Abstract
Compelling evidence shows that the frequency of T cells in the tumor microenvironment correlates with prognosis as well as response to immunotherapy. However, considerable heterogeneity exists within tumor-infiltrating T cells, and significance of their genomic and transcriptomic landscape on clinical outcomes remains to be elucidated. Signaling lymphocyte activation molecule 6 (SLAMF6) is expressed on intra-tumoral progenitor-exhausted T cells, which exhibit the capacity to proliferate, self-renew and produce terminally-exhausted T cells in pre-clinical models and patients. Here, we investigated the impact of SLAMF6 expression on prognosis in two immunologically different tumor types using publicly available databases. Our findings demonstrate that high SLAMF6 expression is associated with better prognosis, expression of TCF7 (encoding T-cell factor 1), and increased gene signatures associated with conventional type 1 dendritic cells and effector function of T cells in melanoma and breast cancer. Single-cell profiling of breast cancer tumor microenvironment reveals SLAMF6 expression overlaps CD8 T cells with a T-effector signature, which includes subsets expressing TCF7, memory and effector-related genes, analogous to progenitor-exhausted T cells. These findings illustrate the significance of SLAMF6 in the tumor as a marker for better effector responses, and provide insights into the predictive and prognostic determinants for cancer patients.
Similar content being viewed by others
Introduction
Type, density and location of immune cell populations play a critical role in prognosis of various solid malignancies1. Particularly, high CD8+ T-cell density is one of the most commonly recognized predictive factors of prognosis as well as response to anti-cancer therapy such as chemotherapy2 and immune checkpoint inhibitor (ICI) therapy3. As ICI therapy becomes the standard-of-care treatment for various cancers; however, we have begun to understand that not only the quantity but also the quality of CD8+ tumor-infiltrating lymphocytes (TILs) is critical predictive and prognostic determents. Indeed, whereas PD-1 expression is the hallmark of ‘exhaustion’, a state of dysfunction such as decreased proliferative capacity and effector function4, studies have revealed a remarkable degree of heterogeneity with distinct predictive value amongst PD-1+ CD8+ TILs5,6. Of these, two phenotypically and functionally distinct subsets of exhausted T cells have been identified in pre-clinical and human tumors: ‘progenitor-exhausted’ T cells and ‘terminally-exhausted’ T cells. It has been demonstrated that progenitor-exhausted T cells exhibit stem cell-like properties with the capacity to proliferate, self-renew, and produce terminally-exhausted T cells during ICI treatment7. Accordingly, the presence of progenitor-exhausted T cells, but not terminally-exhausted T cells, in the tumor microenvironment (TME) is essential to the favorable response to ICIs7,8,9,10.
Progenitor-exhausted T cells are characterized by the expression of T-cell factor 1 (TCF1, encoded by TCF7) and an intermediate level of PD-1 expression, whereas terminally-exhausted T cells do not express TCF1 but harbor high expression of PD-110. TCF1 was found to be expressed by progenitor-exhausted T cells; however, it is also expressed in naïve T cells11,12. Hence, it would be difficult to distinguish progenitor-exhausted T cells and other subsets of T cells only with the use of expression level of TCF1 and PD-1.
Signaling lymphocyte activation molecule 6 (SLAMF6, encoded by Slamf6 in mice, SLAMF6 in humans) is a homophilic receptor belonging to the superfamily immunoglobulin (Ig) domain-containing molecules, expressed on hematopoietic cells including T, natural killer (NK), and B cells13,14 and has emerged as a potential marker for progenitor-exhausted T cells7,8,15,16,17. In preclinical models of melanoma, Slamf6 was identified as a cell-surface marker that distinguished progenitor-exhausted CD8+ T cells from terminally exhausted CD8+ T cells, and CD8+ T cells co-expressing TCF1 and SLAMF6 retained polyfunctionality, produced IFN-γ, TNF, and/or IL-2 and persisted long term upon adoptive transfer, thereby contributing to long-term tumor control compared to terminally-exhausted T cells8. Furthermore, our recent work employing single cell (sc) RNA-seq analysis of the TME of murine breast cancer treated with effective multimodal intralesional therapy revealed that Slamf6 was not expressed on naive or terminally-exhausted T cells16, suggesting that Slamf6 might be a valuable marker to predict response to immunotherapy. However, to date, there has been no study investigating the impact of intra-tumoral SLAMF6 expression on immunological status of the tumor and clinical outcome.
