Abstract
Gastrointestinal stromal tumors (GISTs) are typically characterized by activating mutations of the KIT proto-oncogene receptor tyrosine kinase (KIT) or platelet-derived growth factor receptor alpha (PDGFRA). Recently, the neurotrophic tyrosine receptor kinase (NTRK) fusion was reported in a small subset of wild-type GIST. We examined trk IHC and NTRK gene expressions in GIST. Pan-trk immunohistochemistry (IHC) was positive in 25 (all 16 duodenal and 9 out of 16 small intestinal GISTs) of 139 cases, and all pan-trk positive cases showed diffuse and strong expression of c-kit. Interestingly, all of these cases showed only trkB but not trkA/trkC expression. Cap analysis of gene expression (CAGE) analysis identified increased number of genes whose promoters were activated in pan-trk/trkB positive GISTs. Imbalanced expression of NTRK2, which suggests the presence of NTRK2 fusion, was not observed in any of trkB positive GISTs, despite higher mRNA expression. TrkB expression was found in duodenal GISTs and more than half of small intestinal GISTs, and this subset of cases showed poor prognosis. However, there was not clear difference in clinical outcomes according to the trkB expression status in small intestinal GISTs. These findings may provide a possible hypothesis for trkB overexpression contributing to the tumorigenesis and aggressive clinical outcome in GISTs of duodenal origin.
Similar content being viewed by others
Introduction
Gastrointestinal stromal tumor (GIST) is the most common soft tissue sarcoma of the digestive tract, with a worldwide prevalence of 10–15 cases per million1,2,3. The median age at diagnosis is approximately 60 years, and cases are approximately evenly distributed between sexes1. GIST has been reported to originate from the interstitial cells of Cajal. As intermediates between the autonomic nervous system and smooth muscle cells in the gastrointestinal tract, these cells are involved in the regulation of motility and autonomic function4,5. GISTs arise predominantly in the stomach (55.6%) and small intestine (31.8%), with the rectum (6%), esophagus (0.7%), and various other locations (5.5%) accounting for the remaining cases1.
Diagnosis is based on histological features in addition to the clinical course and tumor location. GISTs also have distinct molecular characteristics, such as immunohistochemical expression of DOG-16. Most cases are characterized by activating mutations of the KIT proto-oncogene receptor tyrosine kinase (KIT, 75–80%) or platelet-derived growth factor receptor alpha (PDGFRA, 10%)3.
Surgery is the initial treatment for primary and localized cases, and drug therapy is the second-line treatment for more advanced cases7. Imatinib is a selective tyrosine kinase inhibitor (TKI) and contributes to improving the prognosis of advanced GISTs8,9. However, due to the poor efficacy of imatinib in some cases involving PDGFRA mutations and KIT and PDGFRA wild-type GISTs, it is necessary to develop new targets for therapy3. Recently, the existence of the neurotrophic tyrosine receptor kinase (NTRK) fusion gene has been reported in a small subset of wild type GISTs, suggesting the effectiveness of a new TKI in this tumor10,11,12. However, there are no other reports, and controversy remains13. In this study, we analyzed NTRK expression and its clinical significance in GISTs.
Materials and methods
Case selection
We examined 139 cases of GIST. Prognostic information was collected from files in the Department of Human Pathology, Juntendo University Hospital, Tokyo, Japan. All patients were treated at the Juntendo University Hospital between 2008 and 2020. These cases were diagnosed using the WHO classification system for soft-tissue tumors and classified using the modified Fletcher classification14. Diagnoses were confirmed by immunohistochemical analysis of DOG1 and c-kit expression. Clinicopathologic data of the 139 patients are shown in Table 1. The follow-up periods ranged from 0.1 to 182 months (mean: 61.3 months). Patients were treated with surgical resection without a pre-adjuvant treatment, such as that with imatinib. In 137 cases, the tumors were completely resected. Tissue microarray (TMA) blocks, each consisting of 2 mm cores, were made for these cases. All experiments were performed in accordance with relevant guidelines and regulations of the institution and the Declaration of Helsinki.
