a second-strand cDNA synthesis. The synthesized cDNA was subjected to end-repair and was 3′ adenylated. Adapters were ligated to the ends of the 3′ adenylated cDNA fragments followed by amplification. The PCR products were purified with Ampure XP Beads (AGENCOURT), and dissolved in EB solution. Library was validated on the Agilent Technologies 2100 bioanalyzer. The double-stranded PCR products were heat denatured and circularized by the splint oligo sequence. The single-strand circular DNA (ssCir DNA) molecules were considered as the final library. The library was then amplified with phi29 to make DNA nanoball (DNB), which had more than 300 copies of one molecule. The DNBs were loaded into the patterned nanoarray paired-end reads 100/150 bp were generated. The sequencing was carried out at the Beijing Genomics Institute (BGI) Tech Solutions, Hong Kong. The sequencing data can be accessed from NCBI-SRA under the accession PRJNA748027.
Analysis of transcriptome sequencing data and differential gene expression
Around 200 Gb of paired-end RNA sequencing data was checked for quality before and after trimming using FastQC v0.10.1 [https://www.bioinformatics.babraham.ac.uk/projects/fastqc/]. The RNA-seq raw data was trimmed for low quality reads based on base-quality scores and read length, using Trimgalore 0.6 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). A minimum quality score of 20 and read length of 50 was considered for trimming. The trimmed and filtered RNA-seq reads were aligned to the assembled genome35 and transcriptome using TopHat version 2.1.036 with default parameters. Quantification of the genes was performed using Cufflinks version 2.2.137 with default parameters. The differential expression analysis of the genes was performed using Cuffdiff package within Cufflinks37, with default parameters. Genes differentially expressed with a |log2-fold ≥ 1| and FDR P value < 0.01 were considered significant in each comparison.
Functional analysis of differentially expressed genes
The significantly differentially expressed genes were considered for functional analysis, performed using Blast2GO tool38. The sequences corresponding to significant genes were mapped to ‘nr’ protein database using blastx with an E-value of 1.0E-5, word size of 6 and HSP length cut-off of 33. The gene sequences were blasted against “Metazoa (tax id: 33208)” sequences in the ‘nr’ database. The blast alignment results were then used for Gene Ontology (GO) mapping (GO database version 2021-11) and annotation. Further, InterproScan analysis was performed using the alignment results.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
For experimental validation of the RNA-seq data, 12 differentially expressed genes were selected for RT-qPCR experiments. Genes for RT-PCR were selected based on their high expression levels determined through RNA sequencing data. Beta-actin was considered as one of the reference gene. We selected another control gene from the list of un-differential genes showing the highest p value and lowest logFC across all treatment conditions. Primers for the genes were designed using the Ex-Ex Primer tool39. A list of primers used is provided in Supplementary file 1. RT-qPCR was performed using TB Green Premix Ex Taq II (Tli RNase H Plus) in a StepOnePlus RealTime PCR System (Applied Biosystems). The thermal cycling conditions were set as follows: sample holding at 95 °C for 10 min; 40 cycles of denaturation step at 95 °C for 20 s; annealing at 63 °C for 20 s; extension at 72 °C for 20 s. The Ct value was calculated with auto threshold using the default option. The calculation of the fold change was performed as previously described40. The fold change of the genes was calculated using the 2^(-DeltaDeltaC(T))/comparative C(T) method. For both control and KCO-WAF exposed samples, the expression level was calculated using the following formula: “2^-(Ct (gene of interest − reference gene)”, and the fold change for each gene was derived by dividing its expression in the KCO-WAF exposed samples with that of in control samples.
Results
Transcriptome sequencing of KCO-WAF treated and untreated S. hasta larval samples
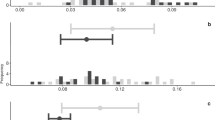
A total of 35 samples across six treatment and control conditions were sequenced to obtain approximately two billion paired-end reads. On average, 60 million raw reads were obtained per sample, and ~ 58 million good-quality reads per sample were retained after quality filtering. Except for 24 h KCO-WAF treated sample, all samples were sequenced in triplicate. The quality filtering of the reads resulted in approximately 96.3% of good-quality reads (Table 1). The number of the raw and filtered reads obtained for each replicate along with the read length are listed in Supplementary File 2. The filtered reads were aligned to an assembled genome35 containing 41201 genes. Differential expression analysis between treatment and control conditions resulted in varying numbers of genes across different treatment groups (Fig. 1). There was no specific trend in the number of genes expressed differently across the increasing treatment durations. The 96 h treatment resulted in least number of differentially expressed genes, whereas 6 h KCO-WAF treatment resulted in most differential genes (Table 2 and Supplementary File 3). Overall, the number of downregulated genes was more than that of the upregulated genes across the treatment groups.
Functional analysis of significantly differentially expressed genes
The significantly differentially expressed genes with a log2-fold change ≥ 1 and FDR P value < 0.01 at each treatment time point were considered for functional analysis using Blast2GO. The results revealed that > 90% of the differentially expressed genes had a significant BLAST match in case of both upregulated (Fig. 2A) and downregulated (Fig. 2B) lists. Furthermore, the two most common matching species based on BLAST alignment were Acanthopagrus latus and Sparus aurata. Approximately 50% of the S. hasta genes matched to A. latus. In addition, approximately 65% of the significantly differentially expressed genes had Gene Ontology annotations, while > 35% of the S. hasta genes had KEGG pathway annotations across various treatments. It is noteworthy that a higher fraction of the genes regulated at early and late treatments encoded for enzymes than those regulated during intermediate treatment. Furthermore, 50% of the enzymes encoded by the up/down-regulated genes belonged to transferase or translocase classes, whereas < 10% of the enzymes belonged to lyases, ligases and isomerases together (Fig. 3). Additionally, a higher percentage of the hydrolases (28%) were encoded by the downregulated genes than that of the upregulated genes (17%).
