Abstract
In the ongoing global fight against coronavirus disease 2019 (COVID-19), the sample preparation process for real-time reverse transcription polymerase chain reaction (rRT-PCR) faces challenges due to time-consuming steps, labor-intensive procedures, contamination risks, resource demands, and environmental implications. However, optimized strategies for sample preparation have been poorly investigated, and the combination of RNase inhibitors and Proteinase K has been rarely considered. Hence, we investigated combinations of several extraction-free protocols incorporating heat treatment, sample dilution, and Proteinase K and RNase inhibitors, and validated the effectiveness using 120 SARS-CoV-2 positive and 62 negative clinical samples. Combining sample dilution and heat treatment with Proteinase K and RNase inhibitors addition exhibited the highest sensitivity (84.26%) with a mean increase in cycle threshold (Ct) value of + 3.8. Meanwhile, combined sample dilution and heat treatment exhibited a sensitivity of 79.63%, accounting for a 38% increase compared to heat treatment alone. Our findings highlight that the incorporation of Proteinase K and RNase inhibitors with sample dilution and heat treatment contributed only marginally to the improvement without yielding statistically significant differences. Sample dilution significantly impacts SARS-CoV-2 detection, and sample conditions play a crucial role in the efficiency of extraction-free methods. Our findings may provide insights for streamlining diagnostic testing, enhancing its accessibility, cost-effectiveness, and sustainability.
Similar content being viewed by others
Introduction
The Coronavirus disease 2019 (COVID-19) has brought considerable challenges to the global public health system, as well as notable environmental consequences due to increased plastic usage associated with diagnostic procedures1,2,3,4. While the COVID-19 pandemic triggered remarkable progress in the field of molecular diagnostics, particularly in the context of detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection, the sample preparation for real-time reverse transcription polymerase chain reaction (rRT-PCR) continues to be a bottleneck in the overall diagnostic testing workflow5,6,7,8,9. The preparation process typically involves extracting and purifying viral RNA from patient samples, which introduces additional steps and time into the testing process. This step is critical for eliminating potential inhibitors and ensuring reliable results. Various commercial kits and methods are available for RNA extraction, and laboratories and diagnostic centers have developed protocols to perform this procedure efficiently10,11,12. However, manual extraction methods used in many laboratories can be time-consuming and require skilled technicians for accurate execution. In addition, the use of plastic materials associated with RNA extraction kits and reagents contributes to environmental concerns and escalates the overall cost of diagnostic testing13,14,15. To tackle this issue, efforts have been made to address bottlenecks and streamline sample preparation. Automated and high-throughput systems have been developed to increase efficiency and reduce the labor required for RNA extraction. These systems can simultaneously process multiple samples and may incorporate robotic platforms or magnetic bead-based technologies to improve throughput16,17,18,19. Furthermore, researchers are exploring alternative methods of RNA extraction that are more cost-effective and less labor-intensive. Some of these methods include the use of magnetic beads20,21, solid-phase extraction22,23, and microfluidic devices24,25,26, which simplify the extraction process and reduce the associated costs.
In addition to technical advancements, initiatives have been made to optimize protocols and develop extraction-free methods27,28,29,30,31,32. Extraction-free rRT-PCR methods aim to simplify and expedite the testing process by bypassing the RNA extraction step, which is time-consuming, labor-intensive. These methods often involve the direct addition of specific reagents or buffers to the patient sample to inactivate potential inhibitors and facilitate the direct amplification of viral RNA. Additionally, one of the practical advantages is their cost-effectiveness compared to extraction kit. For example, the average cost incurred per sample using extraction kits falls within the range of 6–12 USD, while the cost associated with the application of specific reagents such as Proteinase K and RNase inhibitors in the conducted experiments amounts to merely 1.15 USD per sample33. The performance of extraction-free rRT-PCR methods can vary depending on the protocol and patient sample characteristics34,35,36.
