Abstract
The study presents the first to characterize novel Erucastrum canarianse Webb and Berthel (or Can) sterile cytoplasm-based CMS lines in Indian cauliflower (Brassica oleracea var. botrytis L.) and investigating their commercial suitability. Eleven Can-based CMS lines were examined for 12 agro-morphological and yield traits,18 floral traits, four seed yield traits together with three each of the Ogura (source: wild Japanese Radish) and Tour (Source: Brassica tournefortii) cytoplasms. All of the recorded floral and seed traits showed significant (P > 0.05) differences between the CMS lines of each group. Agro-morphological and yield traits in CMS lines and their maintainers, however, were non-significantly different. All the Can- and Ogura-based CMS lines showed flowering and appropriate seed formation by natural cross-pollination. Only two Tour cytoplasm-based CMS lines, Tour (DC-41-5) and Tour (DC-67), produced the smallest malformed flowers and stigma. The highest seed yield per plant in CMS lines was in Ogu (DC-98-4) and the lowest in Tour (DC-67). P14 and P15, two polymorphic mtDNA markers, were discovered for the Can CMS system for early detection. Five primers (ITS5a-ITS4, atpF-atpH, P16, rbeL and trnL), along with their maintainers, were sequenced and aligned to detect nucleotide changes including as additions and or deletions at different positions. The newly introduced E. canariense sterile cytoplasm-based CMS system in cauliflower is the subject of the first comprehensive report, which emphasises their potential as a further stable and reliable genetic mechanism for hybrid breeding.
Similar content being viewed by others
Introduction
Cytoplasmic male sterility (CMS) is a robust, easy-to-maintain and produces satisfactory hybrid seed yield which are essential features for a genetic mechanism to use in commercial hybrid seed production. It is the maternal inheritance of the inability of a plant to produce a viable pollen grain and is governed by mitochondrial genome1,2. Introgression of sterile cytoplasm from different species or genus creates new nulceo-cytoplasmic interactions in the converted CMS lines. However, the level of impact needs to be investigated which may vary depending upon donor source (species or genus) and recipient backgrounds (different genotypes).
There are different male sterile cytoplasms available for Brassica crops, namely ‘Ogu’ from wild Japanese radish3,‘Can’ of Erucastrum canariense Webb and Berthel4, ‘Tour’ of Brassica tournefortii Goun5, nap’ of Brassica napus L.6 ,‘kosena’ of ‘Raphanus sativus L.7 , ‘mur’ of Diplotaxis muralis (L.) DC8 , ‘Pol’ of B. napus9,‘hau’ of Brassica juncea (L.) Czern10 ,‘mori’ of Moricandia arvensis L.11 ,‘fruti’ of Brassica fruticulosa Cirillo12 ,‘inap’ of Brassica napus L. + Isatis indigotica Fort.13 and Tour-Stiewe of B. tournefortii Goun (somatic cell fusion)14. The present study elaborates on alloplasmic CMS lines developed using two sets of combinations: first, two species of the same genus i.e. cytoplasm of Brassica tournefortii (Tour cytoplasm) and nuclear genome of cauliflower (Brassica oleracea var. botrytis L.) and second, two different genera either ‘Raphanus sativus L.’ (Ogura cytoplasm) or E. canariense (Can cytoplasm) and nuclear genome of cauliflower. The Ogura cytoplasm is the only commercially exploited male sterility in Brassica oleracea L. for hybrid breeding.
Cauliflower (Brassica oleracea var. botrytis L.; 2n = 2x = 18) is one of the most important vegetable crops worldwide. It is being grown in an area of 1.36 million ha with production of 25.53 million tonnes15. With a contribution of around 33% in area and 34.5% in global production, India ranks second behind China. In India, area and production of cauliflower are 0.473 million ha and 9.22 million tonnes, respectively16. Cauliflower is being grown for white curds which have unique texture, flavour and taste. Fresh curd portion has 1.9% protein, 5% carbohydrate, 95% water, 48.2 mg vitamin C, and 199 mg potassium17. It contains health-beneficial glucosinolates particularly sinigrin, glucobrassicin and 4 hydroxy-glucobrassicin18.
Cauliflower originated in the Island of Cyprus and travelled to Europe via trade routes. The European type varieties of cauliflower were first introduced to the Indian sub-continent in 1822 at Company Bagh, Saharanpur in United Province (currently Uttar Pradesh, India) by a British botanist named Dr. Jemson19. The newly introduced cauliflower could only be grown in winter due to India’s higher temperature during summer and rainy seasons20. However, with staggered planting, human and natural selection pressure and crossing-recombination occurrences, the introduced cauliflowers evolved into ‘local types’ that were better adapted to high temperature and humidity. It formed curd at higher temperature (20–27 °C) than the source European type varieties (10–16 °C or less). The ‘local’ or ‘Indian’ type cauliflower produced satisfactory seeds with a lesser requirement of cool temperature which was prevalent during winters in North Indian plains19,21,22. Based on temperature requirement for curd formation, the Indian cauliflower is grouped into Early (20–27 °C), Mid-early (16–20 °C) and Mid-late (12–16 °C) maturity groups23. Mid-September to mid-November, mid-November to mid-December and mid-December to mid-January, respectively are the contiguous seasons for these temperatures. Late or snowball cauliflower (10–16 °C) is also common for harvesting during February–March in North Indian plains; nevertheless, due to temperature considerations, its seed production is feasible in the middle to higher altitude mountainous regions24,25.
The F1 hybrids have been crucial in increasing yield of cauliflower while also improving crop uniformity and product quality, both of which attracted farmers and retailers. The F1 hybrids are advantageous over the open-pollinated varieties (OPV) for uniformity, productivity, quality and tolerance to various biotic and abiotic stresses26. Therefore, the primary breeding goal for cauliflower is to develop F1 hybrids using genetic mechanisms for economic hybrid seed production. The earlier efforts were confined to self-incompatibility (SI) for hybrid development due to lack of alternative usable genetic mechanisms. In India, the SI system has been utilized for breeding three hybrids namely Pusa Hybrid-2 (1998), Pusa Kartik Sankar (2004) and Pusa Cauliflower Hybrid-101 (2021). However, the SI system has issues of poor stability at higher temperature, sib seeds and cumbersome maintenance26,27. The breeders tried for alternative genetic mechanism(s) with robust phenotype and easy maintenance. Genic male sterility (GMS) was identified in Brassica oleracea crops such as cauliflower (ms-4, ms-5) and other Cole vegetables namely ms-128,29, ms-2, ms-4, ms-5 and ms-630, DGMs79-399-331 in broccoli and Ms-cd1 in cabbage32, but unfit for commercial use in hybrid breeding25. The identification of Ogura sterile cytoplasm in wild Japanese radish3, its deployment in Brassica oleracea L.33 and refinements for chlorosis defects34 has opened a new era of hybrid breeding. The cytoplasmic male sterility (CMS) owing to its robustness and easy maintenance was deployed in different nuclear backgrounds of cauliflower including genotypes from all the maturity groups22. The Ogura system imparts complete male sterility which facilitated its use in commercial hybrid seed production. E. canariense (Can system) and B. tournefortii (Tour system) are two other sterile cytoplasms deployed in cauliflower4,35. Cauliflower breeders at ICAR-IARI, New Delhi transferred these three CMS systems in different genetic backgrounds in cauliflower36. Singh et al.22 studied the Ogura, Can and Tour CMS cytoplasm deployed CMS lines in Indian cauliflower for floral and yield traits and indicated for better prospect of Ogura- and Can- based CMS systems in hybrid breeding. They reported that Tour cytoplasm-based CMS lines of cauliflower have ‘gripping phenomena’ wherein stigma gets trapped in a sepal which hinders insect pollination. Ogura cytoplasm has been well investigated for its interaction with different genetic backgrounds22,25,37,38 and for their heterotic potential7,39,40. It impact floral traits in snowball cauliflower41 and Indian cauliflower22. The Can cytoplasm is the most recently deployed sterile cytoplasm in different genotypes of the early group of Indian cauliflower at ICAR-IARI, New Delhi. These Can-based CMS lines need to be investigated for the impact of Can cytoplasm on agro-morphological and floral traits in nuclear backgrounds of cauliflower genotypes and also their potential in F1 hybrid seed production.