Immunohistochemistry (IHC)
IHC staining was performed for all cases using the antibodies described in Supplementary Table 1. The expression of tropomyosin receptor kinase (TRK) A, trk B, and trk C encoded by either NTRK1, NTRK2 or NTRK3 was examined by IHC.
RNA extraction
For NanoString analysis, RNA was extracted from formalin-fixed paraffin-embedded (FFPE) samples using the RNeasy FFPE Kit (QIAGEN, Hilden, Germany). For CAGE analysis, RNA was extracted from fresh-frozen samples using the RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany). RNA concentration was measured using Nanodrop2000 (Thermo Fisher Scientific, Inc. MA).
DNA extraction
DNA was extracted from tumoral and corresponding non-tumoral tissues using the QIAamp DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocols. Samples were treated with RNase A accordingly, and DNA concentration was measured using Nanodrop2000.
Nanostring-based mRNA imbalance analysis
Nanostring (NanoString Technologies, Inc., Seattle, WA, USA) analysis was performed (probe set described in Supplementary Table 2) to target a total of 32 genes as previously described15. This assay can estimate the formation of fusion genes by comparing the mRNA expression levels at the 5ʹ-side and 3ʹ-side. Briefly, 250–400 ng of ribonucleic acid (RNA) was hybridized to the probes (a reporter probe and a capture probe) at 65 °C for 18–24 h using a thermal cycler. Samples were added into the nCounter Prep Station for 3 h to remove excess probes, purify, and immobilize the sample on the internal surface of the cartridge. Finally, the sample cartridge was transferred to the nCounter Digital Analyzer, where color codes were counted and tabulated for each target molecule. The expression number for the base sequence of the probe part was analyzed using nSolver Analysis Software Version 4.0 (https://www.nanostring.com/products/analysis-software/nsolver).
NanoString-based copy number variation (CNV) analysis
NanoString-based CNV analysis was performed using nCounter (NanoString Technologies, Seattle, WA, USA) as previously described16. The probe list used in this assay is described in Supplementary Table 3. Three probes were prepared for each of the total 24 genes. According to the sample status, 150–300 ng of DNA was processed for the NanoString nCounter CNV analysis according to the manufacturer’s protocol (NanoString Technologies, Seattle, WA, USA). The CNV for the base sequence of the probe part was analyzed using nSolver Analysis Software Version 4.0. For GIST clinical samples, each data point was normalized by dividing each score from the tumoral DNA by the score from the corresponding non-tumoral DNA. The mean of the three normalized scores for each gene was then calculated. The cut-off for amplification was defined as 2.0.
Quantitative polymerase chain reaction for trk ligands
Each trk has specific ligands. Various neurotrophins (NT), including NT-4 (NT-5), NT-3, nerve growth factor (NGF), and brain-derived neurotrophic factor (BDNF) have been reported as ligands. BDNF, NT-4 (NT-5), and NT-3 are known to bind to trkB and activate downstream trkB signaling17,18. We examined the expression of ligands BDNF, NT-4, and NT-3 using qPCR. All quantitative real-time PCR (qPCR) was performed with TaqMan Fast Advanced Master Mix (Applied Biosystems) on an Applied Biosystems Step One Plus Real Time PCR System in accordance with standard protocols. qPCR was performed using predeveloped TaqMan assays (20× Primer Probe mix; Applied Biosystems, CA, USA) for BDNF (Assay ID Hs02718934_s1), NT3 (Assay ID Hs00267375_s1), NT4 (Assay ID Hs01921834_s1), and GAPDH (Assay ID Hs02786624_g1). The amount of each target gene relative to the GAPDH housekeeping gene was determined using the comparative threshold cycle (Ct) method.