Here, we hypothesize that high expression of SLAMF6 in the tumor correlates with higher immune activities and better clinical outcome. We investigated the association between expression of SLAMF6 and immunological status in the tumor and survival in the cohort of breast cancer and melanoma. Using the publicly available data of The Cancer Genome Atlas (TCGA) and single-cell (sc) profiling of breast cancer (GSE161529), our data demonstrate that high expression of SLAMF6 in the tumor correlates with better patient survival and elevated immune activity in breast cancer and melanoma.
Results
High SLAMF6 expression is associated with longer progression free interval and overall survival than low SLAMF6 expression in breast cancer and melanoma
First, we evaluated the prognostic relevance of SLAMF6 expression on patient outcomes using TCGA database. We analyzed the progression free interval (PFI) and overall survival (OS) of breast cancer and melanoma patients according to the expression level of SLAMF6. In the breast cancer cohort, PFI and OS was significantly longer in the high SLAMF6 group than in the low SLAMF6 group (PFI; p = 0.0011, OS; p = 0.0019) (Fig. 1A). In patients with primary melanoma, similarly to the data in breast cancer, the high SLAMF6 group showed significantly longer PFI and OS than the low SLAMF6 group in (PFI; p = 0.00055, OS; p = 0.0032) (Fig. 1B). As for metastatic melanoma, there was a trend that the high SLAMF6 group had a longer PFI than in the low SLAMF6 group (p = 0.11), while OS was significantly longer in the high SLAMF6 group than in the low SLAMF6 group (p = 0.00039) (Fig. 1C). Collectively, these findings suggest that higher intra-tumoral SLAMF6 expression correlates with better prognosis in melanoma and breast cancer patients.
High expression of SLAMF6 correlates with longer PFI and OS in breast cancer and melanoma. (A–C) Progression-free interval (PFI) (left) and overall survival (OS) (right) for high versus low SLAMF6 group in breast cancer (BRCA Primary) (A), primary melanoma (SKCM Primary) (B) and metastatic melanoma (SKCM Metastatic) cohort (C). P values were calculated by a log-rank (Mantel–Cox) test (A–C).
SLAMF6 high tumors are enriched with effector T cell- and cDC1-related gene expression in breast cancer and melanoma
Next, we investigated the relationship between SLAMF6 expression and immune activity in the TME of breast cancer and melanoma. We found that SLAMF6 expression was highly correlated not only with the expression of T-cell markers (CD3E, CD8B, CD8A, and CD4) but also with upregulation of TCF7, PDCD1 encoding PD-1, GZMK, and effector-associated genes including IFNG and PRF1 in breast cancer (Fig. 2A). A previous study has shown that effector T-cell marker gene expression correlates with Batf3-dependent conventional type 1 DC (cDC1) signature in the TME18. Therefore, we evaluated expression of SLAMF6 and cDC1-associated genes (BATF3, IRF8, THBD encoding CD141, CLEC9A, and XCR1). In breast cancer, cDC1-related genes were substantially upregulated in the high SLAMF6 group than in the low SLAMF6 group except for THBD (Fig. 2A). Consequently, CD8+ T-cell effector (Teff) score, comprising gene expression of IFNG, PRF1, CD8A and CD8B, and BATF3-DC score, calculated with the expression level of BATF3, IRF8, THBD, CLEC9A, and XCR118 were markedly increased in the high SLAMF6 group than in the low SL AMF6 group in breast cancer. Similar results were observed in the melanoma cohort both in the primary and metastatic settings (Fig. 2B, C).
SLAMF6 high breast cancer and melanoma exhibits high CD8+ effector T cell and Batf3-DC Scores. (A–C) Normalized expression of SLAMF6, CD3e, CD8b, CD8a, CD4, IFNG, PRF1, GZMK, PDCD1, TCF7, BATF3, IRF8, THBD, CLEC9A, and XCR1 in the SLAMF6 low and high group (left). Right panels show CD8+ effector T cell score (Teff.Score) and Batf3-DC score (BATF3-DCScore) in in the SLAMF6 low and high group. Breast cancer (BRCA) (A), primary melanoma (SKCM Primary) (B), and metastatic melanoma (SKCM Metastatic) (C). P values were calculated by a two-tailed Mann–Whitney U-test (A–C).