Pathways regulated by differentially expressed genes in response to KCO-WAF treatment at different durations
The pathway analysis of the up-and down-regulated genes following KCO-WAF treatment at different durations resulted in a varying number of pathways at each time point (Table 3). Highest number of pathways were represented by the genes differentially regulated at 6 h WAF treatment, and the least number of pathways by the genes regulated at 96 h WAF treatment. Table 4 lists top 10 differentially expressed genes between the treated and untreated samples at each time point along with their annotation details and number of associated pathways. Interestingly, even though a comparatively lower number of genes were differentially expressed at 24 h WAF treatment than that at 3 h treatment, a higher number of pathways were regulated at 24 h indicating the modulation of different pathways by KCO-WAF at 24 h post exposure. Supplementary file 4 lists all the pathways represented by the differentially expressed genes at each time point.
Furthermore, the genes differentially expressed at 3 h WAF treatment were found to be involved in various signaling pathways (Fig. 4A,B, and Supplementary file 4). Among these, ‘MAPK signaling’ pathway was represented by highest number of upregulated genes (~ 15%), whereas, only 1% of the downregulated genes were involved in this pathway. Interestingly, in addition to various signaling pathways, neurological physiology related pathways were represented by the genes differentially expressed at 6 h (Fig. 4C,D) and 24 h (Fig. 5A,B) post exposure to WAF. These include ‘pathways of neurodegeneration’, ‘axon guidance’, ‘glutamatergic synapse’, ‘Alzheimer’s disease’, ‘GABAergic synapse’, and ‘serotonergic synapse’. The ‘pathways of neurodegeneration’ was represented by the highest number of upregulated genes by both 6 and 24 h post exposure to WAF. The pathways ‘protein digestion and absorption’ and ‘pancreatic secretion’ were represented by highest number of upregulated genes (~ 21%), whereas, ‘retrograde endocannabinoid signaling’ and ‘glutamatergic synapse’ were represented by most number of downregulated genes (~ 17%) at 48 h post exposure to WAF (Fig. 5C,D). Furthermore, 72 h post exposure to WAF showed upregulated pathways, related to ‘neuroactive ligand-receptor interaction’, ‘dopaminergic synapse’, and ‘spinocerebellar ataxia’, while downregulated pathways, such as ‘focal adhesion’, and ‘protein digestion and absorption’ (Fig. 6A,B). The ‘tight junction’ pathway was represented by highest number of upregulated genes (~ 29%), and ‘phagosome’ and ‘drug metabolism’ pathways were represented by most number of downregulated genes (25%) at 96 h WAF treatment (Fig. 6C,D).
Pathways represented by the early-responsive genes following KCO-WAF treatment of S. hasta larva. Top 10 pathways represented by the genes up and downregulated at 3 h (A, B, respectively) or 6 h (C, D, respectively) WAF exposure. Pathways shared by early responsive up- (E) or downregulated (F) genes.
Pathways represented by the intermediate-responsive genes following KCO-WAF treatment of S. hasta larva. Top 10 pathways represented by the genes up and downregulated at 24 h (A, B, respectively) or 48 h (C, D, respectively) WAF exposure. Pathways shared by intermediate-responsive up- (E) or downregulated (F) genes.
Pathways represented by the late-responsive genes following KCO-WAF treatment of S. hasta larva. Top 10 pathways represented by the genes up and downregulated at 72 h (A, B, respectively) or 96 h (C, D, respectively) WAF exposure. Pathways shared by late-responsive up- (E) or downregulated (F) genes.
Common and exclusive pathways regulated by early-, intermediate-, and late-responsive genes following KCO-WAF treatment
Furthermore, we identified pathways that were common and exclusive to early, intermediate, or late responsive genes. The intermediate responsive genes (differentially regulated at 24 and 48 h post exposure to WAF) had highest percentage of shared pathways with 21.7% upregulated and 58.6% downregulated (Fig. 5E,F) followed by early responsive genes (differentially regulated at 3 and 6 h post exposure to WAF) with 55.1% upregulated and 19.8% downregulated pathways (Fig. 4E,F). The late responsive genes (differentially regulated at 72 and 96 h WAF treatment) shared 27.9% upregulated and 14.8% downregulated (Fig. 6E,F) pathways. Supplementary file 5 provides a list of common and exclusive pathways regulated by early-, intermediate-, and late-responsive genes expressed in response to exposure to KCO-WAF.
The genes upregulated at 3 and 6 h WAF treatment regulated various signaling pathways. A few of these pathways include, MAPK, cAMP, ErbB, TNF, PI3K-Akt, oxytocin, and glucagon signaling. The pathways exclusively represented by the upregulated genes at 3 h treatment include ‘glycolysis/gluconeogenesis’, ‘bisphenol degradation’ and that represented by upregulated genes at 6 h treatment include ‘proteoglycans in cancer’, ‘lysine degradation’, ‘insulin signaling pathway’. Similarly, the genes downregulated at 3 and 6 h treatment represented various signaling pathways. Genes downregulated at 6 h WAF treatment represented 268 exclusive pathways (Fig. 4F), including ‘Hippo signaling pathway’, ‘regulation of actin cytoskeleton’, ‘protein digestion and absorption’, ‘fatty acid degradation’, and ‘endocytosis’ (Supplementary file 5).