CAGE analysis protocol
We analyzed the promoter activity profiles in 10 GISTs, using the CAGE protocol. The 10 cases consisted of eight gastric, one duodenal, and one small intestinal GISTs, with the duodenal and small intestinal GIST being trkB-positive. CAGE libraries were prepared and sequenced in K.K.DNAFORM. The reads were mapped to the reference genome (GRCh38) by STAR v2.7.1019. The aligned reads were counted on regions of GENCODE transcription start sites ± 300 base pairs (GENCODE v41). Count data were normalized as counts per million by edgeR v3.34.0 and the subsequent analysis was performed by R v4.1.0.
KIT mutational analysis for GISTs with pan-trk/trkB expression
Information of genotype of GIST cases were obtained from each medical record where available. KIT mutational analysis was performed for remaining GIST cases with pan-trk/trkB expression as described previously20. Several cases were excluded from this analysis due to the short of materials or inadequate sample quality.
Statistical analysis
Categorical variables were analyzed using Fisher’s exact or chi-square test. Column variable was analyzed using the Mann–Whitney test. To determine prognosis, Kaplan–Meier survival analysis was performed. The date of surgical resection was set as the starting point and the date of death, date of recurrence, or last date of follow-up was used as the end point. Statistical analyses were performed using GraphPad Prism® software version 9.4.0 (GraphPad, San Diego, CA, USA). p value of < 0.05 was considered statistically significant.
Ethical standards
This study was reviewed and approved by the Juntendo University School of Medicine Institutional Review Board (#21-079). The informed consents were obtained from all subjects and/or their legal guardian(s).
Results
Clinicopathological analysis
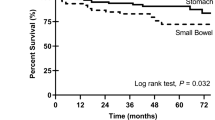
Clinicopathologic characteristics of the 139 patients are summarized in Table 1. Briefly, there were 3 esophageal GISTs (2%), 103 gastric GISTs (74%), 16 duodenal GISTs (11.5%), 16 small intestinal GISTs (11.5%), and 1 vaginal case. Seventeen cases (12%) were classified as very low risk, 58 cases (42%) as low risk, 29 cases (21%) as intermediate risk, and 35 cases (25%) as high risk. Primary site (p < 0.0001), larger tumor size (p = 0.0153), higher mitotic figure (p < 0.0001), higher modified Fletcher classification (0 < 0.0001) and higher MIB-1 index (p < 0.0001) were significantly associated with poor prognosis. Interestingly, the duodenal GIST showed poor prognosis (p < 0.0001) than other GISTs (Fig. 1). Age and sex were not associated with prognosis.
Pan-trk expression in GIST
TMA-based pan-trk IHC identified positively staining in 25 out of 139 GIST cases (Fig. 2A, Table 2). Pan-trk IHC showed diffuse cytoplasmic and membranous staining (Fig. 2A). Furthermore, all pan-trk IHC positive GISTs showed only trkB expression encoded by NTRK2, however, trkA and trkC expression was not observed (Fig. 2B–D).
Clinicopathological characteristics of pan-trk positive GISTs
Clinicopathological characteristics of pan-trk positive GISTs are summarized in Table 2. Of the primary sites for tumors with pan-trk expression, 16 were duodenal and nine were small intestinal. Interestingly, all of the duodenal cases and more than half of the small intestinal cases (at least seven cases were of jejunal origin) showed pan-trk positive staining (p < 0.0001, Table 2). In addition, all pan-trk positive cases showed diffuse and strong expression of c-kit and harbored KIT mutations where information was available. None of pan-trk-positive cases had clinical signs of type 1 neurofibromatosis. By risk classification, 13 out of 16 duodenal GISTs showed very low- or low-risk groups, and recurrence was observed in one case each from low- and high-risk groups. On the other hand, seven out of nine small intestinal GISTs with pan-trk expression were high-risk, and the remaining two cases were discovered incidentally on histological examination of other surgically resected malignant tumors. No recurrence was observed for the small intestinal GISTs with pan-trk expression, despite of their High-grade natures (Fig. 3). Pan-trk IHC-positive GISTs tended to have a poor prognosis, although there was no statistically significant difference between pan-trk IHC-positive GISTs and IHC-negative GISTs (Fig. 4A). None of the patients with pan-trk positive GIST experienced recurrence. In pan-trk IHC-positive GISTs, primary site, age, sex, modified Fletcher classification, and size were not significantly associated with poor prognosis (Fig. 4B), whereas mitotic figure (p < 0.0001) and MIB-1 index (p < 0.0001) were significantly associated with poor prognosis (Fig. 4B).
