Abstract
Among the most prevalent neurodevelopmental disorders, Autism Spectrum Disorder (ASD) is highly diverse showing a broad phenotypic spectrum. ASD also couples with a broad range of mutations, both de novo and inherited. In this study, we used a proprietary SNP genotyping chip to analyze the genomic DNA of 250 Vietnamese children diagnosed with ASD. Our Single Nucleotide Polymorphism (SNP) genotyping chip directly targets more than 800 thousand SNPs in the genome. Our primary focus was to identify pathogenic/likely pathogenic mutations that are potentially linked to more severe symptoms of autism. We identified and validated 23 pathogenic/likely pathogenic mutations in this initial study. The data shows that these mutations were detected in several cases spanning multiple biological pathways. Among the confirmed SNPs, mutations were identified in genes previously known to be strongly associated with ASD such as SLCO1B1, ACADSB, TCF4, HCP5, MOCOS, SRD5A2, MCCC2, DCC, and PRKN while several other mutations are known to associate with autistic traits or other neurodevelopmental disorders. Some mutations were found in multiple patients and some patients carried multiple pathogenic/likely pathogenic mutations. These findings contribute to the identification of potential targets for therapeutic solutions in what is considered a genetically heterogeneous neurodevelopmental disorder.
Similar content being viewed by others
Introduction
Autism spectrum disorder
Autism spectrum disorder (ASD) is known to be a heritable and heterogeneous group of neurodevelopmental phenotypes that manifests itself via numerous social, emotional, communicative, and behavioral challenges1. The origin of ASD is not fully understood, and evidence suggests that both genetic and environmental factors are involved2,3. Although most everyone in the general population carries almost all the genetic risk factors for autism, the disorder, as per its modern terminology, lies on a spectrum or continuum, with only the extreme tail of individuals expressing its defined signs to a more notable degree4. Antaki et al. have postulated that the phenotypic spectrum of ASD is correlated to multiple genetic factors, parental factors, and gene-by-sex effects. The authors also underscored that their research showcases the importance that rare and common variations, in combination, yield greater contributions to the development of ASD than alone5. Individuals with ASD can display various aspects of cognitive difficulties, with the prevalence of intellectual disability in conjunction with ASD estimated to be 3.8 in 1000 in an Australian sample for births from 1993 to 20056.
Prevalence among east asians
Globally, the World Health Organization7 estimates that 1 in 100 children have ASD with a consistently higher prevalence in males8. In 2018, researchers in the U.S. estimated that 1 in 44 children aged 8 years had ASD. Among Asian Americans, the prevalence of ASD appears to be on the rise due to increased visibility and public awareness of the disorder9.
Often, the prevalence of ASD has been reported to be lower in East Asia than in Europe or North America. However, this discrepancy may be due to methodological differences between studies. In fact, modernized research in China indicates that the prevalence rates are comparable between the geographic locations10. More broadly, it’s estimated that the prevalence of ASD in East Asia stands at 0.51% and is likely increasing11.
Diagnostic and therapeutic challenges for East/Southeast Asians
Numerous aspects of East Asian culture are thought to influence clinical diagnosis and treatment for children with ASD; as well as the way the clinician might interact with an East Asian parent in the context of ASD12. Clinicians must consider the culture and the stigma that may be associated with autism and how parents of autistic children might view the disorder.
Many of these same, and other, cultural factors can also affect parental care and well-being. For instance, stress among parents with children diagnosed with ASD may partly stem from unique cultural thoughts (parental stress due to cultural factors, parents’ psychopathological symptoms, problem behaviors in ASD children, caregiver burden and more were identified) present in East Asia and Southeast Asia that are not found in other cultures to the same degree13. Furthermore, we see an increase in diagnostic rate in more developed countries in the regions such as Singapore, Philippines (1–1.1%) compared to Vietnam, Thailand (~ 0.7%) and Laos, Cambodia, Brunei (~ 0.5%). This rate does not correlate geographically, indicating that among East Asian and Southeast Asian, there can be a lack of proper understanding of the diagnosis and treatment of children with ASD starting from parental recognition to start doctor’s visit14.