Materials and methods
Plant material and morphological observations
Seventeen CMS lines (11 Can CMS; 3 Ogura CMS; 3 Tour CMS) and their 11 maintainers were developed and being maintained by Division of Vegetable Science, IARI, New Delhi (Table 1). The original introgression Can CMS line ‘NIPB-1’ was developed and maintained by ICAR-NIPB, New Delhi and maintainer ‘PM’ by ICAR-IARI, New Delhi. Three maintainers namely DC 98-4, DC-41-5 and DC-67 were kept common for all three sterile cytoplasms (i.e. Can, Ogura and Tour) carrying CMS lines. The experiment was carried out at Vegetable Research Farm, Division of Vegetable Science, ICAR-IARI, New Delhi (28.08 0N and 77.12 0E, above mean sea level being 228.61 m). The seedlings were raised on nursery beds of 3 m × 0.5 m × 0.20 m size. Sowing was done on 20.07.2021 and transplanted on 25.08.2021 at 45 × 45 cm spacing in complete randomized block design (RBD) with 3 replications. Standard crop management practices and plant protection measures were adopted for raising the trial crop23.
The CMS lines and their maintainers were observed for 12 agro-morphological and curd yield traits, 18 floral traits and four seed yield traits. Plant height (PH), plant spread (PS), leaf length (LL), leaf width (LW), number of leaves per plants (NLP), gross plant weight (GPW), marketable curd weight (MCW), net curd weight (NCW), root length (RL) and root width (RW) were recorded from five random plants in each plot at curd maturity or harvesting stage. Marketable curd yield (MCY, t/ha) was calculated from plot yield. The harvest index (HI) was obtained by using the flowing formula: HI = MCW (g)/GPW (g).
The 18 floral traits were recorded each of 10 random plants at the flowering stage in CMS lines and their maintainers grown in replicated trials with three replications. The observations were taken using 25 to 30 fully developed floral buds (for floral bud traits) or naturally opened flowers (for flower traits) from each plant. Flower length (FL), flower diameter (FD), stalk length (StaL), petal length (PL), petal width (PW), sepal length (SL), sepal width (SW), bud length (BL) and bud width (BW) were recorded using standard rural scale. Stigma length (StiL), long stamen length (LSL), short stamen length (SSL), long filament length (LFL), short filament length (SFL), anther length (AL), nectary length (NL) and nectary width (NW) were taken using Discovery.18 SteREO Zoom Microscope (Carl Zeiss Microimaging Gmbh, Germany) in Division of Floriculture and Landscaping, IARI, New Delhi. Fully opened flowers from already covered inflorescence branches were taken and nectar was collected in calibrated capillary tubes (Sigma-Aldrich, Missouri, USA). The nectar volume (NV, μl/10 flowers) was calculated by dividing the length of the nectar column in the capillary tubes by the total length of the capillary tube42.
For seed parameters, all the 17 CMS lines were grown alongwith fertile pollen-parent in 1:1 ratio in three replications at two isolation blocks namely Mela Gate Farm (MG) and Vegetable Research Farm (VRF) located ≈1000 m apart in ICAR-IARI, New Delhi. Pollen parent DC-67 and DC-98-4 were planted at MG and VRF site, respectively. The seed parameters namely siliqua length (SqL), number of seeds (NS) per siliqua, seed yield (SY) per plant and 1000-seed weight or test weight (TW) were recorded from five randomly tagged plants.
DNA extraction and PCR analysis
The DNA of CMS lines and their maintainers was extracted from fresh mature leaf samples from healthy young plants by using CTAB protocol43 and stored at −80 °C for further molecular work. The sampling for DNA extraction was done from five plants of each genotype (CMS and maintainers). The DNA quality was checked using agarose gel and DNA concentration was measured using Nanodrop (Eppendorf). The DNA was diluted with 0.1 X TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8.0) to yield a working concentration of 30 ng/μl. An equal volume of this working DNA sample from each of the five plants was mixed and made a representative sample for the respective genotype.
A set of 25 mt-DNA primers available in the public domain were synthesised by Integrated DNA Technologies, (IDT, Coraville, Iowa, USA) and screened for amplification using polymerase chain reaction (PCR) (Biometra Tone 96G, Analytik Jena, Germany). The details of the primers and their sequence information are given in Table S1. These were synthesized from Integrated DNA Technologies, Inc (IDT, USA). All the primers were amplified using 10 μl PCR mixture volume with 2X Master mix (GeneDireX® OnePCR™, Forward and Reverse Primer 1.0 μL (0.5 μL each, 25 μM), Template DNA (1.0 μL, 30 ng/μL) and Sterile double distilled water (3.0 μL, 75 ng/μL) using thermocycler. The PCR steps were: Initial denaturation at 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55–65 °C for 1 min (varied with primer), extension at 72 °C for 1 min; final extension at 72 °C for 10 min and hold at 4 °C for 30 min followed by storing at 4 °C for further processing.
Electrophoresis and fragment detection
The amplified PCR products were resolved on 3% agarose gel in 100 mL 1X TAE buffer using 1 μg/mL of ethidium bromide florescent dye in gel electrophoresis (BioRad, USA). DNA bands were visualized and photographed under UV light in gel documentation unit (Alpha imager Cell Biosciences, Santa Clara, CA). The molecular markers showing variation in banding pattern among the genotypes were considered as polymorphic while rest were counted as monomorphic. Amplicon size was measured using the DNA ladder (GeneDireX Inc., 50 bp) as a reference.
Nucleotide sequencing
The PCR products were obtained from selected eight genotypes representing three different sterile cytoplasms namely Can, Ogura and Tour. These included five CMS lines, namely Can (DC-23), Can (DC-98-4), Can (DC-41-5), Ogu(DC-41-5) and Tour (DC-41-5) and three maintainers, namely DC-23, DC-98-4 and DC-41-5. The PCR products from five selected primers (i.e., ITS5a-ITS4, atpF-atpH, trnL, P16 and rbeL) were custom sequenced at Agri-Genome Pvt. Ltd., Hyderabad. The forward and reverse sequences, thus, obtained were assembled to obtain contigs using Bioedit sequence alignment editor version 7.0.5.3. The contigs were aligned using ClustalW multiple alignment option of BioEdit software and output was obtained under graphic view and exported as rich text with 50 residues per row and in blocks of ten residues.