Risk classification of trkB positive GISTs. In duodenal GISTs, 13 out of 16 cases are classified as either very low- or low-risk by modified Fletcher classification (A). In small intestinal GISTs, at least 7 out of 12 jejunal GISTs show trkB expression (B). In contrast, 7 out of 9 small intestinal GISTs with trkB expression are classified as high-grade (C).
Disease-free survival rate is inferior in pan-trk/trkB positive-GISTs compared with that in pan-trk/trkB negative-cases, although this was not statistically significant (A, left). This trend was not clear in small intestinal cases (A, right). Survival analysis in pan-trk/trkB positive-GIST reveals that disease-free survival was affected only by mitotic activity and MIB-1 LI (B).
NanoString assay for the pan-trk IHC positive GISTs
Next, to examine whether trkB expression reflected the presence of NTRK2 fusion, we performed NanoString imbalance assay for 23 out of 25 cases with trkB expression in 30 tyrosine kinase genes including NTRK1-315,21,22. Two cases were not available for adequate tissue for this analysis. Imbalanced expression of NTRK1-3 was not observed in any of the analyzed GISTs (Supplementary Fig. 1, Supplementary Table 4). Interestingly, NanoString-based mRNA imbalance analysis revealed constantly high expression of KIT in all the cases with trkB expression, in line with the c-kit IHC findings. High expression of NTRK2 was observed in all the cases with trkB expression, but not for NTRK1 and NTRK3 (Supplementary Table 5, Fig. 5). NanoString-based CNV analysis revealed that one case (Case#76) showed NTRK1 amplification (× 2.3; 4.6 copies), but no case showed NTRK2 amplification (Supplementary Table 6). These findings suggested that high expression of NTRK2 in GIST with trkB expression could be due to transcriptional activation of NTRK2.
Ligand expression of NTRK2
There was no difference in the expression of examined ligands between trkB positive GISTs and trkB negative GISTs. Furthermore, NT-4, the major NTRK2 ligand, was not expressed even in trkB positive GISTs (Supplementary Fig. 2).
Profiles of genome-wide promoter activities in GIST
We analyzed the promoter activity profiles in eight gastric, one duodenal, and one small intestinal GISTs. This analysis clustered 10 GISTs into three groups according to Pfetin and pan-trk/trkB IHC status (Fig. 6A,B). Pfetin, found to be expressed in approximately 80% of the GISTs, is reported to be a prognostic factor in GIST, and Pfetin negative cases show a poor prognosis23. GIST#104 and #122 were of duodenal and small intestinal origin and were positive for pan-trk/trkB and pfetin. GIST#95 and #107 were of gastric origin and were characterized by negative staining for Pfetin/pan-trk/trkB. GIST#100, #105, #106, #108, #109 and #112 were of primary gastric GISTs and were positive for Pfetin and negative for pan-trk/trkB.
MDS plot and cluster dendrogram reveals that GIST samples with trkB expression (Cases#104, #122) were separately clustered (A,B). Furthermore, GIST samples without trkB expression are subdivided according to pfetin expression (A,B). Cases#95 and #107 are pfetin-negative cases. GISTs are clearly separated by differentially expressed genes according to trkB expression. The number of genes with promoter activation is higher in GIST samples with trkB expression than in those without (C).