The most straightforward extraction-free method is to dilute the sample to reduce inhibitory substances that might interfere with the PCR. While the optimal dilution for extraction-free methods may vary depending on the specific assay and sample type, several studies have reported successful detection using a 1:1 dilution33,37,38. Other extraction-free methods, such as thermal or direct lysis, may also be used in combination with or as alternatives to dilution32,39,40,41. These methods involve the disruption of cell membranes through heat, allowing nucleic acid molecules to be accessible for PCR amplification without the need for a separate extraction step. The impact on reducing the occurrence of invalid results and minimizing the risk of contamination in unextracted rRT-PCR through the utilization of Proteinase K and RNase inhibitors has also been reported28,33,42. The summary of extraction-free protocols and their corresponding sensitivity in previous studies can be found in Table S1 in the supplementary information. However, there has been a lack of study on optimized strategies that combine sample dilution with Proteinase K and RNase inhibitors along with heat treatment. Validation and evaluation studies are necessary to ensure that these methods yield reliable and accurate results, aligning with the performance standards of traditional rRT-PCR, especially when using samples having diverse viral concentrations.
Herein, we validated the use of heat treatment and sample dilution as extraction-free methods for detecting SARS-CoV-2. Proteins can interfere with the extraction process, and the use of Proteinase K improves viral nucleic acid accessibility by dissolving sample proteins33,42,43. Ribonucleases are enzymes that can degrade RNA, and the use of RNase inhibitors helps inhibit their activity, preserving the integrity of viral RNA during sample preparation. Therefore, we also incorporated Proteinase K and RNase inhibitors for improving viral nucleic acid detection. Combining methods, including heat treatment, sample dilution, Proteinase K, and RNase inhibitors, we aimed to develop an optimal extraction-free method for viral nucleic acid detection. This approach simplifies the sample preparation process and may improve the efficiency and scalability of diagnostic testing, particularly during situations like a pandemic, where large-scale testing is required. By streamlining the RNA extraction process, laboratories can enhance their testing capacities, reduce turnaround times, and make diagnostic processes more accessible and cost-effective.
Result and discussion
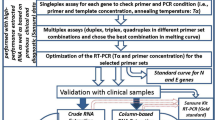
The efficiency of each extraction-free protocol was evaluated by comparing differences in Ct values (cycle threshold) with those obtained using the standard methodology using the extraction step (Group I). The experimental groups consisted of variations in the extraction-free protocols, denoted as Groups II–VI. Specifically, the groups were categorized as follows: Group I (standard protocol), Group II (extraction-free protocol based on heat treatment), Group III (extraction-free protocol based on heat treatment and sample dilution), Group IV (extraction-free protocol based on heat treatment, sample dilution, and addition of Proteinase K), Group V (extraction-free protocol based on heat treatment, sample dilution, and addition of RNase inhibitors), and Group VI (extraction-free protocol based on heat treatment, sample dilution, and addition of Proteinase K and RNase inhibitors). Detailed information can be found in the “Methods” section and Table S2 of the supplementary information. Overall, 170 samples—108 positives and 62 negatives—were included in our evaluation with the standard methodology using the extraction step recommended by the manufacturer. Among positive samples, 46 (46/108, 42.6%) showed Ct values < 20, 36 (36/108, 33.3%) had intermediate Ct values (20 ≤ Ct ≤ 30), and 26 (26/108, 24.1%) showed low Ct values (Ct > 30) for the ORF1ab gene. Similarly, for the N gene, 54 (54/108, 50%) showed Ct values < 20, 34 (34/108, 31.5%) had intermediate Ct values (20 ≤ Ct ≤ 30), and 20 (20/108, 18.5%) showed low Ct values (Ct > 30). The Ct values obtained from the experimental groups using different extraction-free protocols correlated with those obtained with the standard methodology (Fig. S1 in the supplementary information). However, false negatives were also observed for each protocol.
Figure 1 comprehensively summarizes the Ct value distributions across experimental groups. In Group II, regardless of the target genes, no targets were detected for samples with Ct values above 30 (Fig. 1A). The median in the Ct values for the ORF1ab and N gene varied across experimental conditions: Group II (30.48, 32.11), Group III (25.53, 24.49), Group IV (24.44, 24.03), Group V (25.49, 24.77), and Group VI (24.14 23.77). These findings indicate that sample dilution notably affected nucleic acid detection. Other factors, namely, treatment with Proteinase K and RNase inhibitors contributed to only a marginal improvement compared with Group III, in which samples were treated with a combination of dilution and heating and did not yield statistically significant differences in Ct values (Fig. 2). Bland–Altman plots were used to compare the extraction-free protocols with the standard methodology using the extraction step (Fig. S2 in supplementary information). The differences in the Ct values across extraction-free protocols compared with the standard protocol are shown in Table 1. In the context of sample enrichment and purification, the typical nucleic acid extraction step can lead to a four-fold increase in sample concentration, wherein 200 µL of the sample is concentrated to 50 µL of eluate. During the extraction process, various inhibitory substances can be co-purified along with RNA, potentially interfering with downstream PCR. In contrast, the extraction-free protocol involves a 1:1 dilution of the sample, resulting in a two-fold dilution or 0.5 times the concentration of the original sample. As a result, a significant eight-fold variation in amplification results may be observed when comparing identical samples. Considering the logarithmic scale of the Ct values of quantitative PCR, a mere ten-fold difference between samples corresponds to a 3.3 Ct difference under the assumption of 100% PCR efficiency. Interestingly, in our study, Group VI exhibited a mean increase in Ct value of + 3.8 compared to Group I, supporting the expected variation in results.