SLAMF6 identifies the immune enriched TME of human breast cancer and melanoma
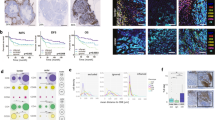
We next examined differentially expressed genes between SLAMF6 high and low tumors to further elucidate the impact of SLAMF6 expression on immunological status in the TME. Volcano plots showed that there were a number of differentially expressed genes between SLAMF6 high and low tumors (Fig. 3A). In addition to the genes presented in Fig. 2A, SLAMF6 high tumors exhibited high expression of genes and transcripts associated with T-cell activation, effector function, cytotoxicity, exhaustion and memory (CD27, GZMB, CD69, PDCD1, TOX, EOMES, LAG3, HAVCR2, IL7R, and CD28) compared with SLAMF6 low tumors. In contrast, Th2 signature gene, GATA3 was down regulated in SLAMF6 high tumors.
Melanoma and breast cancer with high SLAMF6 expression are associated with gene expression profiles suggestive of elevated immune activity. (A–C) Volcano plots (left) showing enrichment differentially expressed genes between the high and low SLAMF6 group in breast cancer (BRCA) (A), primary melanoma (SKCM) (B), and metastatic melanoma (SKCM Metastatic) (C). Each red and blue dot denotes an individual gene with Benjamini–Hochberg-adjusted P value < 0.05 and log fold change > 0.25. Enrichment correlation plots of gene set enrichment analysis (GSEA) between high and low SLAMF6 expression in the GOBP_LYMPHOCYTE_ACTIVATION (red) HALLMARK_INFLAMMATORY_RESPONSE (green), and HALLMARK_INTERFERON_GAMMA_RESPONSE (blue).
We next performed Gene set enrichment analysis (GSEA) of select differential gene sets from three pathway databases (the Hallmark, Canonical pathways, and GO Biological Processes Ontology collections) compiled from the Molecular Signatures Database (MSigDB)19. Gene sets associated with inflammatory response, immune response, T-cell activation, and cytokine and IFN signaling pathways were substantially upregulated in breast cancer (Fig. 3A, Supplementary Fig. 1). Similar results were consistently obtained not only in the cohort of primary melanoma but also in that of metastatic melanoma cohort. (Fig. 3B, C, Supplementary Fig. 1).
Single-cell profiling of the TME of breast cancer identifies SLAMF6+ T cells with a genomic signature resembling progenitor-exhausted T cells
To validate the significance of SLAMF6 expression in the TME which was obtained from the TCGA database, we next examined SLAMF6 expression in the scRNA-seq database (GSE161529) of breast cancer20. Clustering analysis and subsequent cell type annotation by the human primary cell atlas21 revealed 7 clusters (Epithelial cells, T, NK cells, Endothelial cells, Macrophages, Fibroblasts, B cells, and Monocytes) and 2 small clusters of tissue stem cells and CD34(-) Pre-B cells (Fig. 4A). The expression of representative genes (CD19, CD3E, NCR1, FCGR3A, ITGAM, EPCAM, and ITGA2) in each cluster are shown in Supplementary Fig. 2A and B. SLAMF6 was highly expressed in T cells, NK cells, and B cells, but not in epithelial cells, which is concordant with the previous studies13,14,15,17,22,23. This finding indicates that SLAMF6 is exclusively expressed in immune cells but not in cancerous cells in the TME.
Single cell profile in breast cancer reveals SLAMF6+ cells have high capacity of immune response. (A) UMAP plots of total cells in breast cancer, and expression plots of indicated genes in T and NK cell population. (B) Expression plots of indicated genes in tumor-infiltrating immune cells. Expression levels are color-coded: gray, not expressed; purple, expressed. (C) UMAP plots of Teff Signature. Expression levels are color-coded: gray, not expressed; orange, expressed. (D) Correlation heatmap showing the Pearson coefficient between SLAMF6 and select gene or Teff signature expressions. Colors range from red for positive correlations, to blue, for negative correlations.