The heatmap with 208 differentially expressed promoter (FDR < 0.05) showed different promoter activation patterns between pan-trk positive and negative GISTs (Fig. 6C). Interestingly, mRNA level of trkB ligands by qPCR analysis showed almost the same trend when they were examined in detail using RNA extracted from FFPE samples, although mRNA expression level of NTRK by Nanostring analysis was significantly higher in pan-trk/trkB positive group (Fig. 5, Supplementary Fig. 2) Furthermore, CNV analysis did not reveal amplification of NTRK2 in any of GIST cases (Supplementary Table 6). The promoter activity of NTRK2 was higher in pan-trk/trkB positive group, compared to pan-trk negative group, although this difference was not statistically significant (FDR = 0.15, p = 0.0067; Not shown). On the other hand, regardless of pan-trk IHC results, there was no difference in the promoter activity of KIT (Not shown) as well as KIT mRNA expression (Fig. 5). In the promoter activity profiles, as with mRNA expression, there was no difference in the ligand expressions between trkB positive GISTs and trkB negative GISTs (Supplementary Fig. 2). Interestingly, vascular developments including VEGFA-VEGFR2 signaling pathway appeared as one of the differentially activated pathways in pan-trk/trkB positive group by CAGE analysis (Supplementary Fig. 3).
Discussion
The NTRK gene encodes the neurotrophic-tropomyosin receptor tyrosine kinase, and NTRK1, NTRK2, NTRK3 encode trkA, trkB, and trkC proteins, respectively. NTRKs are involved in the survival and proliferation of nerve cells24,25. Several molecules have been reported as trk ligands, including NGF, BDNF, NT-3 and NT-4 (NT-5). When bound, the ligands directly activate downstream effectors of trks17. In oncology, it has been reported that this induces tumorigenesis, including differentiation, growth, and apoptosis, thereby showing potential as therapeutic targets in malignant tumors across systemic organs26,27. NTRK-fusions are known to be the most effective therapeutic targets among tyrosine kinase fusions. Selective TRK inhibitors show high antitumor effects on tumors carrying the NTRK fusion28. Although NTRK fusion is generally quite rare, being detected in less than 1% of cancers, it is reported to occur frequently in a small subset of cancers such as secretory carcinoma of the parotid gland and infantile fibrosarcoma17. Furthermore, the significance of NTRK amplification or trk overexpression on cancer progression remains unclear. Recently, overexpression of NTRK1 is shown in 20% of breast cancers, and its involvement in tumorigenesis and susceptibility to selective TRK inhibitors is reported29. Thus, it is increasingly important to find NTRK/trk alterations other than NTRK fusions.
The relationship between GIST and NTRK fusion/expression remains controversial, with few reports still scattered. Wang et al. reported that the expression of the NTRK-like family member 3 (SLITRK3), a member of the Slitrk family of structurally related transmembrane proteins that are involved in controlling neurite outgrowth, is associated with malignancy including recurrence and metastasis of GISTs30. Recently, ETV6-NTRK3 fusion has been reported within a subset of wild-type GISTs10,11. Furthermore, Shi, E et al. report the efficacy of TRK inhibitors for this type of GISTs11. However, NTRK fusion genes are almost always restricted to gastrointestinal mesenchymal tumors other than GISTs, characterized by the lack of DOG1, with c-kit and NTRK fusion tumors being distinct13. In this study, duodenal and some small intestinal GISTs showed trkB expression encoded by NTRK2, at both the mRNA and protein levels. However, Nanostring analysis did not show imbalanced expression of NTRK2 in any of these trkB positive GISTs, suggesting the absence of NTRK2 fusion in these tumors. This finding is consistent with strong c-kit expression and KIT mutations in these GISTs (Supplementary Table 5), since the reported NTRK fusion in GISTs is restricted to wild-type cases10,11.