This points to two things. First, there is a need to utilize, in concert with existing methodologies, standardized genetic screening techniques for the diagnosis of ASD. Techniques that may have been previously underutilized due to cultural and socioeconomic differences, even among Asian populations. Second, to increase the rate of appropriate diagnosis and treatment, there is a need to properly educate clinicians and parents on ASD in the context of specific Asian cultural factors.
Methods
Sample collection
Parents of autistic children were present during sample collection at Hue Central Hospital. Children in this study were diagnosed using the DSM-IV criteria (American Psychiatric Association (2000)-Diagnostic and statistical manual of mental disorders (4th edition)). Participant’s screening results are in supplemental Table 3. Saliva samples were stored at room temperature and subjected to automated DNA extraction and processing. A total of 254 saliva samples were collected using the Oragene•ONE ON-600 (Dnagenotek, Canada).
Ethics declaration
Prior to the start of this study and enrollment of research participants, the Ethics committee of Hue Central Hospital in Vietnam approved this study. All the methods were performed in accordance with the relevant local guidelines and regulations.
Prior to collection of the saliva samples, written informed consents were signed by parents of 254 children diagnosed with Autism Spectrum Disorder allowing the authors of this study to use the saliva samples and all the related medical data for research, publications, and biobanking purposes.
DNA extraction
The genomic DNA was extracted from the collected saliva using Chemagic Prime™ Robot. The process is entirely automated using chemagen patented M-PVA Magnetic Bead technology for DNA and RNA purification with liquid handling to provide high throughput automated isolation of ultra-pure nucleic acids. The process is monitored in accordance with the Quality Control of ISO/IEC 17025.
Genotyping with The GFWv3 custom high-resolution arrays
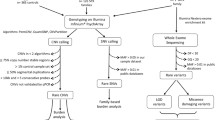
The Axiom workflow was used on GeneTitan instruments (Manufacturer: Thermo Fisher Scientific, catalog number: 00-0373, Model: GeneTitan MC) with wrappers for Analysis Power Tools (APT)–Genotype–quality control tools (apt-geno-qc) and genotype calling tools (apt-probeset-genotype). A tool for SNP metric calculations and a tool to convert the output into Plink format for downstream genomics analyses were used. Samples were registered in a custom file format for a batch of 96 “.CEL” files from a single Axiom plate and a few auxiliary file formats specific to APT tools to facilitate the file selection process in Galaxy. The Galaxy workflow starts with receiving its input of “.CEL” and “.ARR” files from the instrumentation computer. It proceeds with extracting the “.CEL” files and executing the quality control tool with a user-specified Dish-Quality Control (QC) threshold (by default 0.82). The names of the samples that have passed the QC are passed on to the genotyping tool, along with the “.CEL” dataset, for the first round of genotyping. The output from this first round contains, among other metrics, the call rates for each sample. The samples with a call rate above a user-specified threshold (by default 97%), along with the “.CEL” dataset again, is input for the second iteration of genotyping. The final genotype calling report is then annotated with the phenotype data and converted into Plink format, and simultaneously processed by the SNP metrics tool to calculate such statistics as Call Rate (CR), Fisher’s linear discriminant (FLD), FLD calculated for the homozygous genotype clusters (HomFLD), Minor Allele count.
The GFWv3 custom array is a High-resolution Affymetry SNP array consisting of 2.5 million (2.5 M) probes to assay for SNPs and CNVs with 800.000 direct targets and two million more with imputation. We designed and validated this array by both inter-assays (reproducibility > = 99.8%) and intra-assays (reproducibility > = 99.8%). Manufacturer: Thermo Fisher Scientific. There are 3 parts of each kit: Axiom™ GFWv3 96 well Plate: part number 551159; Axiom™ GeneTitan Consumables Kit: part number 901606 and the Axiom™ 2.0 Reagent Kit: part number 901758.