Heat map of Ct values for (A) the ORF1ab gene and (B) the N gene subjected to various experimental conditions compared to a standard methodology using the extraction step. Gray color represents lower Ct values of samples labeled as not detected. (Ct values determined by I, standard methodology using the extraction step; II, extraction-free protocol based on heat treatment; III, extraction-free protocol based on heat treatment and sample dilution; IV, extraction-free protocol based on heat treatment, sample dilution, and addition of Proteinase K; V, extraction-free protocol based on heat treatment, sample dilution, and addition of RNase inhibitors; VI, extraction-free protocol based on heat treatment, sample dilution, and addition of Proteinase K and RNase inhibitors).
The individual and distribution of Ct values of various experimental conditions for (A) the ORF1ab gene and (B) the N gene. The median of each group is presented as the white circle with an interquartile range box and range. Dotted lines show the positive thresholds for ORF1ab and N genes. (Ct values determined by I, standard methodology using the extraction step; II, extraction-free protocol based on heat treatment; III, extraction-free protocol based on heat treatment and sample dilution; IV, extraction-free protocol based on heat treatment, sample dilution, and addition of Proteinase K; V, extraction-free protocol based on heat treatment, sample dilution, and addition of RNase inhibitors; VI, extraction-free protocol based on heat treatment, sample dilution, and addition of Proteinase K and RNase inhibitors).
The detection rates based on Ct values for ORF1ab are summarized in Fig. 3. In the extraction-free protocols, lower detection rates were observed for samples with Ct values above 30 for the ORF1ab gene: Group II (0/26, 0%), Group III (4/26, 15.4%), Group IV (7/26, 26.9%), Group V (8/26, 30.8%), and Group VI (9/26, 34.6%). Similarly, lower detection rates were observed for samples with Ct values above 30 for the N gene: Group II (0/20, 0%), Group III (2/20, 10.0%), Group IV (3/20, 15.0%), Group V (4/20, 20.0%), and Group VI (5/20, 25.0%).
True positive fractionation across all experimental conditions for (A) the ORF1ab gene and (B) the N gene. SARS-CoV-2 RNA loads were categorized as high (Ct values less than 20), intermediate (Ct values of 20–30), or low (Ct values of more than 30) determined by standard methodology using the extraction step (Group I). I, standard methodology using the extraction step; II, extraction-free protocol based on heat treatment; III, extraction-free protocol based on heat treatment and sample dilution; IV, extraction-free protocol based on heat treatment, sample dilution, and addition of Proteinase K; V, extraction-free protocol based on heat treatment, sample dilution, and addition of RNase inhibitors; VI, extraction-free protocol based on heat treatment, sample dilution, and addition of Proteinase K and RNase inhibitors.
The sensitivity and specificity in each experimental group are shown in Table 2. Although the sensitivity of all experimental conditions was notably lower than that of the standard methodology, we observed substantial improvements among different conditions in response to sample dilution. Moreover, when combined with heat treatment, sample dilution resulted in a sensitivity of 79.63%, which represents a marked improvement of 38% compared with the application of heat treatment alone. In addition, the incorporation of Proteinase K and RNase inhibitors in conjunction with sample dilution and heat treatment contributed to a further slight improvement in sensitivity, which can be attributed to a marginal improvement in the low Ct group (Ct > 30). Although the inclusion of Proteinase K and RNase inhibitors has shown benefits in numerous studies, it is imperative to consider certain factors when implementing them in extraction-free PCR methods. First, the concentration and incubation time of these reagents should be optimized to ensure their effectiveness without causing adverse effects on the PCR. Second, the compatibility of these reagents under different sample conditions should be carefully evaluated to ensure consistent and reliable performance. Indeed, our experimental findings clearly showed a consistent occurrence of false negatives in a number of samples across extraction-free protocols that involved heat treatment and sample dilution (Table S3 in the supplementary information). Because the efficiency of extraction-free PCR can be significantly influenced by the condition of the sample, factors such as sample quality, presence of inhibitors, and composition of the sample matrix should be factored in when implementing extraction-free PCR methods to develop strategies for maximizing the accuracy and reliability of this approach. Finally, the cost-effectiveness and scalability of incorporating Proteinase K and RNase inhibitors should be considered, as they may contribute to the overall expenses of diagnostic testing.