TrkB is involved in formation and maintenance within the nervous system and construction of normal lung tissue31,32. The association between neuroblastoma and trkB expression is well known, and its expression is observed in about 30% of these cases33. Furthermore, it has been shown that trkB contributes to the growth and differentiation of neuroblastoma cells, suggesting a relationship to a poor prognosis33,34. In addition, high expression of trkB has been reported as a poor prognostic factor in cancers of the digestive system, ovaries, prostate, and lungs33,35.
In GISTs, tumor size, mitotic rate and tumor location have been reported to be associated with recurrence36. Additionally, duodenal GISTs should be considered for aggressive treatment because of their poor prognosis compared with those of other primary sites37. In this study, all of the duodenal GISTs showed overexpression of trkB. Molecular genetic characteristics that associate aggressive behavior in duodenal GISTs are still unknown. Our findings may provide a possible hypothesis that trkB overexpression contributes to the tumorigenesis or aggressive clinical outcome in GISTs of duodenal origin. In contrast, more than half of the GISTs of small intestinal origin also showed overexpression of trkB, however, small intestinal GISTs with trkB overexpression did not show clinical disadvantage compared to those without. This point needs to be further evaluated in the future.
CAGE and subsequent MDS plots, cluster dendrograms and heatmaps analysis revealed that GIST samples with trkB expression were separately clustered from those without (Fig. 6A,B). Furthermore, GIST samples without trkB expression were subdivided according to pfetin expression (Fig. 6A,B). Pfetin has been shown to be expressed in approximately 80% of GISTs and to be a favorable prognostic factor in GIST23. In addition, our analysis also showed significant differences in gene promoter activity according to pan-trk/trkB status. The number of genes with promoter activation was higher in GIST samples with trkB expression than in those without (Fig. 6C). These findings suggest that the GISTs with trkB expression may have different characteristics from those without. On the other hand, although the presence of ligand is usually required for the activation of the downstream pathways of trk17,38, in this study we could not find any difference in the expression of trkB ligands between pan-trk positive and negative GISTs. A recent study demonstrated that overexpression of trkA in breast carcinoma cells led to growth factor-independent proliferation29. Regarding this point, we found activation of VEGFA/VEGFR2 signaling pathway in pan-trk/trkB positive GIST by CAGE analysis. Interestingly, microvessel density and vascular endothelial growth factor expression have been shown as adverse prognostic factors39. However, trkB involvement in the aggressive behavior of duodenal and small intestinal GISTs needs to be verified in vitro and in vivo, and further analysis is required, together with the accumulation of clinical samples of duodenal and small intestinal GISTs with trkB expression to determine its contribution to aggressive clinical outcomes.
By Nanostring analysis, genetic analysis of NTRK2 in trkB positive GIST did not show amplification of NTRK2 as a mechanism of trkB overexpression. On the other hand, significantly higher mRNA expression levels of NTRK2 in trkB positive GISTs was observed when compared with those without trkB expression. These findings suggested possible transcriptional activation of NTRK2 in this subset of GISTs, although NTRK fusion seemed to be less likely present.
In summary, trkB expression was found in duodenal GISTs and more than half of small intestinal GISTs, and seemed to be associated with poor prognosis. These findings could provide a possible hypothesis for trkB overexpression contributing to the aggressive clinical outcome in GISTs of duodenal origin.
Data availability
The data of this study is available upon reasonable requests. Please contact to Tsuyoshi Saito (email: tysaitou@juntendo.ac.jp).
References
Søreide, K. et al. Global epidemiology of gastrointestinal stromal tumours (GIST): A systematic review of population-based cohort studies. Cancer Epidemiol. 40, 39–46 (2016).
Rubin, B. P., Heinrich, M. C. & Corless, C. L. Gastrointestinal stromal tumour. Lancet 369, 1731–1741 (2007).