A total of 48 pathways were commonly upregulated at 24 and 48 h post exposure to WAF, mostly including signaling related pathways. Among these, ‘cAMP signaling pathway’ was represented by 26% of the pathway annotated upregulated genes at 24 h treatment, whereas ‘pancreatic secretion’ pathway was represented by most number (21%) of pathway annotated upregulated genes at 48 h treatment. Furthermore, the genes upregulated at 24 h treatment represented 136 exclusive pathways, whereas 37 pathways were exclusive to the upregulated genes at 48 h treatment (Fig. 5E). Similarly, 41 pathways were exclusive to genes downregulated at 24 h WAF treatment, whereas 34 pathways were exclusive to the genes downregulated at 48 h treatment (Fig. 5F). Among the exclusive pathways, ‘pathways of neurodegeneration’ was represented by 29% of annotated genes upregulated at 24 h treatment, whereas ~ 21% of the annotated genes were involved in ‘protein digestion and absorption’ among those upregulated at 48 h treatment. When downregulated genes were considered, 106 pathways were commonly regulated by 24 and 48 h WAF treatment, whereas 41 and 34 pathways were exclusively regulated by 24 and 48 h treatment, respectively. ‘Axon guidance’ was exclusively regulated by 8.2% of the pathway annotated genes that were downregulated at 24 h, whereas, each of the ‘retinol metabolism’ and ‘Hippo signaling’ pathways were regulated by 4.8% of the pathway annotated genes that were downregulated at 48 h treatment (Supplementary file 5).
Furthermore, a total of 64 pathways were commonly upregulated by late WAF treatment (72 and 96 h treatment). Among these, ‘neuroactive ligand-receptor interaction’ was represented by 14% of the pathway annotated genes upregulated at 72 h treatment, whereas ‘tight junction’ pathway was represented by most number (29.4%) of pathway annotated genes upregulated at 96 h treatment. In addition, the genes upregulated at 72 h treatment represented 150 exclusive pathways, whereas 15 pathways were exclusive to the upregulated genes at 96 h treatment (Fig. 6E). ‘Dopaminergic synapse’ and ‘spinocerebellar ataxia’ pathways were each exclusively represented by 14% of the pathway annotated genes that were upregulated at 72 h treatment, whereas, ‘phagosome’ was exclusively represented by 23.5% of the pathway annotated genes that were upregulated at 96 h treatment. There were 34 pathways that were commonly regulated by the genes downregulated at 72 and 96 h treatment, while 186 and 10 pathways were exclusive to the downregulated genes at 72 h and 96 h treatments, respectively (Fig. 6F).
RT-qPCR validation of differentially expressed genes
RT-qPCR was performed to validate the RNA sequencing results. The validation was performed by randomly selecting 12 genes among those that were significantly differentially expressed (FDR P value < 0.05) in at least one treatment group based on RNA sequencing. The differential regulation obtained by the RT-qPCR analysis showed complete agreement with that of the RNA sequencing data for the validated genes (Table 5).
Discussion
Anthropogenic activities, such as crude oil contamination, have a significant impact on the marine ecosystem and the health of fish. The release of crude oil into the marine environment can occur through natural disasters, such as oil spills, or through human activities, such as oil exploration and transportation. The impacts of crude oil contamination on the marine ecosystem and fish health can be devastating, and the effects can be long-lasting. It is crucial to investigate the impact of oil pollution on fish species. While, oil pollutants greatly affect the adult fish, their deleterious effects are much more pronounced in the larval stages. Consequently, several studies have focused on examining the damage caused to fish larvae or eggs when exposed to crude oil9,18,19. Here we further explore the impacts of crude oil contamination on the marine ecosystem and fish health, with a focus on the changes in the gene expression which results in alteration in the regulation of genetic pathways during the early developmental stages of fish larvae.
In the current study, we exposed S. hasta larvae to KCO-WAF for varying durations (3, 6, 24, 48, 72, or 96 h) and explored its deleterious effects on the gene expression using RNA sequencing. Additionally, we studied the functional processes and pathways that are modulated at different exposure times.
The use of WAF of crude oil is a well-established method to test the toxicity of petroleum31,41,42. However, it has some limitations mainly due to minor differences in the technique used for WAF preparation which can alter the composition of the WAF in different laboratories. Also, the chemical composition of the crude oil can vary depending on the source and batch, and thus the water WAF composition could vary. The selection of KCO-WAF concentrations in our study was based on established practices in the field of aquatic toxicity testing for petroleum products24,28,43,44,45,46,47,48. Use of WAF has emerged as the method of choice for its simplicity and wide applicability over three decades, allowing for the testing of complex substances like petroleum products in biological systems48. In the aquatic toxicity tests, the exposure concentration primarily represents the bioavailable dissolved fraction of the test material. WAF based toxicology experiments has proven effective across various organisms, including algae, crustaceans, gastropod and bivalve mollusks, and fish, both in freshwater and saltwater environments48. The utilization of WAF concentrations in our study aligns with the well-established and widely adopted methodology in the field of aquatic toxicity assessment for petroleum products.
Earlier reports demonstrated the degree of toxicity of WAF prepared from varying KCO loadings conducted in exposure chambers with fish larvae exposed for 96 h28,49. In this study, the total petroleum hydrocarbon (TPH) concentrations in the KCO-WAF prepared using 0.5 g /L oil loading in seawater was found to be 0.4 ± 0.09 mg/L. Although the WAF exposure experiments with sobaity larvae were conducted at three different KCO oil loading (0.25, 0.5, 1 g/L), the results indicated that 0.5 g/L KCO loading for WAF preparation is optimum for crude oil exposure assays (data not shown). Earlier studies also have highlighted that the acute toxicity in crude oils is primarily attributed to the presence of polycyclic aromatic hydrocarbons (PAHs), with naphthalene being a significant contributor50.