Variant validation
Identified variants were validated by Sanger sequencing and Amplification-refractory mutation system polymerase chain reaction (ARMS-PCR). Each unique mutation found in this study was validated using Sanger sequencing. If there are several samples found to carry the same mutation, we did Sanger sequencing to verify the mutation identified in one sample and in parallel, ARMS PCRs for the other samples carrying the same mutation. This method became a more cost-effective and accurate workflow as we expanded the analyses to more common variants. The principle of validation of identified variants with ARMS-PCR is shown in Supplemental Fig. 1. The list of primers designed for PCR and Sanger sequencing is included in Supplemental Table 1.
These oligos were designed for identified variants by using the Primer-BLAST tool of NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and PRIMER1 on the website http://primer1.soton.ac.uk/primer1.html. The same DNA samples used for genotyping in this study were also used for the validation process. PCR amplification, Sanger sequencing, and ARMS PCR for variant verification.
Results
Demographic and clinical characteristics of the research participants in this study
All 254 children recruited as research participants were from Central Vietnam and are current patients of Hue Central Hospital. We have not collected and analyzed DNA from the parents as the initial results showed all mutations found were heterozygous. The children were diagnosed with ASD using the DSM-IV. The male/female ratio in this study is 5.86, (Table 1). This is higher than previously estimated ratio in the general population which is closer to 48,11,15,16,17. This could be due to several factors such as those willing to participate in the study skewing the ratio to more males than expected. Furthermore, the fact that females often being diagnosed with ASD less or later than males or having different, less obvious symptoms compared to males18 might contribute to this result.
Among the 254 samples collected, four samples were degraded yielding minimal amount of DNA or low-quality DNA. The families decided not to recollect these four samples. For the 250 samples that passed QC, the genotyping results were analyzed using the Axiom analyses Suite 5.1 (Thermo Fisher).
All the children in this study have speech delays characterized by no word being spoken by the age of 16 months and other communication issues such as not responding to their name when being called, avoiding eye contact or avoiding interacting with others, as reported by their parents (Table 1 and Sup. Table 3).
Mutations identified were categorized as pathogenic/likely pathogenic according to Clinvar database (https://www.ncbi.nlm.nih.gov/clinvar/). A total of 23 pathogenic/ likely pathogenic mutations were identified in this study including 12 missense mutations, 5 stop-gained mutations, 3 frameshift mutations, 1 splicing mutation, and two in non-coding transcripts (Table 2). A few variants showed conflicting interpretations (pathogenic to benign). For instance, rs1799990 in the PRNP gene was classified as pathogenic/risk factor/ likely benign. However, based on the high frequency of occurrence of this SNP (also shown in this study), it was classified as a likely benign variant.
Multiple pathogenic mutations were identified in single individuals. Several cases with very severe ASD diagnosis were found to be the carriers for multiple mutations. For instance, AUT4875, is the carrier of 4 different mutations, two in the ZGRF1 gene, one in the SCN9A gene and another in the HCP5 gene; AUT4870 carries mutations in the RIPK1 and SCN9A gene (Table 3).
In the two predominant databases for genes associated with ASD, SFARI and AutDB, only 9 mutations were previously reported among the total of 23 unique pathogenic/likely pathogenic mutations identified. Though, many of the identified genes identified are implicated in ASD and other neurological disorders as discussed below. All 23 of the pathogenic/likely pathogenic SNPs assessed were confirmed via orthogonal methods (i.e. Sanger Sequencing or ARMS PCR). There are variations in genes previously strongly associated with ASD such as SLCO1B126, ACADSB27, TCF428, HCP529, MOCOS30, SRD5A231 , MCCC232, DCC33 and PRKN34. The ratio of males/ females among the samples with identified mutations is 3.93 (Sup. Table 3).