Statistical analysis
The data from agro-morphological, floral and seed yielding traits were recorded and subjected "doebioresearch" package of R software for ANOVA (analysis of variance). Standard deviation and per cent change due to CMS introgression were calculated over the fertile (maintainer) line for agro-moprhological and floral traits using Microsoft Excel 2019. K-means clustering was implemented using “cluster” package of R software. The optimal number of clusters was determined by average silhouette width. Average silhouette width was computed employing “factoextra” package of R software. The "Hmisc" R package was utilized to perform correlation analysis on both agro-morphological and floral traits. Non-significant correlations, determined at a significance level of 5%, have been marked with a strikeout. Principal Component Analysis (PCA) has been employed using the “factominer” package of R Software. Factorial RBD design has been employed using the “doebioresearch” package of R software.
Statement regarding plant guidelines
The use of plant parts in the study complies with international, national, and/or institutional guidelines.
Results
Analysis of variance (ANOVA) for traits
Table 2 provides the results of the analysis of variance for agro-morphological features observed in early cauliflower CMS lines and their maintainers. There were significant differences among the groups (CMS lines and maintainers) for the observed agro-morphological traits except PH. No significant difference observed between the groups.
Variation in agro-morphological and curding traits
The CMS lines and their maintainers showed significant differences for all the observed agro-morphological traits (Table 3). Box plot of these traits from the CMS lines (Fig. 1a) and their maintainers (Fig. 1b) depicts minimum, first quartile (Q1), median, third quartile (Q3) and maximum values of the traits. The box plot analysis showed wide variation in both CMS and maintainer lines for PH, PS, MCW, NCW and GPW. PH in CMS lines was ranged from 53.5 cm in Can (DC-94-2) to 63.6 cm in Ogu (DC-98-4) and in maintainers from 58.1 cm (DC-41-5) to 64.8 cm (DC-63). PS ranged from 55.7 cm in Can (DC-41-5) to 89.8 cm in Tour (DC-98-4) in CMS lines. LL, LW and NLP also showed significant variation in CMS lines and maintainers. LW ranged from 40.0 cm in Can (DC-41-5) to 54.8 cm in Can (DC-63) and LL from 19.1 cm in Can (DC-67) to 23.9 cm in Ogu (DC-98-4), respectively. NLP were maximum in Tour (DC-41-5) (20.3) in CMS lines and DC-63 (22.4) in testers. GPW ranged from 1215.0 g in Can (DC-98-4) to 1766.3 g in Can (DC-209). Among maintainers, the GPW was highest in DC-41-5 (1677.9 g). Can (DC-903) (690.9 g; 567.0 g) in CMS lines and DC-63 (673.8 g; 551.7 g) in testers had maximum MCW and NCW, respectively. MCY was highest in Can (DC-903), ranging from 17.7 to 24.2 t/ha. It was the greatest among the testers in DC-63 (23.6 t/ha). HI ranged from 0.34 to 0.53 with the highest in Can (DC-41-5) in CMS lines. Among maintainers, it was highest in DC-8 (0.42). RL (18.6 cm) and RW (19.1 cm) were the greatest in CMS Can (DC-8) while in maintainers these were maximum in DC-94-2 (20.2 cm) and DC 41-5 (19.4 cm), respectively.
Variation in CMS lines and fertile maintainers for floral traits
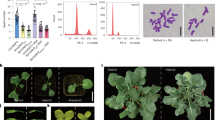
The ANOVA for 18 floral traits in CMS lines and their maintainers is Table 4. Table 5 displays average values of 18 floral traits observed in CMS lines and their maintainers. The box plot of floral traits showed wide range (minimum, maximum), median, first quartile (Q1) and third quartile (Q3) in CMS lines (Fig. 1c) and their maintainers (Fig. 1d). Box plot analysis also revealed large variation in floral traits such as FL, FW, PL, PW, BW, StiL and NL in CMS lines. The cytoplasmic sterility reduced overall flower size and size of reproductive organs (stigma and stamens) in CMS lines (Fig. 2a). The flower parts (sepals, petals, stigma and anthers) of CMS lines and their maintainers of early cauliflower are presented in Fig. 2b. All the observed floral traits showed a significant difference in the CMS lines and maintainers. The tiniest and narrowest flowers were observed in all the three Tour-based CMS lines. FD ranged from 1.23 cm in Tour (DC-67) to 1.76 cm in Can (DC-94-2) in CMS lines and DC-41-5 (2.13 cm) in maintainers. Can (DC-41-5) had the shortest StaL (1.61 cm) in CMS lines while DC-7 had the longest StaL (1.93 cm) in maintainers. PL and PW were ranged from 1.12 cm [Can (DC-18)] to 1.59 cm [Can (DC-7)] and 0.62 cm [Can (DC-209)] to 0.79 cm [Can (DC-67)], respectively. DC-63 (1.62 cm) and DC-98-4 (1.93 cm) had the highest values of these two petal parameters. SL and SW were ranged from 0.69 to 0.78 cm and 0.31 to 0.40 cm in CMS lines. DC-67 had maximum SL (0.91 cm) while DC-63 had the widest sepals (0.47 cm). BW ranged from 0.39 to 0.56 cm and BL from 0.53 cm to 0.70 cm.
StiL in CMS lines was ranged from 0.65 to 1.03 cm which was significantly less than the maintainers (0.69–1.05 cm). Observations on length of stamens (i.e. LSL and SSL) and anther filament (LFL and SFL) showed significant differences in CMS lines and their maintainers. NL was maximum in Ogu(DC-67) (1.77 mm) and NW in Ogu (DC-41-5) (1.83 mm) while in maintainers, the observed maximum values were 2.06 mm (DC-63) and 1.95 mm (DC-8), respectively. NV also varied significantly in CMS lines (0.34–0.46 µL/10 flowers) and maintainers (0.48–0.56 µL/10 flowers).
Can cytoplasm-associated changes in agro-morphological and floral traits
In comparison to their maintainers, the introgression of nuclear genome of Indian cauliflower genotypes in sterile cytoplasms had significant impact on all 12 of the observed morphological traits (Fig. 3). PL changed in range of −16.1% in Can (DC-94-2) to 9.1% in Can (DC-98-4) and PS reduced the greatest in Can (DC-8) (−20.8%). LL and LW also influenced by sterile cytoplasms in the ranges of -26.2% to 6.1% and -17.7% to 11.2%, respectively. Except for Can (DC-23), all Can based CMS lines showed reduction in NLP. Impact of Can cytoplasm was noticed on GPW (-21.7 to 16.6%), MCW (-19.6 to 12.0%) and NCW (up to 13.4%). The changes in RL and RW ranged from -23.3% to 15.0% and -22.1% to 19.1%, respectively. The sterile cytoplasms also affected MCY, which was decreased in eight CMS lines while slightly increased in 3 CMS lines by 9.6 to 14.0%. Can (DC-98-4) experienced the greatest decline (19.5%). HI was also influenced in both the directions.