Environmental changes due to anthropogenic activities can have detrimental affect on the aquatic organisms. Scott and Sloman discussed how various toxicants disrupt complex fish behaviors essential for fitness and survival, often at lower exposures than those causing mortality20. In this context, crude oil pollution of aquatic habitat has been shown to exhibit profound negative effects on fish species especially at the early larval stages. Incardona et al. found that exposure to crude oil-derived PAHs caused specific dose-dependent defects in cardiac function in pelagic fish species, indicating potential mortality and malformations51.
Understanding the early changes in gene expression in fish larvae exposed to crude oil toxicity represents a crucial area of research with several notable gaps. While there is a growing body of literature examining the impacts of crude oil exposure on fish development, there is a paucity of comprehensive studies focused specifically on the gene expression changes in the early developmental stages of larvae. Understanding the molecular responses during this critical period is essential for elucidating the mechanisms underlying developmental abnormalities and impaired growth observed in fish exposed to crude oil. Additionally, there is a need for more extensive investigations into the specific genes and pathways that are modulated early on, as this information could provide valuable insights into the biomarkers indicative of oil-induced stress and potential targets for mitigation strategies.
Studying the immediate effects of crude oil on fish larvae, ranging from 3 to 96 h of acute exposure, allows us to capture the early molecular responses triggered by crude oil exposure, providing insights into the immediate changes in gene expression and signaling pathways. The acute exposure period is critical for understanding the initial impact of crude oil on biological systems, as it helps identify the immediate stress responses and potential adaptive mechanisms activated by the organisms. The differential expression analysis of genes indicated that WAF treatment at 6 h had a profound effect on the fish larvae showing higher number of differentially expressed genes. These findings were further corroborated by pathway analysis, which showed modulation of most pathways by the genes deregulated at 6 h treatment. Most of the top 10 genes were annotated to known genes using BLAST, however, very few of these had pathway annotations, which indicates that the differencially expressed genes in response to KCO-WAF need to be further investigated in terms of their role. When we grouped the treatments into early (3 and 6 h), intermediate (24 and 48 h) and late (72 and 96 h) exposure, the genes modulated by the intermediate exposure were found to be similar in function based on the pathways they shared, whereas, a higher percentage of pathways were unique during the late exposure. Thus, different sets of pathways are regulated by KCO-WAF exposure in a time-dependent manner.
The activation of multiple genes associated with crucial cellular signaling pathways (Fig. 4), such as MAPK (Mitogen-Activated Protein Kinase), cAMP (cyclic adenosine monophosphate), and PI3K-Akt (Phosphoinositide 3-kinase-Akt), within a short period of 3–6 h post crude oil exposure suggests a rapid and dynamic cellular response to environmental stress. These signaling pathways play pivotal roles in various cellular processes, including cell proliferation, survival, and response to external stimuli. Crude oil exposure can lead to oxidative stress and dysfunction in various organisms. Earlier reports suggests uprgulation of CYP450 in response to crude oil exposure43,46,47,52,53,54. Cytochrome P450 is a xenobiotic-metabolizing enzyme, and its upregulation suggests a detoxification response to the presence of crude oil contaminants. Members of the CYP450 family, belonging to 1A and 1B groups are induced by aryl hydrocarbon receptors (AhRs) upon binding of PAH ligands and results in enhanced metabolism of xenobiotics55. Rapid upregulation of Cytochrome P450 1B1 gene was detected in our study at 3 h of post exposure to WAF (Table 4). The effectiveness of the detoxification response in mitigating the toxic effects of crude oil is still a subject of research. In earlier studies it has been shown that exposure to elevated crude oil concentrations induces a lethal syndrome of heart failure in fish embryos, primarily attributed to cardiotoxic polycyclic aromatic hydrocarbons (PAHs) found in petroleum. Even low concentrations of oil during embryonic development result in sublethal toxicity, which persists and is not mitigated by Cytochrome P450 induction56. The findings could have profound implications beyond fish populations, indicating potential impact of such pollution for other vertibrates including human56.
The MAPK pathway is a well-known signaling cascade that regulates cellular responses to a variety of stimuli, including environmental toxins57. The rapid activation of MAPK genes implies that fish larvae are mobilizing intracellular signaling mechanisms to adapt to the stress induced by crude oil exposure58,59,60. This early response could be indicative of the early protective mechanisms or repair processes. The upregulation of genes associated with cAMP suggests that fish larvae are orchestrating a complex intracellular communication network in response to crude oil exposure. This could involve modulation of various cellular processes, including gene expression, metabolism, and stress responses as cAMP is a key messenger that mediates cellular responses to extracellular signals61,62,63,64. The involvement of the PI3K-Akt pathway further highlights the complexity of the cellular response. This pathway is central to regulating cell survival and growth, and its activation may indicate an effort to maintain cellular homeostasis under the stress of crude oil exposure65,66,67,68,69. The upregulation of genes such as synaptosomal-associated protein and cytochrome P450 1B1 indicates its possible role in synaptic vesicle trafficking and neurotransmitter release, suggesting that crude oil exposure may influence neural processes in fish larvae.
In summary, the changes in gene expression indicates an orchestrated molecular response in sobaity fish larvae to crude oil exposure. Understanding these early changes at the genetic level provides valuable insights into the mechanisms of adaptation and potential stress-induced impacts on various cellular processes. Further research into the functional consequences of these gene activations can help unravel the intricacies of the biological response to environmental stressors.