Discussion
To date, hundreds of potential genetic alterations have been identified as associated with autism though with no defining cohort. What has been determined is that there is likely shared pathophysiology for neurodevelopmental disorders and that autism is along a continuum between intellectual disability and schizophrenia35. Since ASD is multigenic and heterogeneous and can occur in conjunction with other neurological conditions, it is difficult to discern the genes that are responsible for the disease phenotypes36. Previous studies showed consistent results of two classes of proteins, those involved in synapse formation and those involved with transcriptional regulation and chromatin-remodeling pathways37.
From 250 Vietnamese children diagnosed with varying degrees of autism spectrum disorder, our high-resolution SNP array data has identified both rare and common SNPs previously known to associate with ASD, we then validated these data and provided information regarding frequency among Southeast Asians.
Of the confirmed SNPs, there were 7 SNPs that were shared among several of the children tested, these were SNPs in HCP5, SRD5A2, PRNP, ZGRF1, SCN9A and LOC107987057. rs2395029 in HCP5 has not previously been identified as an autism-associated variant though there is increasing evidence that immune-related genes, such as HCP5, and immune dysregulation are associated with neurodevelopmental disorders29. Recent studies have shown that HCP5 rs2395029 is in complete linkage disequilibrium with HLA-B*570119, which is a risk allele of intellectual disability20. Several alleles of HLA genes have been reported to be associated with autism, intellectual disability, schizophrenia20. Using Fisher Exact test, frequency of this mutation among the cases in this study is significantly higher than its frequency in the East Asian population (p-value = 0.000046) (Sup. Table 4). In terms of SRD5A2, a recent study found that ASD boys with rs9282858 mutation in this testosterone metabolism-related gene showed higher levels of restricted and repetitive behaviors31. In addition, multiple recent studies published on Clinvar have consistently concluded SRD5A2 rs9332964 as pathogenic/likely pathogenic. Frequency of this mutation among the cases in this study is also significantly higher than its frequency in the general East Asian population (p-value = 0.00013) (Sup. Table 4). Recently studies showed that key proteins in the metabolism of ROS were downregulated in autistic people, including PRNP, marking PRNP as a potential biomarker of autism for early diagnostic purposes38.
ZGRF1 encodes a protein with functions related to motor praxis and highly expressed in the cerebellum, raising the possibility that disrupted ZGRF1 may interfere with cerebellar function39,40. Two ZGRF1 variants detected as compound heterozygotes in 7 ASD patients in this study, rs61745597 and rs76187047, have been identified as the potential genetic causality of childhood apraxia of speech (CAS), which is prevalent in approximately 25–30% of children with ASD41. Both variants result in missense mutations, thereby it is logical to expect disruption of the ZGRF1 protein in both instances. While CAS is a complex disorder, it is unlikely that ZGRF1 would be the sole causative gene target, it is one more piece in the puzzle when narrowing down potential therapeutic targets for ASD and its associated disorders40. Mutations in the primary central nervous system sodium channels are associated with neurological, psychiatric, and neurodevelopmental disorders including autism. SCN9A has been indicated to be important for normal brain function and variants in this gene are involved in familial autism42. LOC107987057 or C9orf72 variants have been linked to several neurological disorders, including ASD43. However, Chi-square analyses showed that the frequencies of these 4 variants LOC107987057 rs2814707, ZGRF1 rs61745597, ZGRF1 rs76187047 and SCN9A rs12478318 observed in our study are not significantly different from the East Asian population in the 1000 Genome Project 30x (p-value = 0.27, 0.82, 0.82, and 0.21 respectively) (Sup. Table 4). In addition, given the high frequency of the minor allele, the association of these variants to ASD should be reconsidered.
Confirmed variants in the following GJB2 were seen in five or six children, respectively. GJB2 is most known for its linkage in children with non-syndromic genetic sensorineural hearing loss and has not been identified previously as having an association with ASD22.