Contrary to agro-morphological traits, the sterile cytoplasms had highly significant and negative impact on all the 18 observed floral traits (Fig. 4). Eight floral parameters namely SL and SW, StaL, LSL, SSL, AL, NL, NW and NV showed negative impact only. The reduction in FL ranged from -36.5% in Can (DC-67) to 11.4% in Can (DC-7). Similar trend was observed for FD and StaL. Notable impact was observed on petals (PL: −30.2 to 14.3%; PW: −36.0 to 13.4%), sepals (SL: −1.2 to −27.4%; SW: −31.1 to 2.7%) and floral buds (BL: −29.4 to 25%; BW: −41.5 to 21.7%). Significant reduction was observed in StiL (−1.9 to −35.8%), LSL (−24.7 to -33.70%) and SSL (−4.8 to −23.3%). The CMS introgression reduced NL, NW and NV in range of −18.9 to −8.2%, −16.0 to −8.2% and −32 to −6.2%, respectively.
Impact of sterile cytoplasms on agro-morphological traits
Nine CMS lines developed through introgression of Tour, Ogura and Can cytoplasms in nuclear backgrounds of DC-98-4, DC-67 and DC-41-5 were observed for SqL, NS/siliqua, SY (g/plant) and TW (g). The sterile cytoplasms influenced seed traits and the greatest reduction was observed in Tour-based CMS lines (Fig. 5a–d). While, the TW was found to be in the same range, SqL and NS/siliqua showed significant different. SY was significantly affected due to CMS introgression particularly due to Tour sterile cytoplasm.
Principal component, bi-plot and correlation analyses
Principal component analysis (PCA) for agro-morphological traits (Fig. 6a, b) and floral traits (Fig. 6c, d) explains prominence of PCA1 (65.3%) and PCA2 (22.5%) combining of 87.8% in explaining variances in CMS lines and PCA1 (36.5%), PCA2 (28.3%) and PCA3 (17.3%) in maintainers (Fig. S1a, b). For floral traits, PCA1 (38.3%) and PCA2 (23.8%) together explained maximum part of variance in CMS lines while only PCA1 could explain 59.7% variance in maintainers (Fig. S1c, d).
Contribution of variables of agro-morphological traits in variation in CMS lines and maintainers is presented in (Fig. S2a, b). and their floral traits in (Fig. S2c, d). RL, GPW, HI and PS are important determinant show similarity in CMS lines while GPW, HI, NCW, MCY and MCW in maintainers .SL, FL, StaL, SW, NL, LFL, StiL, PL, FW and BL in CMS lines and SSL, LSL, LFL, SFL, FW, BW, NV, NL and StaL were contributors in variance in maintainers.
Bi-plot analysis for agro-morphological traits (Fig. 6a, b) and floral traits (Fig. 6c, d) in CMS lines and maintainers in early group of Indian cauliflower using "FactoMiner" R Package. This plot interpreted the similarity in most of the agro-morphological traits. HI, RL and GPW showed similarity in CMS lines whereas all the traits showed similarity in maintainers except GPW. For floral traits, LSL and SSL showed similarity in CMS lines whereas all the traits showed similarity in maintainers except SL. Cluster analysis of CMS lines and their maintainers for agro-morphological trait (Fig. 7a, b) and floral traits (Fig. 7c, d) could reveal three and two clusters, respectively.
Correlation analysis between agro-morphological traits in CMS lines and maintainers is presented in Fig. 8a, b and their floral traits in Fig. 8c, d. A significant positive correlation was observed among the traits such as NCW with MCW and MCY. RL positively correlated with HI and NL positively correlated with RW while GPW negatively correlated with PS and PS negatively correlated with LL in CMS lines. In maintainers MCW positively correlated with MCY and NCW positively correlated with MCW while PS negatively correlated with HI. In CMS lines, RL was strongly correlated with NLP, RW and HI. In floral traits, LFL has strong correlation with FW and NW. StiL significantly correlated with StaL and SL in CMS lines. In maintainers, LFL was strongly correlated with NV, SSL, SFL and FW (Fig. 8c, d). SSL significantly correlated with NV and SFL and BW with NL. These correlation studies are significant at 5% level of significance.
Variation in CMS lines for F1 hybrid seed traits
Table 6 lists the performance of 17 CMS lines of Indian cauliflower for F1 hybrid seeds production in two isolations using DC-67 and DC-98-4 fertile inbred lines as male parents. The SqL was ranged from 4.7 cm [Tour (DC-41-5)] to 5.9 cm [Tour (DC-67)]. Can (DC-23) had maximum NS (14.5/siliqua) while Tour-based CMS lines namely Tour (DC-41-5) and Tour (DC-67) had minimum NS (4.8/siliqua). Among CMS lines, SY was maximum in Ogu (DC-98-4) (28.0 g/plants) followed by Can (98-4) (24.2 g/plant) and minimum in Tour (DC-67) (1.5 g/plant). TW was ranged from 1.9 g [Can (DC-18)] to 3.6 g [Tour (DC-41-5)]. Satisfactory flowering and seed settings were observed in all the Can and Ogura cytoplasm-based CMS lines.
Mitochondrial specific DNA markers for CMS lines
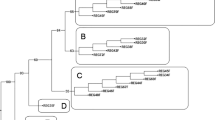
The observations for amplification and level of polymorphism in CMS lines with 25 mtDNA markers are presented in Fig. S3. Two markers, namely P14, P15 and P16 were polymorphic in CMS and fertile maintainers. The marker P14 amplified fragments of 200 bp in CMS lines (lane 1–6, 8,9) and that of 250 bp in fertile maintainers (lane 15–20), whereas P15 amplified distinct bands of 200 bp in CMS lines (lane 3–8) and 250 bp in fertile maintainers (lane 13–18), respectively. P14 showed polymorphism between CMS lines carrying Ogura (lane 21, 22, 24) and canarianse (lane 1–6, 8, 9) CMS systems and their fertile counterparts. It also generated a unique band (175 bp) for Tour cytoplasm (lane 23), separating it from Ogura and canarianse cytoplasms. P15 could distinguish canarianse (lane 3–8) from Ogura (lane 21, 22, 24)) and Tour (lane 23) cytoplasms, however, it could not differentiate between the latter two systems. Notably, intra-CMS system variation was also observed among the Can cytoplasm carrying CMS lines such as Can (DC-209) (lane 7) and Can (DC-63) (lane 10) which amplified an amplicon of 250 bp while rest of the eight Can based CMS lines generated 200 bp with P14 primers. Similar observations were made from P15 which could generate three different bands in Can based CMS lines. P16 revealed a polymorphic band (450 bp) for Ogura (DC-45) (lane 24) while its maintainer DC-41-5 (lane 18) amplified 500 bp amplicon.