Furthermore, our functional analysis showed that many of the differentially expressed genes matched to transferase or translocase enzyme classes. Transferases are the class of enzymes that catalyze the transfer of specific functional groups from one molecule to another70. These classes of enzymes are involved in various biochemical pathways and integral to most of the important biological processes, e.g., translation71. Furthermore, translocases are class of enzymes that catalyze the movement of ions or molecules across membranes72. These classes of enzymes are involved in process, such as oxidative phosphorylation, electron transport chain, and fatty acid oxidation70,71,72,73,74. Thus, a higher percentage of the genes coding for such enzymes being differential clearly indicates the modulation of various metabolic pathways in fish larvae following WAF exposure.
The differentially expressed genes modulated various pathways, including those related to signaling, neurodegeneration, nervous system, and different diseases. These pathways are fundamental to the intricate network of molecular interactions that regulate critical aspects of an organism's development, physiological functions, adaptations and stress tolerance. By modulating these pathways, cells can respond to various internal and external cues, ensuring the proper progression of developmental stages and the coordination of essential physiological activities during growth and development. Understanding the intricacies of these pathways is important to study the molecular mechanisms that underline stress tolerance in S. hasta larvae exposed to chemical pollutants including WAF of crude oil.
Disruption of calcium signaling pathways has been known to result in cardiac morphology abnormalities75,76, and leads to defects in cardiac rhythm and contractility, as well as cardiomyocyte proliferation51,77. Our results revealed that differentially expressed genes at the 24-h and 48-h treatment time points were associated with the 'calcium signaling pathway,' suggesting that WAF exposure impacts cardiac functions in S. hasta fish larvae (Fig. 5). At 24 h post-exposure, a higher percentage of genes associated with 'calcium signaling pathway,' were downregulated, whereas at 48 h, a higher percentage of genes were found to be up regulated (Fig. 5). Furthermore, we found a higher number of genes to be associated with neurodegenerative disorders and other neurology-related pathways when fish larvae were exposed to WAF for longer duration. Thus, the crude oil exposure for extended duration may result in neurological damages in developing fish larvae. Our findings are in agreement with the previous whole transcriptome studies, which demonstrated that exposure to crude oil components result in physiological changes in the brain tissue, neurotransmitter regulation, and locomotory behaviour in fish species, indicating a neurotoxic potential of the petroleum constituents78,79,80. Furthermore, Xu et al. demonstrated time-dependent transcriptomic and physiological changes in embryos/larvae of mahi-mahi exposed to WAF of weathered (slick) oil47. The predominant transcriptomic responses upon slick oil exposure included alteration of signaling, ribosome biogenesis, steroid biosynthesis, and activation of the Cytochrome P450 pathway, corroborating with the findings from the current study. Additionally, 96 h of exposure to WAF resulted in significant perturbations in the eye and peripheral nervous system which was in agreement with our findings. A transcriptomic study on mahi-mahi indicated a switch in the developmental progression based on the enrichment of different biological processes and pathways while progressing from early embryonic stage to late developmental stages81.
Earlier studies predominantly focused on juvenile or mature fishes, leaving a critical gap in understanding how gene expression changes over time in the early life stages of fish. This lack of knowledge is particularly significant as fish larvae represent a vulnerable stage, and their responses to environmental stressors may differ substantially from those of adults. An ecotoxicogenomic study on juvenile rainbow trout exposed to crude oil, temporal changes in gene expression were examined, revealing unique patterns in gill and liver responses82. While the gill exhibited significant alterations in the expression of several genes during exposure, the liver showed delayed responses, with concentration-dependent effects82. A high number of 1137 genes were differentially regulated within 24 h of exposure in the gill tissues. The observed rapid response of larvae at early developmental stages aligns with findings in the study on juvenile rainbow trout exposed to crude oil, where a high number of pathways/genes were differentially regulated during initial exposure at 24 h. There number of pathways gradually decreased at later stages (72 and 96 h post-exposure) in S. hasta larvae. The late-responsive genes identified in our study were particularly those associated with the nervous system, phagosome, and dopaminergic synapse, parallel the neurodegenerative/nervous system-related pathways. Prolonged 96 h exposure to KCO-WAF resulted in the manifestation of symptoms such as abnormal and slow swimming, musculoskeletal deterioration leading to impaired mobility and body control which typically mirrors the deleterious effects of crude oil exposure as shown in previous work by others28,32,33,49,83. Our study also showed an enrichment of different sets of pathways in S. hasta larvae following WAF treatment in a time-dependent manner, showing that WAF exposure for longer durations have significant deleterious effects on fish larvae.
The current study used RNA sequencing to reveal the effects of KCO-WAF on the developmental phases of S. hasta. Different sets of genes were found to be differentially expressed following WAF exposure at 3, 6, 24, 48, 72, or 96 h. WAF exposure at 6 h had the most significant effect on fish larvae, as evident from the number of differentially expressed genes. Functional analysis of the differentially expressed genes clearly indicated modulation of various physiological and developmental processes in fish larvae following WAF exposure. The pathways that were found to regulated by the differentially expressed genes at different time points included primarily signaling, cardiac, and nervous system related pathways. Thus, our results indicate that WAF exposure has deleterious effects on cardiac physiology and nervous system during the developmental stages of S. hasta.
Deposited data and information to the user
The complete sequences, which were used for the genome assemblies and annotations, have been deposited in public data repositories. The DNA libraries used in draft genome assembly for S. hasta have been deposited in the NCBI sequence read archive (SRA: SRR17438565) under the Project ID: PRJNA794279. The RNA sequence data is available under the BioProject accession: PRJNA748027.
References
Aronson, R. B., Thatje, S., McClintock, J. B. & Hughes, K. A. Anthropogenic impacts on marine ecosystems in Antarctica. Ann. N. Y. Acad. Sci. 1223, 82–107 (2011).
Sørhus, E. et al. Untangling mechanisms of crude oil toxicity: Linking gene expression, morphology and PAHs at two developmental stages in a cold-water fish. Sci. Total Environ. 757, 143896 (2021).