From the initial analysis of total cells, we isolated T and NK cells to evaluate the relation between SLAMF6 expression and various markers (Fig. 4B), and the T-effector signature defined by the expression level of CD8A, CD8B, IFNG, and PRF1 (Fig. 4C)24. SLAMF6 expression was predominantly observed in CD8A+ cells with a high T-effector signature, overlapping with the expression of GZMA, GZMB, TBX21 encoding T-bet, PRF1, and IFNG, in agreement with the results from TCGA database (Figs. 2, 3). Yet, SLAMF6 expression partially overlapped with the expression of MKI67, GZMK, TCF7, memory T cell (SELL, CCR7, and IL7R)-, and exhaustion-related genes (PDCD1, LAG3, and HAVCR2).
In line with these findings, while the expression of effector-related genes was markedly associated each other, the correlation between these genes and SLAMF6 was moderate (Fig. 4D). In addition, the expression of SLAMF6 was only moderately correlated with exhaustion-related genes. These findings suggest that heterogeneity might exist within SLAMF6-expressing T cells in the TME, including subsets co-expressing SLAMF6, TCF7, memory- and exhaustion-related genes analogous to progenitor-exhausted T cells observed in pre-clinical and human melanomas7,8,9.
Discussion
In this study, we demonstrate the immunological relevance and prognostic significance of SLAMF6 expression in the TME of human breast cancer and melanoma. High expression of SLAMF6 is associated with increased expression of Teff-related genes and TCF7 which marks tumor-infiltrating T cells with stem cell-like properties7,8. Furthermore, high expression of SLAMF6 is associated with the elevation of anti-cancer immune response represented by the enrichment of gene sets associated with proinflammatory, IFN-γ and T-cell response, and improved survival in patients with breast cancer and melanoma. scRNA-seq analysis of existing human breast cancer dataset demonstrates that subsets of T cells co-expressing SLAMF6, TCF7, memory- and exhaustion-related genes comparable to progenitor-exhausted T cells identified in pre-clinical models and human melanomas7,8,9. Melanoma is known to harbor one of the highest somatic mutation frequencies among human solid malignancies25. Subsets of somatic mutations can create tumor-specific antigens which are recognizable by the immune system. Hence, melanoma has been considered immunogenic tumors, and the targets of immunotherapy. High mutational burden of melanoma is associated with enriched T-cell infiltration, and such T-cell inflamed tumors are referred as immunologically ‘hot’ tumors26. In contrast, breast cancer is characterized with infrequent somatic mutation or poor T-cell infiltration25,27, and is often called as immunologically ‘cold’ tumors. The results of this study showed that SLAMF6 expression was associated with elevated anti-cancer immune activity and better prognosis both in the ‘cold’ tumor (breast cancer) and the ‘hot’ tumor (melanoma). Although we could not investigate the association between SLAMF6 expression and response to ICIs, SLAMF6 expression might be a promising biomarker that can be useful in predicting response to ICIs. Further studies to address this point are warranted.
A large body of evidence has shown the critical roles of the cross-presenting DCs to elicit tumor-specific T-cell immunity, increase TILs and improve the treatment outcome of ICIs in mouse melanoma models and patients with this disease18,28,29,30. A previous study also showed the key role of tumor-residing cDC1s in facilitating trafficking of effector T cells into the TME via chemokine/chemokine receptor axis18. Furthermore, a recent study demonstrated that cDC1s could traffic to tumor-draining lymph nodes, and become a continual source of generating Tcf1+ CD8+ T cells31. In accordance with this scenario, our studies recently demonstrated that induction and activation of tumor-residing cDC1s could promote generation of tumor-specific T cells, convert poorly T cell-infiltrated tumors into infiltrated TMEs, facilitate an influx of Slamf6+ Tcf1+ T cells into tumors, induce not only the regression of primary but also untreated distant lesions, and overcome resistance to ICIs using mouse models of melanoma and breast cancer16,32,33,34.