In this study, we aimed to optimize the extraction-free approach, simplify the sample preparation process, and potentially increase testing efficiency and scalability. To this end, we validated the use of extraction-free protocol based on heat treatment and sample dilution while incorporating Proteinase K and RNase inhibitors to enhance nucleic acid extraction efficiency. Our results demonstrate that Group VI, which included all parameters (extraction-free protocols based on heat treatment, sample dilution, and addition of Proteinase K and RNase inhibitors), consistently showed the lowest average Ct values for all target genes. In contrast, Group II, which involved only heat treatment protocol, had the highest average Ct value. Importantly, most false negatives observed using the extraction-free protocols incorporating sample dilution exhibited Ct values above 30, regardless of the target gene. However, the extraction-free protocol based solely on heat treatment exhibited lower detection rates even for samples with a Ct value of less than 30 in the standard protocol. Our findings highlight the significant influence of sample conditions on the efficiency of extraction-free methods. Sample quality, presence of inhibitors, and composition of the sample matrix are among the factors that can remarkably affect amplification efficiency. However, we found that it is crucial to further validate these methods to ensure their reliability and comparability, particularly in the context of massive sample testing. Furthermore, additional prospective studies are required to substantiate the effect of sample conditions on the efficiency of extraction-free methods. We believe the findings of this study contribute to the ongoing efforts to streamline diagnostic testing, improve efficiency, enhancing its accessibility, cost-effectiveness, and sustainability by minimizing plastic waste and chemical reagent usage, especially in large-scale testing scenarios such as pandemics.
Conclusion
In conclusion, this study emphasizes the potential of extraction-free methods for SARS-CoV-2 detection. By optimizing the sample preparation process and incorporating specific treatments and reagents, we demonstrated the feasibility of an extraction-free approach for enhancing nucleic acid detection. Our study revealed that sample dilution had a notable impact on the testing process, whereas the influence of Proteinase K and RNase inhibitors was found to be insignificant. Additional large-scale prospective studies are required to substantiate the effect of sample conditions on the efficiency of extraction-free methods.
Methods
Ethics statement
The study protocol was reviewed and approved by the Institutional Review Board of Samsung Medical Center, Seoul, Republic of Korea (IRB No. SMC 2023-03-031) and conducted in accordance with relevant guidelines and regulations. The requirement for informed consent was waived by the Institutional Review Board of Samsung Medical Center because of the retrospective nature of the study and the use of anonymized patient data.
Preparation of clinical specimens
A total of 182 respiratory SARS-CoV-2 nasopharyngeal swab (NPS) specimens (120 positive and 62 negative samples) collected in a viral transport medium (VTM; Noble Bioscience, Inc., Hwaseong, Republic of Korea) were de-identified to ensure patient anonymity. Each sample was aliquoted into individual microcentrifuge tubes based on the required volume for RNA extraction and extraction-free methods and stored at − 80 °C until further experiments. All sample preparation, processing, and PCR settings were performed within a class 2 Biosafety Cabinet located in a Biosafety Level 2 (BSL2) facility.
RNA extraction
Nucleic acid extraction was conducted using the MagMax viral/pathogen nucleic acid isolation kit and the KingFisher Flex purification system (Thermo Fisher Scientific Inc., Massachusetts, United States) following the manufacturer’s instructions. Briefly, 200 μL of each sample was mixed with 530 μL of a binding solution containing an appropriate concentration of guanidine thiocyanate for effective virus inactivation. This mixture was then supplemented with 20 μL of magnetic beads and 10 μL of Proteinase K to increase the extraction yield. Subsequently, the purification system automatically carried out the subsequent steps, including washing, elution, and tip comb plate preparation.