Kelly, C. M., Gutierrez Sainz, L. & Chi, P. The management of metastatic GIST: Current standard and investigational therapeutics. J. Hematol. Oncol. 14, 2 (2021).
Hirota, S. et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 279, 577–580 (1998).
Miettinen, M. & Lasota, J. Gastrointestinal stromal tumors: Review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch. Pathol. Lab. Med. 130, 1466–1478 (2006).
Miettinen, M., Wang, Z. F. & Lasota, J. DOG1 antibody in the differential diagnosis of gastrointestinal stromal tumors: A study of 1840 cases. Am. J. Surg. Pathol. 33, 1401–1408 (2009).
Nishida, T., Blay, J. Y., Hirota, S., Kitagawa, Y. & Kang, Y. K. The standard diagnosis, treatment, and follow-up of gastrointestinal stromal tumors based on guidelines. Gastric Cancer 19, 3–14 (2016).
Demetri, G. D. et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 347, 472–480 (2002).
Heinrich, M. C. et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 21, 4342–4349 (2003).
Brenca, M. et al. Transcriptome sequencing identifies ETV6-NTRK3 as a gene fusion involved in GIST. J. Pathol. 238, 543–549 (2016).
Shi, E. et al. FGFR1 and NTRK3 actionable alterations in “Wild-Type” gastrointestinal stromal tumors. J. Transl. Med. 14, 339 (2016).
Castillon, M. et al. Fluorescent in situ hybridization must be preferred to pan-TRK immunohistochemistry to diagnose NTRK3-rearranged gastrointestinal stromal tumors (GIST). Appl. Immunohistochem. Mol. Morphol. 29, 626–634 (2021).
Atiq, M. A. et al. Mesenchymal tumors of the gastrointestinal tract with NTRK rearrangements: A clinicopathological, immunophenotypic, and molecular study of eight cases, emphasizing their distinction from gastrointestinal stromal tumor (GIST). Mod. Pathol. 34, 95–103 (2021).
Joensuu, H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum. Pathol. 39, 1411–1419 (2008).
Kurihara, T. et al. Nanostring-based screening for tyrosine kinase fusions in inflammatory myofibroblastic tumors. Sci. Rep. 10, 18724 (2020).
Sasa, K. et al. Establishment of rapid and accurate screening system for molecular target therapy of osteosarcoma. Technol. Cancer Res. Treat. 21, 15330338221138216 (2022).
Okamura, R. et al. Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: Implications for NTRK-targeted therapeutics. JCO Precis. Oncol. 2018, 1–20 (2018).
Cocco, E., Scaltriti, M. & Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 15, 731–747 (2018).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Yamamoto, H. et al. c-kit and PDGFRA mutations in extragastrointestinal stromal tumor (gastrointestinal stromal tumor of the soft tissue). Am. J. Surg. Pathol. 28(4), 479–488 (2004).
Suehara, Y. et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin. Cancer Res. 18, 6599–6608 (2012).
Suehara, Y. et al. Identification of a novel MAN1A1-ROS1 fusion gene through mRNA-based screening for tyrosine kinase gene aberrations in a patient with leiomyosarcoma. Clin. Orthop. Relat. Res. 479, 838–852 (2021).
Suehara, Y. et al. Pfetin as a prognostic biomarker of gastrointestinal stromal tumors revealed by proteomics. Clin. Cancer Res. 14, 1707–1717 (2008).
Klein, R., Jing, S. Q., Nanduri, V., O’Rourke, E. & Barbacid, M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell 65, 189–197 (1991).
Kawamura, K. et al. Brain-derived neurotrophic factor promotes implantation and subsequent placental development by stimulating trophoblast cell growth and survival. Endocrinology 150, 3774–3782 (2009).
Nakagawara, A. Trk receptor tyrosine kinases: A bridge between cancer and neural development. Cancer Lett. 169, 107–114 (2001).