Bukola, D., Zaid, A., Olalekan, E. I. & Falilu, A. Consequences of anthropogenic activities on fish and the aquatic environment. Poult. Fish. Wildl. Sci. 3, 1–12 (2015).
Barron, M. G., Vivian, D. N., Heintz, R. A. & Yim, U. H. Long-term ecological impacts from oil spills: Comparison of Exxon Valdez, Hebei Spirit, and Deepwater Horizon. Environ. Sci. Technol. 54, 6456–6467 (2020).
Mearns, A. J. et al. Effects of pollution on marine organisms. Water Environ. Res. 92, 1510–1532 (2020).
Murawski, S. A., Hollander, D. J., Gilbert, S. & Gracia, A. Deepwater oil and gas production in the Gulf of Mexico and related global trends. In Scenarios and Responses to Future Deep Oil Spills: Fighting the Next War, 16–32 (2020).
Passow, U. & Stout, S. A. Character and sedimentation of “lingering” Macondo oil to the deep-sea after the Deepwater Horizon oil spill. Mar. Chem. 218, 103733 (2020).
Ali, A. O. et al. The effects of oil exposure on peripheral blood leukocytes and splenic melano-macrophage centers of Gulf of Mexico fishes. Mar. Pollut. Bull. 79, 87–93 (2014).
Magalhães, K. M. et al. Polycyclic aromatic hydrocarbons (PAHs) in fishery resources affected by the 2019 oil spill in Brazil: Short-term environmental health and seafood safety. Mar. Pollut. Bull. 175, 113334 (2022).
Yaghmour, F. et al. Oil spill causes mass mortality of sea snakes in the Gulf of Oman. Sci0 Total Environ. 825, 154072 (2022).
Bender, M. L. et al. Combined effects of crude oil exposure and warming on eggs and larvae of an arctic forage fish. Sci. Rep. 11, 8410 (2021).
Booman, C. & Føyn, L. In Produced Water 2: Environmental Issues and Mitigation Technologies (eds Mark Reed & Ståle Johnsen) 149–162 (Springer, US, 1996).
Bird, E. Encyclopedia of the World’s Coastal Landforms (Springer, New York, 2010).
Alqattan, M. E. & Gray, T. S. Marine pollution in Kuwait and its impacts on fish-stock decline in Kuwaiti waters: Reviewing the Kuwaiti government’s policies and practices. Front. Sustain. 2, 667822 (2021).
Edmonds, N. et al. Kuwait’s marine biodiversity: Qualitative assessment of indicator habitats and species. Mar. Pollut. Bull. 163, 111915 (2021).
Fodrie, F. J. et al. Integrating organismal and population responses of estuarine fishes in Macondo spill research. BioScience 64, 778–788 (2014).
Hjermann, D. Ø. et al. Fish and oil in the Lofoten-Barents Sea system: Synoptic review of the effect of oil spills on fish populations. Mar. Ecol. Prog. Ser. 339, 283–299 (2007).
Sørhus, E. et al. Unexpected interaction with dispersed crude oil droplets drives severe toxicity in Atlantic haddock embryos. PLoS One 10, e0124376 (2015).
Langangen, Ø. et al. The effects of oil spills on marine fish: Implications of spatial variation in natural mortality. Mar. Pollut. Bull. 119, 102–109 (2017).
Scott, G. R. & Sloman, K. A. The effects of environmental pollutants on complex fish behaviour: Integrating behavioural and physiological indicators of toxicity. Aquat. Toxicol. 68, 369–392 (2004).
Meier, S. et al. Development of Atlantic cod (Gadus morhua) exposed to produced water during early life stages: Effects on embryos, larvae, and juvenile fish. Mar. Environ. Res. 70, 383–394 (2010).
Debruyn, A. M. et al. In situ experimental assessment of lake whitefish development following a freshwater oil spill. Environ. Sci. Technol. 41, 6983–6989 (2007).
Incardona, J. P. et al. Unexpectedly high mortality in Pacific herring embryos exposed to the 2007 Cosco Busan oil spill in San Francisco Bay. Proc. Natl. Acad. Sci. U. S. A. 109, E51-58 (2012).
Zhu, L., Qu, K., Xia, B., Sun, X. & Chen, B. Transcriptomic response to water accommodated fraction of crude oil exposure in the gill of Japanese flounder, Paralichthys olivaceus. Mar. Pollut. Bull. 106, 283–291 (2016).
Bayha, K. M. et al. Crude oil impairs immune function and increases susceptibility to pathogenic bacteria in southern flounder. PLoS One 12, e0176559 (2017).
Zamora-Briseño, J. A. et al. Gill and liver transcriptomic responses of Achirus lineatus (Neopterygii: Achiridae) exposed to water-accommodated fraction (WAF) of light crude oil reveal an onset of hypoxia-like condition. Environ. Sci. Pollut. Res. Int. 28, 34309–34327 (2021).
Jantzen, C. E., Annunziato, K. A., Bugel, S. M. & Cooper, K. R. PFOS, PFNA, and PFOA sub-lethal exposure to embryonic zebrafish have different toxicity profiles in terms of morphometrics, behavior and gene expression. Aquat. Toxicol. 175, 160–170 (2016).
Karam, Q. et al. The response of sobaity sea bream Sparidentex hasta larvae to the toxicity of dispersed and undispersed oil. Pol. J. Environ. Stud. 30, 5065–5077 (2021).
OECD. Test No. 236: Fish Embryo Acute Toxicity (FET) Test (2013).
OECD. Organisation for Economic Co-operation and Development (OECD). Fish Embryo Toxicity (FET) Test (OECD Guideline for the Testing of Chemicals, 2006).