Confirmed variants in RIPK144, CAPN345, KAT6A46, TACR347, GJB248, FAM98C24, PRKN, SLC3A1, CUBN, and PYGM were seen in one or two children in this study. Interestingly, rs751037529 in PRKN has been identified as a pathogenic variant associated with early-onset Parkinson’s disease49. Also, PRKN knock-out mice show autistic-like behaviors, giving weight to PRKN as a potential candidate gene for ASD34. Previous studies have also shown that the disruption of genes that encode large amino acid transporters, like SLC3A1, increases the risk of ASD50. These abnormalities in large amino acid transporters can affect the utilization of certain amino acids and their availability during brain development, resulting in an increased risk of ASD. Along that line, rs143944436 in the CUBN gene identified in this study results in a premature stop codon resulting in a non-functional protein. The CUBN gene provides instructions for making a protein called cubilin which is involved in the uptake of vitamin B12 from food into the body, linking vitamins and their bioavailability as potential treatments for individuals with ASD.
Considering the vast number of inherited, common, and rare genetic variants that have been associated with ASD, the etiology is complicated, to say the least. This study specifically assessed a cohort of Southeast Asian children with varying degrees of ASD compared to a control population to identify those variants which may be potential diagnostic or therapeutic targets.
This study provides an initial step towards understanding the genetic underpinnings of ASD in Southeast Asian populations. We view these data as a contribution towards identifying the loci which contribute to ASD and we anticipate that some of these loci will eventually have sufficient evidence to become established robust ASD risk loci.
Some genetic variants correlate to a very high risk of disease while most do not. In its simplest terms, a polygenic risk score (PRS), sometimes called a polygenic score (PGS) or a genetic/genomic risk score (GRS), reflects the overall genetic predisposition to a disease based on the sum of all known and common variants linked to that disease51. This study has focused on pathogenic/likely pathogenic mutations. The next step, the study will be furthered with analysis of more samples, not only for SNPs but copy number variants and also the attribution of more common variants using the Polygenic Risk Score.
Data availability
Data generated and analyzed in this study are included in this article and its Supplementary Information. Our data are also deposited in ClinVar under accession numbers: SCV004013880—SCV004013903.
References
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
McElhanon, B. O., McCracken, C., Karpen, S. & Sharp, W. G. Gastrointestinal symptoms in autism spectrum disorder: A meta-analysis. Pediatrics 133, 872–883 (2014).
Colvert, E. et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry 72, 415–423 (2015).
Robinson, E. B. et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat. Genet. 48, 552–555 (2016).
Antaki, D. et al. A phenotypic spectrum of autism is attributable to the combined effects of rare variants, polygenic risk and sex. Nat. Genet. 54, 1284–1292 (2022).
Bourke, J., de Klerk, N., Smith, T. & Leonard, H. Population-based prevalence of intellectual disability and autism spectrum disorders in Western Australia: A comparison with previous estimates. Medicine 95, 3737 (2016).
Asia, W. H. O. R. O. for S.-E. WHO South-East Asia regional strategy on autism spectrum disorders. (World Health Organization. Regional Office for South-East Asia, 2017).
Zeidan, J. et al. Global prevalence of autism: A systematic review update. Autism Res. 15, 778–790 (2022).
Maenner, M. J. et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 Sites, United States, 2018. MMWR Surveill. Summ. 70, 1–16 (2021).
Sun, X. et al. Autism prevalence in China is comparable to western prevalence. Mol. Autism 10, 7 (2019).
Qiu, S. et al. Prevalence of autism spectrum disorder in Asia: A systematic review and meta-analysis. Psychiatry Res. 284, 112679 (2020).
Bernier, R., Mao, A. & Yen, J. Psychopathology, families, and culture: Autism. Child Adolesc. Psychiatr. Clin. N. Am. 19, 855–867 (2010).
Ng, C. S. M., Fang, Y., Wang, Z. & Zhang, M. Potential factors of parenting stress in chinese parents of children with autism spectrum disorder: A systematic review. Focus Autism Dev. Disabil. 36, 237–248 (2021).