Sequence analysis
The results of alignment of amplicons produced by the eight genotypes employed in the study using ITS 5a-ITS 4 primer are shown in Fig. S3a. The aligned sequences were filtered from both the ends to remove the gaps and the central 614 residues were finally analyzed. In the aligned sequences, the sequences bearing similarity are shaded with the same colour e.g. A is shaded green, T red, C blue and G black. The unshaded residues are indicating that the residue in question is variable in the compared genotypes. There was not much variation in the analysed sequence up to 460 bases. In this portion, the sequences were observed to be same in all the genotypes except DC-23 and DC-98-4. It was observed that there was C in place of A at 95th position, G in place of T at 163rd position, C in place of T at 402nd, C in place of T at 416th position in DC-23 and DC-98-4. At the same time genotype Can (DC-41-5) showed presence of G at this position (416th). At position 432nd there was C in Can (DC-41-5) while G in rest of the genotypes. Similar analysis of the sequences revealed that only DC-23, DC-98-4 and Can (DC-41-5) showed different pattern as compared to the rest of the genotypes. The results of alignment of amplicons obtained using atpF-atpH primer from the eight genotypes (5 CMS lines and 3 fertile maintainers), two maintainers were common for four CMS lines (Fig. S3b). The central 429 residues were finally analysed. In the aligned sequences, the sequences bearing similarity are shaded with the same colour as in case of ITS 5a-ITS4. The lines showed variation at different positions for nucleotides. Can (DC-23) had deletions of A at two positions i.e. at 166th position A is deleted and T is deleted at 299th position. Five conversions were also observed at 178th, 258th, 318th, 370th and 429th positions. The sequence analysis revealed variation among the sterile cytoplasms and also in fertile inbreds for the observed sequences. The deletion was also observed in Can (DC-98-4) at 299th position but not in Can (DC-41-5) indicating change in sequences. At 323th position, the sequence alignment of atpF-atpH showed difference in the CMS lines namely Can (DC-41-5), Ogu (DC-41-5) and Tour (DC-41-5) and their maintainers DC-41-5. These CMS lines had T, while maintainer DC-41-5 had A. The sequence alignment of amplicons obtained using P16 primer from the eight genotypes (Fig. S3c) revealed no variation between Can (DC-23) and DC-23, while Can (DC-98-4) and Can (DC-41-5) showed variation due to addition and deletion of nucleotides at different positions. In case of rbeL primer (Fig. S3d). The central 663 residues were finally analyzed. In the aligned sequences no variation was detected between Can (DC-23) and DC-23 while Can (DC-98-4) and Can (DC-41-5) showed variation due to the addition and deletion of nucleotides at different positions. The sequence alignment of amplicons obtained using trnL primer from eight genotypes (Fig. S3e) could show addition and deletion of nucleotides in Can (DC41-5). It has additions at 14th, 15th, 60th, 74th, and 80th positions. Can (DC-41-5) had one deletion of nucleotides at 22nd, 32nd, 42nd, 48th, 50th and 57th positions.
Discussion
In terms of agro-morphological features, the CMS lines carrying Can and Tour sterile cytoplasms are on par with the CMS lines carrying Ogura sterile cytoplasm. However, there was a significant difference in floral traits between the cauliflower CMS lines carrying these three sterile cytoplasms. Tour cytoplasm-based CMS lines namely Tour (DC-41-5) and Tour (DC-67) showed a considerable reduction in floral traits.
Variations in agro-morphological, floral and seed traits
Cauliflower is a crop that was introduced to the Indian subcontinent, and the original genotypes had a temperate flowering habit meaning that they needed a protracted period of cold temperature for proper bolting and flowering for seed production. Newly evolved cauliflower had varied morphologies in terms of stalk length, leaf orientation, leaf size and colour, curd size and colour and plant frame19. ‘Indian cauliflower’ or ‘tropical cauliflower’ was the name given to the evolved ecotype particularly to the early and mid-early groups. The cytoplasmic male sterility is widely used genetic mechanism for hybrid breeding in Brassica crops. It arises as a result of a genomic conflict between the mitochondrial and the nuclear genomes. The Ogura sterile cytoplasm was first deployed in European cauliflowers35 and later on introgressed in Indian cauliflowers26,38. Still it is a robust and stable genetic system that ensures complete cross-pollination across the temperature range. The modification or alteration in the mitochondrial genome that causes male sterility may substantially impair CMS system performance. This has been documented in the ‘Polima’ cytoplasm where partial restoration of fertility has occurred as a result of higher temperature44. As hybrid breeding in cauliflower currently solely depends on the Ogura CMS system, this will be a major calamity, if a Polima-like situation occurs. Thus, two additional sterile cytoplasms namely E. canariense (Can) and B. tournefortii Goun (Tour)4,35. This CMS introgression process might had an impact on other non-target but important traits such as agro-moprphological and floral traits. Because, normal size of flowers, properly developed petals, normal stigma and style and well-developed functional nectaries are required for drawing honey bees for pollination activities. The previous researchers have showed the negative effects of Ogura sterile cytoplasm on floral traits including flower size, petal length and width, stamens, stigma and nectary size as well as nectar volume37,45,46,47. These defects/deformities included reduced flower size, petaloid anthers, carpeloid stamens, malformed nectary, twisted stigma and split anthers in Brassica crops. A reduction in floral traits in CMS lines of Indian cauliflower carrying Ogura, Can and Tour sterile cytoplasm was also reported22,38. Size of functional nectaries is correlated with nectar volume38,48. And, the pollinator insects in Brassica crops are attracted to flowers based on the amount of nectar they produce 49,50,51.
In current investigation, the pattern of the per cent change in plant morphology, curd yield and floral traits revealed varied results. The parameters of PH, PS and leaf attributes showed a variable pattern. Maximum reduction in PH was recorded in DC 98-4 (−16.1%), while overall gain was seen in Ogu (DC-98-1) and Can (DC-7) in Ogura and Can cytoplasm-based CMS lines, respectively. Three CMS lines namely Can (DC-903) (12.0%), Can (DC-209) (7.5%) and Can (DC-18) (3.5%), which were considerably greater than other two CMS systems, demonstrated good gains in MCW. Dey et al.37 and Singh et al.38 both made similar observations. The MCY from Can-based CMS lines (21.8 t/ha) was nearly equal to that of the CMS lines carrying Ogura (21.8 t/ha) and Tour (21.4 t/ha) cytoplasm. This might be explained by nuclear–cytoplasmic interactions and cooperative functions between particular nuclear backgrounds of cauliflower and alien cytoplasms (i.e. sterile cytoplasm from other species). These alloplasmic lines, which carry cytoplasms namely Can, refined Ogura and Tour from three different sources—E. canariense (complete cytoplasm), Raphanus sativus L. (only mitochondria) and B. tournifortti (complete cytoplasm), respectively. Refined Ogura based CMS lines carry chloroplast of B. oleracea, a unique functional combination in plant system.
The conversion procedure from fertile inbred lines to CMS lines had impact on floral features. Both FL, FD, PL and PW were decreased significantly. The findings are consistent with earlier studies in broccoli46, snowball cauliflower37,48 and Indian cauliflower22. The conversion process significantly decreased the size of the nectaries (NL, NW). The findings align with earlier reports22,48. NV also affected due to CMS trait. This might be the reason for a negative effect on seed yield in honey bee pollinated crops. The reduction in floral characteristics agreed with the findings of Dey et al.41 In comparison to their fertile lines, they reported a considerable decline in floral characteristics in refined Ogura based CMS lines of snowball cauliflower. However, all the Can and refined Ogura carrying CMS lines had decent size of flowers for honey bee visits (based on field observations of the senior author). Among the Tour-based CMS lines, only Tour (DC98-4) had a proper flower while the remaining Tour (DC-67) and Tour (DC-41-5) had most malformed flowers with ‘gripping phenomenon’ as also noted by Singh et al.22 The CMS phenotype is associated with abnormal recombination of mitochondria genome and CMS genes (i.e. orf138 in Ogura CMS) encodes transmembrane proteins (ORF138) which bind to the mitochondrial membrane, affecting the hydrogen ion concentration gradient, thus affecting ATP synthesis. This leads to energy deficiency and excess accumulation of reactive oxygen species (ROS). The ROS play key role in cell signaling and their excess accumulation causes damage to lipids, proteins and DNA, inhibit enzyme activity, activate the programmed cell death (PCD) pathway and ultimately affect plant growth and development, particularly reproductive parts52.