In the present study, in order to examine whether SLAMF6 expression might correlate with the presence of cDC1s in the tumor, we evaluated the expression of SLAMF6 and BATF3 scores18. Our findings showed that expression of cDC1-related genes were significantly increased in the high SLAMF6 group than in the low SLAMF6 group, indicating that SLAMF6 expression in the tumor correlates with cDC1s density in the tumor. These findings are concordant with the results from the previous studies that tumor-residing cDC1 plays a critical role in trafficking as well as generation of tumor-specific T cells to the TME16,18,29,35. Given that expression of TCF7 and Teff score were significantly elevated in the high SLAMF6 group than in the low SLAMF6 group, it is conceivable that there might be a correlation of cDC1 and progenitor-exhausted T cells expressing SLAMF6. Of note, these prior results were obtained from preclinical model or melanoma patients18,29,31. Hence, this study provided the novel insight into the correlation between the cDC1 signature and markers of progenitor-exhausted T cells even in the immunologically ‘cold’ tumors.
Although ICI therapy has become a standard treatment in the management of a variety of solid malignancies including melanoma and breast cancer, many patients do not respond. PD-L1 expression in the TME correlates with increased response to PD-1/PD-L1 blockade therapy36; however, PD-L1 expression status alone does not appear to be a useful biomarker to select patients for the treatment because PD-L1 negative tumors often show dramatic response. Discovery of additional pre-treatment tumor biomarkers may lead to better identify patients who could get benefit from ICI therapy. TCF1 and SLAMF6 are both recognized as markers for progenitor-exhausted T cells8,10, and hence it would be expected that the expression of TCF7 is positively correlated with that of SLAMF6. Indeed, analysis of TCGA data demonstrated that TCF7 expression was higher in the SLAMF6 high tumors than in the SLAMF6 low tumors both in breast cancer and melanoma. This was consistent with the finding from our scRNA-seq analysis which identified SLAMF6+ cells expressing TCF7, effector- and memory-related genes, compatible to the subset of progenitor-exhausted T cells identified in the TME of melanoma patients7,8,9. We further found that heterogeneity within the SLAMF6+ subsets might exist, which include a population expressing high levels of MKI67 resembling Slamf6+ Ki67+ cells displaying vigorous expansion in response to viral infection in pre-clinical models37,38. Collectively, these findings align with previous studies showing that SLAMF6+cells are poised for better effector responses7,8.
Although SLAMF6 was found to be expressed in mouse and human progenitor-exhausted T cells in the TME, and associated with enhanced effector responses7,8,9,15,16,17, SLAMF6 may hold an inhibitory function. Previous studies have shown that anti-SLAMF6 antibody reduced the tumor burden in preclinical models of leukemia and melanoma through the activation of CD8 T cells17, and knock-out of Slamf6 in anti-melanoma CD8+ T cells improved therapeutic efficacy of adoptive T cell therapy23. Furthermore, Eisenberg et al. reported that the use of the soluble ectodomain of SLAMF6 which enhances receptor dephosphorylation reduced activation-induced cell death, increased IFN-γ production and cytotoxicity in tumor-specific CD8 T cells, resulting in improved tumor control in preclinical models of melanoma39. Therefore, accumulating evidence indicates that SLAMF6 may act as T-cell inhibitory receptor, and is not only expressed in progenitor-exhausted T cells but also various states of T cells, which is in line with our findings from scRNA-seq analysis. In our study, we found the association of SLAMF6 expression with augmented effector signature and improved prognosis. However, the exact mechanisms of how SLAMF6+ T cells contribute to these effects remain to be elucidated. Indeed, it's essential to understand the precise intracellular signaling pathways induced by SLAMF6 engagement in the tumor microenvironment, especially if therapeutic targeting is considered. To this end, further mechanistic studies that can elucidate the signaling dynamics of SLAMF6 are warranted.
This study had several limitations. First, this is a retrospective study using a previously collected cohort of patients, and selection bias in patient background should be noted. TCGA database contains patients who underwent various treatments. Hence, it should be noted that the link between SLAMF6 expression and improved survival is not associated with specific therapy. Furthermore, it is unclear whether the patients in our cohorts were treated with ICIs. Thus, it remains unknown whether SLAMF6 expression is associated with response to ICI therapy. Second, the analyses in our study were based on mRNA expression data, which is not fully concordant with protein expression. Further study is warranted to investigate the protein expression of SLAMF6 using immunohistochemistry. Lastly, the sample size in GSE161529 is relatively small, which potentially limits the generalizability of our findings. Furthermore, pooled analysis using 8 patients signals would not properly reflect the individual-level association. Further validation including individual-based analysis in a larger cohort would strengthen our findings.