Khotskaya, Y. B. et al. Targeting TRK family proteins in cancer. Pharmacol. Ther. 173, 58–66 (2017).
Drilon, A. et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl. J. Med. 378, 731–739 (2018).
Kyker-Snowman, K. et al. TrkA overexpression in non-tumorigenic human breast cell lines confers oncogenic and metastatic properties. Breast Cancer Res. Treat. 179, 631–642 (2020).
Wang, C. J. et al. SLITRK3 expression correlation to gastrointestinal stromal tumor risk rating and prognosis. World J. Gastroenterol. 21, 8398–8407 (2015).
Soppet, D. et al. The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the trkB tyrosine kinase receptor. Cell 65, 895–903 (1991).
García-Suárez, O. et al. TrkB is necessary for the normal development of the lung. Respir. Physiol. Neurobiol. 167, 281–291 (2009).
Desmet, C. J. & Peeper, D. S. The neurotrophic receptor TrkB: A drug target in anti-cancer therapy? Cell Mol. Life Sci. 63, 755–759 (2006).
Nakagawara, A., Azar, C. G., Scavarda, N. J. & Brodeur, G. M. Expression and function of TRK-B and BDNF in human neuroblastomas. Mol. Cell Biol. 14, 759–767 (1994).
Sinkevicius, K. W. et al. Neurotrophin receptor TrkB promotes lung adenocarcinoma metastasis. Proc. Natl. Acad. Sci. U.S.A. 111, 10299–10304 (2014).
Dematteo, R. P. et al. Tumor mitotic rate, size, and location independently predict recurrence after resection of primary gastrointestinal stromal tumor (GIST). Cancer 112, 608–615 (2008).
Han, I. W. et al. Clinicopathologic analysis of gastrointestinal stromal tumors in duodenum and small intestine. World J. Surg. 39, 1026–1033 (2015).
Thiele, C. J., Li, Z. & McKee, A. E. On Trk—The TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin. Cancer Res. 15, 5962–5967 (2009).
Imamura, M. et al. Prognostic significance of angiogenesis in gastrointestinal stromal tumor. Mod. Pathol. 20, 529–537 (2007).
Acknowledgements
This work was carried out in part at the Intractable Disease Research Center, Juntendo University and K.K.DNAFORM.
Funding
This study was supported by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) KAKENHI (JSPS: Grant Numbers 19K16753 to K.A, 18K15329 to T.O, 19H03789 and 19K22694 to Y.S., 20K07415 and 17K08730 to T.S.).
Author information
Authors and Affiliations
Contributions
K.S.: Investigation, Formal analysis, Writing—Original Draft, R.S.: Formal analysis, Writing—Review & Editing, A.O.: Formal analysis, Writing—Review & Editing, K.A.: Mutational analysis for KIT, N.H.: Writing—Review & Editing, D.K.: Writing—Review & Editing, Y.S.: Funding acquisition, Writing—Review & Editing, T.T.: Writing—Review & Editing, T.O.: Funding acquisition, Writing—Review & Editing, K.A.: Funding acquisition, Writing—Review & Editing, K.S.: Resources, Writing—Review & Editing, M.T.: Resources, Writing—Review & Editing, K.S.: Resources, Writing—Review & Editing, T.H.: Resources, Writing—Review & Editing, S.M.: Resources, Writing—Review & Editing, T.F.: Resources, Writing—Review & Editing, M.I.: Writing—Review & Editing, T.H.: Writing—Review & Editing, T.Y.: Writing—Review & Editing, Y.M.: Formal analysis, Writing—Review & Editing, T.S.: Validation, Writing—Review & Editing, Supervision, Project administration. All authors had approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sasa, K., Son, R., Oguchi, A. et al. NTRK2 expression in gastrointestinal stromal tumors with a special emphasis on the clinicopathological and prognostic impacts. Sci Rep 14, 768 (2024). https://doi.org/10.1038/s41598-024-51211-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-51211-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.