The high seed yield in natural crossing conditions is essential for economic use of CMS system in hybrid breeding. The CMS trait impact most of floral traits which reflects in reduced seed yield also, however, in comparison, certain CMS systems have better performance than others53. The seed yield per plant in the refined Ogura-based CMS lines of snowball cauliflower namely Ogu1A, Ogu2A, and Ogu3A was reported to be 30.9 g, 21.4 g and 26.8 g per plant, respectively54. They reported that the seed yield per plant for these CMS lines was 24.3%, 29.3% and 10.1% less than its maintainer, respectively. Kucera et al.55 reported a reduction in seed yield per plant by 79.5% and 85.7% in FT CMS and BR CMS, respectively. It was attributed to fewer siliqua per plant and a smaller number of seeds per siliqua than their maintainers. The observed decrease in seed yield due to CMS trait in present study was consistent with the earlier findings37,55. However, this is the first in-depth account of changes brought on by Can sterile cytoplasm in agro-morphological and floral features of cauliflower. Similar to this, it was found that 11 Can-based CMS lines with the two pollen parents DC-67 and DC-98-4 produced hybrid seeds on average at rates of 19.5 g and 12.8 g/plant, respectively. Three Ogura based CMS lines with same pollen parents produced hybrid seeds 14.7 g and 18.5 g/plant, respectively. The Tour-based CMS lines, particularly Tour (DC-41-5) and Tour (DC 67) were worst performers for hybrid seed yield, mainly due to gripping phenomena of stigma38. High yield of hybrid seeds in present study could be due to genotype, open condition and nicking of CMS and pollen parents. Kucera et al.55 obtained only 2.3 g/plant hybrid seeds from BR CMS line with pollen parent FT13 grown in isolation cages provided with bumble bees. Thus, the study demonstrates the potential of these novel Can sterile cytoplasm carrying CMS lines in production of hybrid seeds.
The PCA and cluster analysis identified key agro-morphological and floral traits which contribute major share of variance in the CMS lines. The potential for trait comprehension for breeding purposes is indicated by the strong association between root and leaf parameters, curd weight and yield characteristics, and stamen length and nectary traits.
mtDNA level molecular variation in CMS lines
At flowering stage, the CMS phenotype is persistent and simple to distinguish from fertile analogues. It is challenging to distinguish them at earlier stages, though. Further, the deployment of several sterile cytoplasms in cauliflower makes it challenging to distinguish these cytoplasms carrying CMS lines at a morphological level. Breeders will therefore benefit from the creation or discovery of DNA markers to differentiate between these several sterile cytoplasms. Additionally, because these markers are stage-independent, they can be applied to any specious mixture during the seedling stage or before flowering. Mitochondrial markers are thought to be a reliable method for identifying sterile and fertile cells as well as the type of cytoplasm that CMS lines carry56,57.
A dominant marker called ISSR3 was used by Wang and Song58 to distinguish fertile counterparts from the CMS lines. According to them, it is possible to distinguish between fertile and sterile lines using an amplicon of 313 bp size using a primer pair P9/P10 for mtDNA marker specific to the CMS cauliflower knxd612. Only two of the 25 mtDNA markers namely P14 and P15 were polymorphic between CMS and male fertile phenotypes. P16 also distinguished Ogura cytoplasm-based CMS line Ogura (DC-41-5) from its maintainer (DC-41-5). Notably, P14 distinguished Tour cytoplasm from Ogura and canarianse CMS systems which was in line with previous reports of Singh et al.22 Additionally, the polymorphism was found among the Can-based CMS lines. This might be the result of an adaptive interaction between the mitochondrial genome of E. canerianse and the nuclear genome of B. oleracea var. botrytis L. (cauliflower), which has changed the genetic makeup of the CMS lines. Such occurrences are uncommon, hence warrant for further studies because the modifications to the mitochondrial DNA do not occur so frequently (i.e. 6 to 7 years of introgression in case of Can—oleracea cytoplasm interaction) as observed from Ogura—Brassica interaction (nearly 25–30 years in existence). Also, further research is warranted to understand the variability among the CMS lines carrying same Can cytoplasm. This could be explained by the lower levels of polymorphism of target locus in the four cytoplasms studied (Oleracea, Eru, Can and Tour). Since these species share a common ancestor and except a few minor variations, their cytoplasmic genomes are still conserved to great extent.
The sequence analysis from five CMS lines including three Can-based [Can (DC-23), Can (DC-98-4), Can (DC-41-5)] and one each with Ogura [Ogu (DC-41-5)] and Tour cytoplasms [Tour (DC-41-5] along with their maintainers revealed insertions and deletions. This could explain the variations among these lines. Sequence analysis of P16 amplicon from Can (DC-98-4) and Can (DC-41-5) CMS lines had distinct bases from their maintainers. Further, sequence analysis of Tour (DC-41-5) CMS line revealed that P16 amplicon at 243rd position and atpF-atpH amplicon at 138th position had T while rest of the lines had C and A, respectively. This indicates that flower deformity (i.e. gripping phenomenon’ could be due to this change in ORF region. However, it needs further investigation to establish the association. Primer atpF-atpH amplicon sequence revealed unique bases at manifold position indicating its distinctness from the other three cytoplasms. Earlier reports also suggest variation among the Ogura-cytoplasm carrying CMS lines of cauliflower25 and broccoli31. Deletions of nucleotide in the ORF coding region of broccoli cytolines was suggested to be associated with carpelloid stamen phenotype31, however, such abnormality was not reported in investigated CMS lines.
Conclusion
The present work provides the first in-depth information on the effect of this sterile cytoplasm in new nucleo-cytoplasmic combination (oleracea-canerianse) lines of Indian cauliflower. Erucastrum canarianse Webb and Berthel is a new CMS system deployed in cauliflower. These lines contain the cauliflower nuclear genome, whereas E. canarianse provides the cytoplasm. Although most agro-morphological features and floral attributes changed as anticipated due to the novel cytoplasm-nuclear interaction, the impact’s size was not averse to the level which can affect pollination drastically. The introgression of Can CMS system in the tropical Indian cauliflower reduced flower parameters equal to those of the Ogura CMS line, but seed-setting capacity is adequate, emphasizing the potential for hybrid seed production. P14 and P15, two polymorphic co-dominant mtDNA markers that were also discovered for Can CMS system, will be helpful for early detection, tracking of change(s) in mtDNA sequences and spotting variation between CMS lines and their maintainers. Similarly, P16 was found to be useful to distinguish Ogu (DC-41-5) from its maintainer DC-41-5. In the instance of sequence analysis, the Can, Ogu and Tour cytoplasm harbouring CMS lines displayed variation. These novels Can CMS lines of Indian cauliflower creates the possibility for immediate use in hybrid breeding or further introgression in other members of B. oleracea group through backcrossing.
Data availability
All the data generated and analysed during this study are included in this published article and its supplementary information. However, additional information can be obtained from corresponding author on reasonable request.
References
Chen, L. T. & Liu, Y. G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606 (2014).