In summary, the results of this study demonstrate that high expression of SLAMF6 in TME correlates with elevated immune activities and better prognosis both in breast cancer and melanoma. Further study may reveal that SLAMF6 expression can be a biomarker predicting response to ICIs.
Methods
Analysis of TCGA melanoma and breast cancer cohorts
TCGA-BRCA and TCGA-SKCM expression and clinical annotations were obtained from the Genomic Data Commons data portal and processed via TCGAbiolinks package in R using TCGAWorkflow guided practices40. Differential expression associated with SLAMF6 expression (SLAMF6-high = top quartile, SLAMF6-low = bottom quartile) within each respective cohort was determined by TCGAbiolinks/edgeR. Gene set enrichment analysis (GSEA) of ranked differential expression was assessed using the clusterProfiler package against gene sets derived from the Hallmark, Canonical pathways, and GO Biological Processes Ontology collections retrieved from the MSigDB41. Enrichment of gene sets reflecting the presence of CD8+ effector T-cells (CD8A, CD8B, IFNG, PRF1) and BATF3 DCs (BATF3, IRF8, THBD, CLEC9A, XCR1) were determined by ssGSEA using the GSVA package42. Progression free interval (PFI) and overall survival (OS) between SLAMF6-high and SLAMF6-low tumors was performed using the survival package.
Analysis of TNBC single-cell RNA-sequencing data
Raw scRNA-seq counts derived from cells captured from 8 TNBC samples20 were obtained directly from the gene expression omnibus (GSE161529). Filtering, normalization, and downstream analyses including variable feature selection, dimensionality reduction (PCA), uniform manifold approximation and projection (UMAP) low-dimensional representation, and kNN based clustering were performed using Seurat (v3)43. Cells were annotated to major cell lineages defined in the human primary cell atlas using singleR21. Cell gene expression was imputed using MAGIC44 implemented via the Rmagic package. T-cells were filtered from total cells for assessment of SLAMF6 in relation to various markers of T-cell phenotype. Single-cell pathway enrichment for CD8+ effector T-cells (CD8A, CD8B, IFNG, PRF1) was performed by UCell, and Teff Signature was calculated24. Associations between SLAMF6 expression and select genes or signatures within T-cells were examined by Pearson correlation analysis.
Data availability
The datasets analyzed during the current study are available at https://portal.gdc.cancer.gov/projects/TCGA-BRCA (TCGA-BRCA) and https://portal.gdc.cancer.gov/projects/TCGA-SKCM (TCGA-SKCM). Raw scRNA-seq counts were obtained directly from the GEO under the accession number GSE161529 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE161529). The data that support the findings of this study are publicly available from the corresponding author upon reasonable request and can be down.
References
Fridman, W. H., Pages, F., Sautes-Fridman, C. & Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012).
Adams, S. et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 32, 2959–2966 (2014).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Wherry, E. J. T cell exhaustion. Nat. Immunol. 12, 492–499 (2011).
Mazzaschi, G. et al. Low PD-1 expression in cytotoxic CD8(+) tumor-infiltrating lymphocytes confers an immune-privileged tissue microenvironment in NSCLC with a prognostic and predictive value. Clin. Cancer Res. 24, 407–419 (2018).
Thommen, D. S. et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 24, 994–1004 (2018).
Siddiqui, I. et al. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195-211.e110 (2019).
Miller, B. C. et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998-1013.e1020 (2018).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
Kratchmarov, R., Magun, A. M. & Reiner, S. L. TCF1 expression marks self-renewing human CD8+ T cells. Blood Adv. 2, 1685–1690 (2018).
Raghu, D., Xue, H. H. & Mielke, L. A. Control of lymphocyte fate, infection, and tumor immunity by TCF-1. Trends Immunol. 40, 1149–1162 (2019).
Bottino, C. et al. NTB-A [correction of GNTB-A], a novel SH2D1A-associated surface molecule contributing to the inability of natural killer cells to kill Epstein-Barr virus-infected B cells in X-linked lymphoproliferative disease. J. Exp. Med. 194, 235–246 (2001).