Su, S., Paynter, J. & Gilmore, L. Chinese Parents’ Understanding of Autism Spectrum Disorder. Int. J. Disabil. Dev. Educ. 68, 414–426 (2019).
Salari, N. et al. The global prevalence of autism spectrum disorder: a comprehensive systematic review and meta-analysis. Ital. J. Pediatr. 48, 112 (2022).
Werling, D. M. & Geschwind, D. H. Sex differences in autism spectrum disorders. Curr. Opin. Neurol. 26, 146 (2013).
Tran, K. T. et al. Genetic landscape of autism spectrum disorder in Vietnamese children. Sci. Rep. 10, 5034 (2020).
Gotham, K. et al. Characterizing the daily life, needs, and priorities of adults with autism spectrum disorder from interactive autism network data. Autism Int. J. Res. Pract. 19, 794–804 (2015).
Nguyen, D. V. et al. The utility of surrogate markers in predicting HLA alleles associated with adverse drug reactions in Vietnamese. Asian Pac. J. Allergy Immunol. 40, 134–140 (2022).
Nudel, R. et al. Immunity and mental illness: findings from a Danish population-based immunogenetic study of seven psychiatric and neurodevelopmental disorders. Eur. J. Hum. Genet. 27, 1445–1455 (2019).
Kikuchi, T., Kimura, R. S., Paul, D. L. & Adams, J. C. Gap junctions in the rat cochlea: Immunohistochemical and ultrastructural analysis. Anat. Embryol. 191, 101–118 (1995).
Adadey, S. M. et al. GJB2 and GJB6 mutations in non-syndromic childhood hearing impairment in ghana. Front. Genet. 10, 841 (2019).
Keino-Masu, K. et al. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell 87, 175–185 (1996).
Ruzzo, E. K. et al. Inherited and de novo genetic risk for autism impacts shared networks. Cell 178, 850–866 (2019).
Pinacho, R. et al. The glial phosphorylase of glycogen isoform is reduced in the dorsolateral prefrontal cortex in chronic schizophrenia. Schizophr. Res. 177, 37–43 (2016).
Medhasi, S. et al. Clinically relevant genetic variants of drug-metabolizing enzyme and transporter genes detected in Thai children and adolescents with autism spectrum disorder. Neuropsychiatr. Dis. Treat. 12, 843–851 (2016).
Kanavin, O. J. et al. 2-methylbutyryl-CoA dehydrogenase deficiency associated with autism and mental retardation: A case report. J. Med. Case Rep. 1, 98 (2007).
Teixeira, J. R., Szeto, R. A., Carvalho, V. M. A., Muotri, A. R. & Papes, F. Transcription factor 4 and its association with psychiatric disorders. Transl. Psychiatry 11, 1–12 (2021).
Needleman, L. A. & McAllister, A. K. The major histocompatibility complex and autism spectrum disorder. Dev. Neurobiol. 72, 1288–1301 (2012).
Féron, F. et al. Olfactory stem cells reveal MOCOS as a new player in autism spectrum disorders. Mol. Psychiatry 21, 1215–1224 (2016).
Lakatošová, S. et al. The relationship of steroid hormones, genes related to testosterone metabolism and behavior in boys with autism in Slovakia. Psychiatry Investig. 19, 73–84 (2022).
Doan, R. N. et al. Recessive gene disruptions in autism spectrum disorder. Nat. Genet. 51, 1092–1098 (2019).
Li, Y. et al. Association between DCC polymorphisms and susceptibility to autism spectrum disorder. J. Autism Dev. Disord. 50, 3800–3809 (2020).
Huo, Y., Lu, W., Tian, Y., Hou, Q. & Man, H.-Y. Prkn knockout mice show autistic-like behaviors and aberrant synapse formation. Iscience 25(7), 104573 (2022).
Anney, R. J. L. et al. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism https://doi.org/10.3389/fncel.2019.00385 (2017).