Pei, X., Jing, Z., Tang, Z. & Zhu, Y. Comparative transcriptome analysis provides insight into differentially expressed genes related to cytoplasmic male sterility in broccoli (Brassica oleracea var. italica). Sci. Hortic. 217, 234–242; https://doi.org/10.1016/j.scienta.2017.01.041 (2017).
Ogura, H. Studies on the new male sterility in Japanese radish, with special references on the utilization of this sterility towards the practical raising of hybrid seeds. Mem Fac Agric Kagoshima Univ. 6, 40–75 (1968).
Chamola, R., Balyan, H.S. & Bhat, S.R. Transfer of cytoplasmic male sterility from alloplasmic Brassica juncea and B. napus to cauliflower (B. oleracea var. botrytis) through interspecific hybridization and embryo culture. Indian J Gen Plant Breed.73, 203–210; https://doi.org/10.5958/j.0975-6906.73.2.030 (2013).
Rawat, D. S. & Anand, I. J. Male sterility in Indian mustard. Indian J Genet Plant Breed. 39, 412–414 (1979).
Liu, Z. et al. A pentatricopeptide repeat protein restores nap cytoplasmic male sterility in Brassica napus. J Exp Bot. 68(15), 4115–4123. https://doi.org/10.1093/jxb/erx239 (2017).
Oshima, M., Kikuchi, R., Imamura, J. & Handa, H. Origin of the CMS gene locus in rapeseed cybrid mitochondria: active and inactive recombination produces the complex CMS gene region in the mitochondrial genomes of Brassicaceae. Genes Genet Syst. 85(5), 311–318. https://doi.org/10.1266/ggs.85.311 (2010).
Shinada, T., Kikuchi, Y., Fujimoto, R. & Kishitani, S. An alloplasmic male-sterile line of Brassica oleracea harboring the mitochondria from Diplotaxis muralis expresses a novel chimeric open reading frame, orf 72. Plant Cell Physiol. 47(4), 549–553. https://doi.org/10.1093/pcp/pcj014 (2006).
Fu, T.D. et al. The discovery, research and utilization of pol cytoplasmic male sterile in Brassica napus. Prog. Nat. Sci. Commun. State Key Lab.5, 287–293 (1995).
Jing, B. et al. A male sterility-associated cytotoxic protein ORF288 in Brassica juncea causes aborted pollen development. J. Exp. Bot. 63(3), 1285–1295;https://doi.org/10.1093/jxb/err355 (2012).
Prakash, S. et al. A Moricandia arvensis–based cytoplasmic male sterility and fertility restoration system in Brassica juncea. Theor Appl Genet. 97(3), 488–492. https://doi.org/10.1007/s001220050921 (1998).
Atri, C. et al. Substituting nuclear genome of Brassica juncea (L.) Czern & Coss in cytoplasmic background of Brassica fruticulosa results in cytoplasmic male sterility. Euphytica.209, 31–40; https://doi.org/10.1007/sr10681-015-1628-4 (2016).
Kang, L. et al. A novel cytoplasmic male sterility in Brassica napus (inap CMS) with carpelloid stamens via protoplast fusion with Chinese woad. Front. Plant Sci.8, 529; https://doi.org/10.3389/fpls.2017.00529 (2017).
Stiewe, G. & Robbelen, G. Establishing cytoplasmic male sterility in Brassica napus by mitochondrial recombination with Brassica tournefortii. Plant Breed. 113(4), 294–304. https://doi.org/10.1111/j.1439-0523.1994.tb00739.x (1994).
F.A.O.S.T.A.T., Food and Agriculture Organization of United Nations. Rome, Italian Republic (2020).
N.H.B. National horticulture board, Government of India, Gurugram, Haryana. (2021).
Gopalan, C. & Rama Sastri, B. V. Nutritive value of Indian foods. (ed. Gopalan, C.) 112–113 (Indian Council of Medical Research, 2011).
Kushad, M. M. et al. Variation of glucosinolates in vegetable crops of Brassica oleracea. J Agric Food Chem. 47(4), 1541–1548. https://doi.org/10.1021/jf980985s (1999).
Swarup, V. & Chatterjee, S. S. Origin and genetic improvement of Indian cauliflower. Econ Bot. 26, 381–393. https://doi.org/10.1007/BF02860710 (1972).
Singh, S. & Kalia, P. Advances in cauliflower (Brassica oleracea var. botrytis L.) breeding, with emphasis on India. (ed. Al-Khayri, J.M., Jain, S.M., Johnson, D.V.). Advances in plant breeding strategies: vegetable crops. 247–301 (Springer, 2021).
Crisp, P. The use of an evolutionary scheme for cauliflowers in the screening of genetic resources. Euphytica. 31(3), 725–734. https://doi.org/10.1007/BF00039211 (1982).
Singh, S. et al. Agro-morphological and molecular diversity analysis of new cytoplasmic male sterile lines in Indian cauliflower for their use in hybrid breeding. Sci. Hortic. 301, 111107; https://doi.org/10.1016/j.scienta.2022.111107 (2022a).
Singh, R. & Sharma, S.R. Textbook of Vegetables Tuber crops and Spices. (ed.N. Singh and Thamburaj) 81–82 (Indian Council of Agricultural Research, New Delhi, 2003).
Verma, T. S. & Sharma, S. C. Producing seeds of biennial vegetables in temperate regions 51–63 (DKMA, 2000).
Singh, S. et al. Elucidating mitochondrial DNA markers of Ogura-based CMS lines in Indian cauliflowers (Brassica oleracea var. botrytis L.) and their floral abnormalities due to diversity in cytonuclear interactions. Front. Plant Sci.12, 631489; https://doi.org/10.3389/fpls.2021.631489 (2021).
Sharma, S.R., Singh, P.K., Chable, V. & Tripathi, S.K. A review in hybrid cauliflower development. J. New Seeds .6(2–3), 151–193 (2005).
Selvakumar, P., Sinha, S.N. & Pandita, V.K. Hybrid seed production in cauliflower (Brassica oleracea var. botrytis subvar. cauliflora). Indian J. Agric. Sci. 77(10), 649–651 (2007).
Anstey, T. H. & Moore, J. F. Inheritance of glossy foliage and cream petals. J. Hered. 45, 39–41; https://doi.org/10.1093/oxfordjournals.jhered.a106433 (1954).
Cole, K. Inheritance of male sterility in green sprouting broccoli. Can. J. Genet. Cytol.1,203–207;https://doi.org/10.1139/g59-028 (1959).
Dickson, M.H. A temperature sensitive male sterile gene in broccoli, Brassica oleracea L. var. italica. J. Am. Soc. Hortic. Sci.95, 13–14; https://doi.org/10.21273/JASHS.95.1.13 (1970).
Shu, J. et al. Detection of the diversity of cytoplasmic male sterility sources in broccoli (Brassica oleracea var. italica) using mitochondrial markers. Front. Plant Sci. 7, 927; https://doi.org/10.3389/fpls.2016.00927 (2016).
Fang, Z. et al. A male sterile line with dominant gene (Ms) in cabbage (Brassica oleracea var. capitata) and its utilization for hybrid seed production. Euphytica. 97(3), 265–268; https://doi.org/10.1023/A:1003026523150 (1997).