Zhong, M. C. & Veillette, A. Control of T lymphocyte signaling by Ly108, a signaling lymphocytic activation molecule family receptor implicated in autoimmunity. J. Biol. Chem. 283, 19255–19264 (2008).
Dragovich, M. A. et al. SLAMF6 clustering is required to augment T cell activation. PLoS ONE 14, e0218109 (2019).
Oba, T. et al. Overcoming primary and acquired resistance to anti-PD-L1 therapy by induction and activation of tumor-residing cDC1s. Nat. Commun. 11, 5415 (2020).
Yigit, B. et al. SLAMF6 as a regulator of exhausted CD8+ T cells in cancer. Cancer Immunol. Res. 7, 1485–1496 (2019).
Spranger, S., Dai, D., Horton, B. & Gajewski, T. F. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711-723.e714 (2017).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Pal, B. et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. Embo J. 40, e107333 (2021).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Hajaj, E. et al. Alternative splicing of the inhibitory immune checkpoint receptor SLAMF6 generates a dominant positive form, boosting T-cell effector functions. Cancer Immunol. Res. 9, 637–650 (2021).
Hajaj, E. et al. SLAMF6 deficiency augments tumor killing and skews toward an effector phenotype revealing it as a novel T cell checkpoint. Elife 9, e52539 (2020).
Andreatta, M. & Carmona, S. J. UCell: Robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19, 3796–3798 (2021).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Topalian, S. L., Weiner, G. J. & Pardoll, D. M. Cancer immunotherapy comes of age. J. Clin. Oncol. 29, 4828–4836 (2011).
Pellegrino, B. et al. A review of immune checkpoint blockade in breast cancer. Semin. Oncol. 48, 208–225 (2021).
Oba, T. et al. A critical role of CD40 and CD70 signaling in conventional type 1 dendritic cells in expansion and antitumor efficacy of adoptively transferred tumor-specific T cells. J. Immunol. 205, 1867–1877 (2020).
Roberts, E. W. et al. Critical role for CD103+/CD141+ dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell 30, 324–336 (2016).
Spranger, S., Bao, R. & Gajewski, T. F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015).
Schenkel, J. M. et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity 54, 2338-2353.e2336 (2021).
Oba, T., Kajihara, R., Yokoi, T., Repasky, E. A. & Ito, F. Neoadjuvant in situ immunomodulation enhances systemic antitumor immunity against highly metastatic tumors. Cancer Res. 81, 6183–6195 (2021).
Yokoi, T., Oba, T., Kajihara, R., Abrams, S. I. & Ito, F. Local, multimodal intralesional therapy renders distant brain metastases susceptible to PD-L1 blockade in a preclinical model of triple-negative breast cancer. Sci. Rep. 11, 21992 (2021).
Patel, A. et al. Multimodal intralesional therapy for reshaping the myeloid compartment of tumors resistant to anti-PD-L1 therapy via IRF8 expression. J. Immunol. 207, 1298–1309 (2021).
Hammerich, L. et al. Systemic clinical tumor regressions and potentiation of PD1 blockade with in situ vaccination. Nat. Med. 25, 814–824 (2019).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Zander, R. et al. CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity 51, 1028-1042.e1024 (2019).
Hudson, W. H. et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51, 1043-1058.e1044 (2019).
Eisenberg, G. et al. Soluble SLAMF6 receptor induces strong CD8+ T-cell effector function and improves anti-melanoma activity in vivo. Cancer Immunol. Res. 6, 127–138 (2018).
Mounir, M. et al. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 15, e1006701 (2019).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A 102, 15545–15550 (2005).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716-729.e727 (2018).
Acknowledgements
This research was funded by National Cancer Institute (NCI) grants, K08CA197966, R01CA255240 (F. Ito), and Uehara Memorial Foundation (T. Oba).
Author information
Authors and Affiliations
Contributions
T.O.: conceptualization, writing, review and editing. M.D.L.: methodology, formal analysis, and writing. K.I.: review and editing. F.I.: conceptualization, supervising, writing, review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oba, T., Long, M.D., Ito, Ki. et al. Clinical and immunological relevance of SLAMF6 expression in the tumor microenvironment of breast cancer and melanoma. Sci Rep 14, 2394 (2024). https://doi.org/10.1038/s41598-023-50062-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-50062-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.