Rylaarsdam, L. & Guemez-Gamboa, A. Genetic causes and modifiers of autism spectrum disorder. Front. Cell. Neurosci. https://doi.org/10.3389/fncel.2019.00385 (2019).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Pangrazzi, L., Balasco, L. & Bozzi, Y. Oxidative stress and immune system dysfunction in autism spectrum disorders. Int. J. Mol. Sci. 21, 3293 (2020).
Guerra, J. & Cacabelos, R. Genomics of speech and language disorders. J. Transl. Genet. Genom. https://doi.org/10.20517/jtgg.2018.03 (2019).
Peter, B. et al. Genetic candidate variants in two multigenerational families with childhood apraxia of speech. PLOS ONE 11(4), 0153864 (2016).
Tager-Flusberg, H. & Kasari, C. Minimally verbal school-aged children with autism spectrum disorder: The neglected end of the spectrum. Autism Res. 6, 468–478 (2013).
Rubinstein, M. et al. Association of rare missense variants in the second intracellular loop of NaV1.7 sodium channels with familial autism. Mol. Psychiatry 23, 231–239 (2018).
Devenney, E. M. et al. Psychiatric disorders in C9orf72 kindreds: Study of 1,414 family members. Neurology 91, e1498–e1507 (2018).
Mifflin, L. et al. A RIPK1-regulated inflammatory microglial state in amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. https://doi.org/10.1073/pnas.2025102118 (2021).
Zhao, Y., Zhao, P., Liang, H. & Zhang, X. Identifying genes associated with autism spectrum disorders by random walk method with significance tests. IEEE Access 8, 156686–156694 (2020).
Murray, C. R. et al. Novel causative variants in DYRK1A, KARS, and KAT6A associated with intellectual disability and additional phenotypic features. J. Pediatr. Genet. 06, 077–083 (2017).
Yonan, A. L. et al. Bioinformatic analysis of autism positional candidate genes using biological databases and computational gene network prediction. Genes Brain Behav. 2, 303–320 (2003).
Wiley, S., Choo, D., Meinzen-Derr, J., Hilbert, L. & Greinwald, J. GJB2 mutations and additional disabilities in a pediatric cochlear implant population. Int. J. Pediatr. Otorhinolaryngol. 70, 493–500 (2006).
Li, T., Kou, D., Cui, Y. & Le, W. Whole exome sequencing identified a new compound heterozygous PRKN mutation in a Chinese family with early-onset Parkinson’s disease. Biosci. Rep. 40, BSR20200534 (2020).
Cascio, L. et al. Abnormalities in the genes that encode Large Amino Acid Transporters increase the risk of Autism Spectrum Disorder. Mol. Genet. Genom. Med. 8, 1036 (2019).
Illumina. Polygenic risk: What’s the score? Nature.
Ye, S., Dhillon, S., Ke, X., Collins, A. R. & Day, I. N. M. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 29, 88 (2001).
Funding
This study is funded by Gene Friend Way.
Author information
Authors and Affiliations
Contributions
D.T.B. and T.A.C. conceived of the project, supervised its execution, analyzed data, and co-wrote the manuscript. D.T.B. initiated and designed most experiments and analyses, D.T.B. and T.A.C. were the lead scientists, performed experiments, analyzed data, and co-wrote the manuscript. A.N.V.T, C.T.D.N., S.H.N., H.K.T., X.T.N. co-designed and took consent forms, performed sample collection, provided information for research participants, and maintained medical records of the research participants. H.N.P., T.N.V.P. provided advice. J.H. provided advice, edited and co-wrote the manuscripts. D.S.T., D.T.B. consolidated and analyzed the raw data. H.T.N., G.L.T.P. performed experiments, did PCR/ Sanger validation for identified SNPs.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bui, D.T., Ton, A.N.V., Nguyen, C.T.D. et al. Pathogenic/likely pathogenic mutations identified in Vietnamese children diagnosed with autism spectrum disorder using high-resolution SNP genotyping platform. Sci Rep 14, 2360 (2024). https://doi.org/10.1038/s41598-024-52777-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-52777-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.