Real-time reverse transcriptase-polymerase chain reaction
Direct clinical specimens treated with the extraction-free protocols and extracted nucleic acids from clinical specimens served as inputs for real-time reverse transcriptase-polymerase chain reaction (rRT-PCR). The STANDARD™ M SARS-CoV-2 (SD BIOSENSOR, Suwon, Republic of Korea) was used for the detection of SARS-CoV-2 RNA, specifically targeting the N (nucleocapsid) and ORF1ab (polyprotein) genes, along with RNaseP (ribonuclease P) as an internal control, in clinical samples, following the manufacturer’s instructions. rRT-PCR was conducted using a CFX96 Real-Time System (Bio-Rad, Hercules, CA, United States). The thermal cycling steps consisted of reverse transcription at 50 °C for 5 min, initial denaturation at 95 °C for 3 min, pre-amplification with 5 cycles at 95 °C for 1 s and 60 °C for 1 s, followed by amplification with 40 cycles at 95 °C for 1 s and 60 °C for 1 s. Positive test results were determined based on the simultaneous detection of both SARS-CoV-2 target genes in accordance with the established guidelines provided by the World Health Organization (WHO). Of the 182 samples, 12 were excluded because of RNA degradation. This decision was made based on discrepancies between the routinely tested results upon arrival at the laboratory and the standard PCR results obtained in this study.
Extraction-free protocols
With the exception of the protocol performed with heat treatment alone, all samples underwent an optimal dilution of 1:1. Additionally, two variations were explored: thermal lysis alone and thermal lysis combined with the addition of RNase inhibitors (Catalog # N8080119, Thermo Fisher Scientific Inc., Massachusetts, United States) and Proteinase K (Catalog # MC5005, Roche Diagnostics GmbH, Mannheim, Germany). For experimental groups II–VI, the samples were heated to 98 °C for 5 min, followed by rapid cooling at 4 °C before the rRT-PCR process. In Groups IV and VI, Proteinase K was directly added to RNase-free water before dilution at a final concentration of 100 μg/mL. Additionally, for this group, a 15-min heat treatment at 55 °C was performed before heating the sample at 98 °C for 5 min. Group V and VI underwent treatment with 1U/μL of RNase inhibitors as per the manufacturer’s protocol.
Statistical analysis
The cycle threshold (Ct) was determined automatically using CFX Manager Software version 3.1 (Bio-Rad). Results were interpreted as positive only if the Ct values obtained for both ORF1ab and N were within the cutoff value, in accordance with the manufacturer's recommendations. The data obtained were described in terms of categorical variables divided into high (Ct values less than 20), intermediate (Ct values between 20 and 30), or low (Ct values greater than 30) categories. The sensitivities and specificities of each condition were assessed based on the results obtained when including the extraction step as the reference standard methodology. An independent t-test was used for the comparison of Ct values between groups, and McNemar’s test was used to compare diagnostic accuracy. A p-value of < 0.05 was considered to be statistically significant. Bland–Altman analysis was used to determine the degree of agreement based on the mean difference and standard deviation (SD) of the positive results.
Data availability
The data that support the findings of this study will be made available from the corresponding author on reasonable request.
References
Loeffelholz, M. J. & Tang, Y. W. Laboratory diagnosis of emerging human coronavirus infections—the state of the art. Emerg. Microbes Infect. 9, 747–756. https://doi.org/10.1080/22221751.2020.1745095 (2020).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 3. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045 (2020).
Wang, Q., Zhang, C. & Li, R. Plastic pollution induced by the COVID-19: Environmental challenges and outlook. Environ. Sci. Pollut. Res. Int. 30, 40405–40426. https://doi.org/10.1007/s11356-022-24901-w (2023).
Shams, M., Alam, I. & Mahbub, M. S. Plastic pollution during COVID-19: Plastic waste directives and its long-term impact on the environment. Environ. Adv. 5, 100119. https://doi.org/10.1016/j.envadv.2021.100119 (2021).
Vogels, C. B. F. et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 5, 1299–1305. https://doi.org/10.1038/s41564-020-0761-6 (2020).
Yu, F. et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 71, 793–798. https://doi.org/10.1093/cid/ciaa345 (2020).
Udugama, B. et al. Diagnosing COVID-19: The disease and tools for detection. ACS Nano 14, 3822–3835. https://doi.org/10.1021/acsnano.0c02624 (2020).