Singer, M. et al. Standardization of the preparation and quantitation of water-accommodated fractions of petroleum for toxicity testing. Mar. Pollut. Bull. 40, 1007–1016 (2000).
Karam, Q. E. Effect of crude oil on early life stages of native fish species of Kuwait (2010).
Karam, Q. et al. A comparative study on the effect of dispersed and undispersed Kuwait crude oil on egg hatching and larval survival of Epinephelus coioides. J. Environ. Biol. 40, 192–199 (2019).
ASTM. Standard Guide for Conducting Acute Toxicity Tests on Test Materials with Fishes, Microinvertebrates, and Amphibians, E729–796 (American Society for Testing and materials (ASTM), 2002).
Karam, Q. et al. De-novo genome assembly and annotation of sobaity seabream Sparidentex hasta. Front. Genet. 13, 988488 (2022).
Kim, D. et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Conesa, A. et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Govindkumar, B. et al. Ex-Ex primer: An experimentally validated tool for designing oligonucleotides spanning spliced nucleic acid regions from multiple species. J. Biotechnol. 343, 1–6 (2022).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Blenkinsopp, S. et al. In International Oil Spill Conference 941–942 (American Petroleum Institute).
Perrichon, P. et al. Toxicity assessment of water-accommodated fractions from two different oils using a zebrafish (Danio rerio) embryo-larval bioassay with a multilevel approach. Sci. Total Environ. 568, 952–966 (2016).
Holth, T. F. et al. Effects of water accommodated fractions of crude oils and diesel on a suite of biomarkers in Atlantic cod (Gadus morhua). Aquat. Toxicol. 154, 240–252 (2014).
Jones, E. R., Martyniuk, C. J., Morris, J. M., Krasnec, M. O. & Griffitt, R. J. Exposure to Deepwater Horizon oil and Corexit 9500 at low concentrations induces transcriptional changes and alters immune transcriptional pathways in sheepshead minnows. Comp. Biochem. Physiol. Part D Genom. Proteom. 23, 8–16 (2017).
Li, X. et al. Phenotypic and transcriptomic consequences in zebrafish early-life stages following exposure to crude oil and chemical dispersant at sublethal concentrations. Sci. Total Environ. 763, 143053 (2021).
Rodgers, M. L. et al. Combined effects of Deepwater Horizon crude oil and environmental stressors on Fundulus grandis embryos. Environ. Toxicol. Chem. 37, 1916–1925 (2018).
Xu, E. G. et al. Time- and oil-dependent transcriptomic and physiological responses to Deepwater Horizon Oil in Mahi-Mahi (Coryphaena hippurus) embryos and larvae. Environ. Sci. Technol. 50, 7842–7851 (2016).
Wheeler, J. R., Lyon, D., Di Paolo, C., Grosso, A. & Crane, M. Challenges in the regulatory use of water-accommodated fractions for assessing complex substances. Environ. Sci. Eur. 32, 1–10 (2020).
Karam, Q., Beg, M., Al-Khabbaz, A., Al-Ballam, Z. & Dakour, S. Effect of wateraccommodated fraction of Kuwait crude oil on developmental stages of orange-spotted grouper hamoor (Epinephelus coicoides). Int. J. Adv. Agric. Environ. Eng 1, 105–512 (2014).
Stefansson, E. S. et al. Acute effects of non-weathered and weathered crude oil and dispersant associated with the Deepwater Horizon incident on the development of marine bivalve and echinoderm larvae. Environ. Toxicol. Chem. 35, 2016–2028 (2016).
Incardona, J. P. et al. Deepwater Horizon crude oil impacts the developing hearts of large predatory pelagic fish. Proc. Natl. Acad. Sci. U. S. A. 111, E1510-1518 (2014).
Kim, H. N. et al. Acute toxic responses of the rockfish (Sebastes schlegeli) to Iranian heavy crude oil: Feeding disrupts the biotransformation and innate immune systems. Fish Shellfish Immunol. 35, 357–365 (2013).
Price, E. R. & Mager, E. M. The effects of exposure to crude oil or PAHs on fish swim bladder development and function. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 238, 108853 (2020).
Rhee, J. S. et al. Whole spectrum of cytochrome P450 genes and molecular responses to water-accommodated fractions exposure in the marine medaka. Environ. Sci. Technol. 47, 4804–4812 (2013).
Kwon, Y. J., Shin, S. & Chun, Y. J. Biological roles of cytochrome P450 1A1, 1A2, and 1B1 enzymes. Arch. Pharm. Res. 44, 63–83 (2021).
Hicken, C. E. et al. Sublethal exposure to crude oil during embryonic development alters cardiac morphology and reduces aerobic capacity in adult fish. Proc. Natl. Acad. Sci. 108, 7086–7090 (2011).
Jing, H., Zhang, Q., Li, S. & Gao, X.-J. Pb exposure triggers MAPK-dependent inflammation by activating oxidative stress and miRNA-155 expression in carp head kidney. Fish Shellfish Immunol. 106, 219–227 (2020).
Holen, E., Winterthun, S., Du, Z. Y. & Krovel, A. V. Inhibition of p38 MAPK during cellular activation modulate gene expression of head kidney leukocytes isolated from Atlantic salmon (Salmo salar) fed soy bean oil or fish oil based diets. Fish Shellfish Immunol. 30, 397–405 (2011).
Huang, X. et al. Roles of stress-activated protein kinases in the replication of Singapore grouper iridovirus and regulation of the inflammatory responses in grouper cells. J. Gen. Virol. 92, 1292–1301 (2011).