Bannerot, H. L., Boulidard, L., Cauderon, Y. & Tempe, J. Transfer of cytoplasmic male sterility from Raphanus sativus to Brassica oleracea. Proc. Eucarpia Meeting – Cruciferae, 52–54 (1974)
Walters, T. W., Mutschler, M. A. & Earle, E. D. Protoplast fusion-derived Ogura male sterile cauliflower with cold tolerance. Plant Cell Rep. 10, 624–628 (1992).
Cardi, T. & Earle, E. D. Production of new CMS Brassica oleracea by transfer of ‘Anand’ cytoplasm from B. rapa through protoplast fusion. Theor. Appl. Genet. 94, 204–212; https://doi.org/10.1007/s001220050401 (1997).
Singh, S. et al. Seventy-five years of research and development in cauliflower and cabbage: A journey from temperate to tropicalization and aristocrats to commoners. Int J Innov Hortic 11(2), 184–197 (2022).
Dey, S.S. et al. Molecular-agronomic characterization and genetic study reveals usefulness of refined Ogura cytoplasm-based CMS lines in hybrid breeding of cauliflower (Brassica oleracea var. botrytis L.). Sci. Hortic. 224, 27–36; https://doi.org/10.1016/j.scienta.2017.05.032 (2017).
Singh, S. et al. Development of Ogura CMS lines in Indian cauliflower and their characterization using agro-morphological traits and mtDNA markers. Sci. Hortic. 291, 110589 (2022).
Verma, V., K. & Kalia, P. Combining ability studies in early and mid-maturity CMS based cauliflower lines. Ind. J. Hort. 68, 503–506 (2011).
Dey, S. S., Dey, R. B., Parkash, C. & Kumar, R. Heterosis and combining ability analysis in snowball cauliflower using indigenously developed CMS lines. Ind. J. Hort. 74(3), 374–381 (2017).
Dey, S.S. et al. Development and characterization of “Ogura” based improved CMS lines of cauliflower (Brassica oleracea var. botrytis L.). Indian J. Genet. Plant Breed. 7, 37–42 (2011).
Cruden, R.W. & Hermann, S.M. Studying nectar? Some observations on the art. The Biology of Nectaries. 223–241 (1983).
Murray, M. G. & Thompson, W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8(19), 4321–4325. https://doi.org/10.1093/nar/8.19.4321 (1980).
Tingdong, F., Guangsheng, Y. & Xiaoniu, Y. Studies on “three line” Polima cytoplasmic male sterility developed in Brassica napus L. Plant Breed. 104(2), 115–120. https://doi.org/10.1111/j.1439-0523.1990.tb00412.x (1990).
Chengkun, H. et al. Preliminary studies on the alloplasmic male sterile materials in cauliflower. Acta Hortic Sin. 26, 125–127 (1999).
Sharma, S.R. & Vinod. Breeding for cytoplasmic male sterility in broccoli (Brassica oleracea. Var. italica Plenck). Indian J. Genet. Plant Breed. 62, 165–166 (2002).
Hu, J. et al. Mitochondria and cytoplasmic male sterility in plants. Mitochondrion.19(B), 282–288; https://doi.org/10.1016/j.mito.2014.02.008 (2014).
Singh, S. et al. Characterization and genetic analysis of OguCMS and doubled haploid based large genetic arsenal of Indian cauliflowers (Brassica oleracea var. botrytis L.) for morphological, reproductive and seed yield traits revealed their breeding potential. Genet. Resour. Crop Evol. 68(4), 1603–1623; https://doi.org/10.1007/s10722-020-01090-4 (2021).
Masierowska, M. & Pietka, T. Variability in nectar and pollen production in flowers of double-row lines of white mustard (Sinapsis alba L.) and their attractiveness to honey bees. Acta Sci. Pol. Hortorum Cultus. 13, 197–209 (2014).
Bertazzini, M. & Forlani, G. Intraspecific variability of floral nectar volume and composition in rapeseed (Brassica napus L. var. oleifera). Front. Plant. Sci.7, 288; https://doi.org/10.3389/fpls.2016.00288 (2016).
Katzer, A. M., Wessinger, C. A. & Hileman, L. C. Nectary size is a pollination syndrome trait in Penstemon. New Phytol. 223(1), 377–384. https://doi.org/10.1111/nph.15769 (2019).
Ding, X. et al. Metabolomics studies on cytoplasmic male sterility during flower bud development in soybean. Int. J. Mol. Sci. 20(12), 2869 (2019).
Kalia, P. & Singh, S. Innovations in varietal development and seed production technologies for effective value chain management. Int. J. Innov. Hort. 8(2), 108–114 (2019).
Dey, S.S. et al. Effect of chloroplast substituted Ogura male sterile cytoplasm on the performance of cauliflower (Brassica oleracea var. botrytis L.). Sci. Hortic. 157, 45–51 ;https://doi.org/10.1016/j.scienta.2013.04.008 (2013).
Kucera, V., Chytilova, V., Vyvadilova, M., & Klima, M. Hybrid breeding of cauliflower using self-incompatibility and cytoplasmic male sterility. Zahradnictvi (Hortic Sci). 33(4), 148–152 (2006).
Wang, Q. et al. Chloroplast and mitochondrial SSR help to distinguish allo-cytoplasmic male sterile types in cabbage (Brassica oleracea L. var. capitata). Mol Breed. 30(2), 709–716; https://doi.org/10.1007/s11032-011-9656-9 (2012).
Zhang, R. J., Hu, S. W., Yan, J. Q. & Sun, G. L. Cytoplasmic diversity in Brassica rapa L. investigated by mitochondrial markers. Genet. Resour. Crop Evol. 60(3), 967–974; https://doi.org/10.1007/s10722-012-9892-9 (2013).
Wang, C. G. & Song, W. Q. Identification of PCR markers associated with cytoplasmic male sterility in Brassica oleracea var. botrytis. Yi Chuan. 27(2), 236–240 (2005).
Acknowledgements
First author thanks ICAR for providing him with a fellowship for his post-graduate study. The senior author thanks ICAR for financial support through CRPHT-Cauliflower (CRPHT-142-12-F) as well as the Director, ICAR-IARI for the experimental facilities and support. We thank Dr. S. R. Bhat, ICAR-National Institute of Plant Biotechnology, New Delhi for sharing BC4 seeds of the Can sterile cytoplasm deployed CMS line (i.e. NIPB-1) in 2015-16.
Author information
Authors and Affiliations
Contributions
MKS performed experiments, wrote draft; SS- conceptualized, generated CMS lines, planned experiments, arranged resources, wrote the manuscript draft, PK- Procurred Can cytoplasm introgressed cauliflower line 'NIPB-1' from ICAR-NIPB, New Delhi, initiated introgression till BC1, reviewed the draft manuscript, MM- assisted in the analysis of genomic sequences, BB- reviewed draft, NS-reviewed draft and interpretation of results, MMR- statistical analysis, MR- manuscript draft review and interpretation of results for Can cytoplasm, BST- designed seed trials, interpretation of data from seed traits.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Manjunath, K.S., Singh, S., Kalia, P. et al. Commercial suitability and characterization of newly developed Erucastrum canariense (Can) sterile cytoplasm based cytoplasmic male sterile (CMS) lines in Indian cauliflower. Sci Rep 14, 2346 (2024). https://doi.org/10.1038/s41598-024-52714-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-52714-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.