Huh, H. J. et al. Surveillance of coronavirus disease 2019 (COVID-19) testing in clinical laboratories in Korea. Ann. Lab. Med. 41, 225–229. https://doi.org/10.3343/alm.2021.41.2.225 (2021).
Kim, Y. J., Sung, H., Ki, C. S. & Hur, M. Response of clinical laboratories to the ongoing COVID-19 pandemic. Ann. Lab. Med. 41, 519–520. https://doi.org/10.3343/alm.2021.41.6.519 (2021).
Dhibika, M., Madhusudhan, N. S., Malini, A. & Natarajan, M. Comparison of manual and automated nucleic acid (RNA) extraction methods for the detection of SARS-CoV-2 by qRT-PCR. Cureus 15, e36773. https://doi.org/10.7759/cureus.36773 (2023).
Chen, S. Y., Chou, Y. C., Huang, J. W. & Er, T. K. Comparison of manual and automated nucleic acid extraction methods in virus transport medium. Clin. Lab. 67, 12. https://doi.org/10.7754/Clin.Lab.2021.210443 (2021).
Chomczynski, P. & Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 1, 581–585. https://doi.org/10.1038/nprot.2006.83 (2006).
Cameron, A., Pecora, N. D. & Pettengill, M. A. Extraction-free methods for the detection of SARS-CoV-2 by reverse transcription-PCR: A comparison with the cepheid Xpert Xpress SARS-CoV-2 assay across two medical centers. J. Clin. Microbiol. 59, 2. https://doi.org/10.1128/JCM.02643-20 (2021).
Kim, T. Y. et al. Performance evaluation of the PowerChek SARS-CoV-2, influenza A & B multiplex real-time PCR kit in comparison with the BioFire respiratory panel. Ann. Lab. Med. 42, 473–477. https://doi.org/10.3343/alm.2022.42.4.473 (2022).
Ji, L. et al. Potential life-cycle environmental impacts of the COVID-19 nucleic acid test. Environ. Sci. Technol. 56, 13398–13407. https://doi.org/10.1021/acs.est.2c04039 (2022).
Fiebelkorn, K. R., Lee, B. G., Hill, C. E., Caliendo, A. M. & Nolte, F. S. Clinical evaluation of an automated nucleic acid isolation system. Clin. Chem. 48, 1613–1615 (2002).
Verheyen, J. et al. Extraction of viral nucleic acids: Comparison of five automated nucleic acid extraction platforms. J. Clin. Virol. 54, 255–259. https://doi.org/10.1016/j.jcv.2012.03.008 (2012).
Thakore, N. et al. A bench-top automated workstation for nucleic acid isolation from clinical sample types. J. Microbiol. Methods 148, 174–180. https://doi.org/10.1016/j.mimet.2018.03.021 (2018).
Kogoj, R. et al. Real-life head-to-head comparison of performance of two high-throughput automated assays for the detection of SARS-CoV-2 RNA in nasopharyngeal swabs: The Alinity m and cobas 6800 SARS-CoV-2 assays. J. Mol. Diagn. 23, 920–928. https://doi.org/10.1016/j.jmoldx.2021.05.003 (2021).
Pham, X. H. et al. Graphene oxide conjugated magnetic beads for RNA extraction. Chem. Asian J. 12, 1883–1888. https://doi.org/10.1002/asia.201700554 (2017).
Pang, Y. et al. Fe3O4@Ag magnetic nanoparticles for microRNA capture and duplex-specific nuclease signal amplification based SERS detection in cancer cells. Biosens. Bioelectron. 79, 574–580. https://doi.org/10.1016/j.bios.2015.12.052 (2016).
Nacham, O., Clark, K. D. & Anderson, J. L. Extraction and purification of DNA from complex biological sample matrices using solid-phase microextraction coupled with real-time PCR. Anal. Chem. 88, 7813–7820. https://doi.org/10.1021/acs.analchem.6b01861 (2016).
Varona, M., Ding, X., Clark, K. D. & Anderson, J. L. Solid-phase microextraction of DNA from mycobacteria in artificial sputum samples to enable visual detection using isothermal amplification. Anal. Chem. 90, 6922–6928. https://doi.org/10.1021/acs.analchem.8b01160 (2018).
Obino, D. et al. An overview on microfluidic systems for nucleic acids extraction from human raw samples. Sens. (Basel) 21, 3059. https://doi.org/10.3390/s21093058 (2021).