Subaramaniyam, U. et al. Effects of microplastics, pesticides and nano-materials on fish health, oxidative stress and antioxidant defense mechanism. Front. Physiol. 14, 1217666 (2023).
Jeffrey, J. D., Gollock, M. J. & Gilmour, K. M. Social stress modulates the cortisol response to an acute stressor in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 196, 8–16 (2014).
Ji, S. et al. cAMP-dependent protein kinase A in grass carp Ctenopharyngodon idella: Molecular characterization, gene structure, tissue distribution and mRNA expression in endoplasmic reticulum stress-induced adipocyte lipolysis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 250, 110479 (2020).
Milton, E. M., Cartolano, M. C. & McDonald, M. D. A multi-targeted investigation of Deepwater Horizon crude oil exposure impacts on the marine teleost stress axis. Aquat. Toxicol. 257, 106444 (2023).
Sandhu, N., Liang, L., McGeer, J., Dores, R. M. & Vijayan, M. M. Cadmium disrupts melanocortin 2 receptor signaling in rainbow trout. Aquat. Toxicol. 209, 26–33 (2019).
Chen, Y., Wu, X., Lai, J., Yan, B. & Gong, Q. Molecular mechanisms of physiological change under acute total dissolved gas supersaturation stress in yellow catfish (Pelteobagrus fulvidraco). Environ. Sci. Pollut. Res. Int. 30, 97911–97924 (2023).
Chen, Y. et al. Integrated metabolomic and transcriptomic responses to heat stress in a high-altitude fish, Triplophysa siluroides. Fish Shellfish Immunol. 142, 109118 (2023).
Wang, J. et al. Difenoconazole causes cardiotoxicity in common carp (Cyprinus carpio): Involvement of oxidative stress, inflammation, apoptosis and autophagy. Chemosphere 306, 135562 (2022).
Zhao, S. S. et al. The transcriptomic responses of blunt snout bream (Megalobrama amblycephala) to acute hypoxia stress alone, and in combination with bortezomib. BMC Genom. 23, 162 (2022).
Zhao, X., Sun, Z., Gao, T. & Song, N. Transcriptome profiling reveals a divergent adaptive response to hyper- and hypo-salinity in the Yellow Drum, Nibea albiflora. Animals 11, 2201 (2021).
Sheehan, D., Meade, G., Foley, V. M. & Dowd, C. A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360, 1–16 (2001).
Green, R. & Noller, H. F. Ribosomes and translation. Ann. Rev. Biochem. 66, 679–716 (1997).
Gruenberg, J. & Clague, M. J. Regulation of intracellular membrane transport. Curr. Opin. Cell Biol. 4, 593–599 (1992).
Raza, H. Dual localization of glutathione S-transferase in the cytosol and mitochondria: Implications in oxidative stress, toxicity and disease. FEBS J. 278, 4243–4251 (2011).
Rasmussen, B. B. & Wolfe, R. R. Regulation of fatty acid oxidation in skeletal muscle. Ann. Rev. Nutr. 19, 463–484 (1999).
Ebert, A. M. et al. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc. Natl. Acad. Sci. U. S. A. 102, 17705–17710 (2005).
Incardona, J. P. Molecular mechanisms of crude oil developmental toxicity in fish. Arch. Environ. Contam. Toxicol. 73, 19–32 (2017).
Sørhus, E. et al. Novel adverse outcome pathways revealed by chemical genetics in a developing marine fish. eLife 6, 20707 (2017).
Gao, D., Wu, M., Wang, C., Wang, Y. & Zuo, Z. Chronic exposure to low benzo[a]pyrene level causes neurodegenerative disease-like syndromes in zebrafish (Danio rerio). Aquat. Toxicol. 167, 200–208 (2015).
Vignet, C. et al. Changes in brain monoamines underlie behavioural disruptions after zebrafish diet exposure to polycyclic aromatic hydrocarbons environmental mixtures. Int. J. Mol. Sci. 18, 560 (2017).
Xu, E. G. et al. Larval Red Drum (Sciaenops ocellatus) sublethal exposure to weathered deepwater horizon crude oil: Developmental and transcriptomic consequences. Environ. Sci. Technol. 51, 10162–10172 (2017).
Xu, E. G. et al. Developmental transcriptomic analyses for mechanistic insights into critical pathways involved in embryogenesis of pelagic mahi-mahi (Coryphaena hippurus). PLoS One 12, e0180454 (2017).
Hook, S. E., Lampi, M. A., Febbo, E. J., Ward, J. A. & Parkerton, T. F. Temporal patterns in the transcriptomic response of rainbow trout, Oncorhynchus mykiss, to crude oil. Aquat. Toxicol. 99, 320–329 (2010).
Sørhus, E. et al. Crude oil exposures reveal roles for intracellular calcium cycling in haddock craniofacial and cardiac development. Sci. Rep. 6, 31058 (2016).
Acknowledgements
The authors gratefully acknowledge Kuwait Foundation for the Advancement of Sciences (KFAS) for partially funding the project (Grant No. PR18-12SL-01). The authors wish to thank Prof. Kshitish Acharya and his team for assisting in bioinformatics work.
Author information
Authors and Affiliations
Contributions
V.K., Q.K.: Conceptualization, sampling, laboratory experiments, manuscript preparation. V.K., A.S.: DNA and RNA isolation, sample processing, data analysis. Q.K.: funding acquisition. S.A., Z.S., S.D.: fish larvae culture, rearing, and supply.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kumar, V., Karam, Q., Shajan, A.B. et al. Transcriptome analysis of Sparidentex hasta larvae exposed to water-accommodated fraction of Kuwait crude oil. Sci Rep 14, 3591 (2024). https://doi.org/10.1038/s41598-024-53408-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-53408-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.