Jin, C. E. et al. Use of dimethyl pimelimidate with microfluidic system for nucleic acids extraction without electricity. Anal. Chem. 89, 7502–7510. https://doi.org/10.1021/acs.analchem.7b01193 (2017).
Wen, J., Legendre, L. A., Bienvenue, J. M. & Landers, J. P. Purification of nucleic acids in microfluidic devices. Anal. Chem. 80, 6472–6479. https://doi.org/10.1021/ac8014998 (2008).
Fomsgaard, A. S. & Rosenstierne, M. W. An alternative workflow for molecular detection of SARS-CoV-2—escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill. 25, 14. https://doi.org/10.2807/1560-7917.ES.2020.25.14.2000398 (2020).
Villota, S. D. et al. Alternative RNA extraction-free techniques for the real-time RT-PCR detection of SARS-CoV-2 in nasopharyngeal swab and sputum samples. J. Virol. Methods 298, 114302. https://doi.org/10.1016/j.jviromet.2021.114302 (2021).
Lownik, J. C., Way, G. W., Farrar, J. S. & Martin, R. K. Extraction-free rapid cycle quantitative RT-PCR and extreme RT-PCR for SARS-CoV-2 virus detection. J. Mol. Diagn. 23, 1671–1679. https://doi.org/10.1016/j.jmoldx.2021.08.004 (2021).
Kim, Y. K. & Chang, S. H. Clinical usefulness of extraction-free PCR assay to detect SARS-CoV-2. J. Virol. Methods 296, 114217. https://doi.org/10.1016/j.jviromet.2021.114217 (2021).
Visseaux, B. et al. Evaluation of three extraction-free SARS-CoV-2 RT-PCR assays: A feasible alternative approach with low technical requirements. J. Virol. Methods 291, 114086. https://doi.org/10.1016/j.jviromet.2021.114086 (2021).
Smyrlaki, I. et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 11, 4812. https://doi.org/10.1038/s41467-020-18611-5 (2020).
Mallmann, L. et al. Proteinase K treatment in absence of RNA isolation classical procedures is a quick and cheaper alternative for SARS-CoV-2 molecular detection. J. Virol. Methods 293, 114131. https://doi.org/10.1016/j.jviromet.2021.114131 (2021).
Vogels, C. B. F. et al. SalivaDirect: A simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med 2, 263–280. https://doi.org/10.1016/j.medj.2020.12.010 (2021).
Mahmoud, S. A. et al. Evaluation of RNA extraction-free method for detection of SARS-CoV-2 in salivary samples for mass screening for COVID-19. Biomed. Res. Int. 2021, 5568350. https://doi.org/10.1155/2021/5568350 (2021).
Wyllie, A. L. et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 383, 1283–1286. https://doi.org/10.1056/NEJMc2016359 (2020).
Freppel, W., Merindol, N., Rallu, F. & Bergevin, M. Efficient SARS-CoV-2 detection in unextracted oro-nasopharyngeal specimens by rRT-PCR with the Seegene Allplex 2019-nCoV assay. Virol. J. 17, 196. https://doi.org/10.1186/s12985-020-01468-x (2020).
Hasan, M. R. et al. Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLOS ONE 15, e0236564. https://doi.org/10.1371/journal.pone.0236564 (2020).
Panpradist, N. et al. Simpler and faster Covid-19 testing: Strategies to streamline SARS-CoV-2 molecular assays. EBioMedicine 64, 103236. https://doi.org/10.1016/j.ebiom.2021.103236 (2021).
Byrnes, S. A. et al. Multiplexed and extraction-free amplification for simplified SARS-CoV-2 RT-PCR tests. Anal. Chem. 93, 4160–4165. https://doi.org/10.1021/acs.analchem.0c03918 (2021).
Bruce, E. A. et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an extraction step. bioRxiv https://doi.org/10.1101/2020.03.20.001008 (2020).
Genoud, V. et al. Extraction-free protocol combining proteinase K and heat inactivation for detection of SARS-CoV-2 by RT-qPCR. PLoS One 16, e0247792. https://doi.org/10.1371/journal.pone.0247792 (2021).
Sung, H. et al. Comparative evaluation of three homogenization methods for isolating middle east respiratory syndrome coronavirus nucleic acids from sputum samples for real-time reverse transcription PCR. Ann. Lab. Med. 36, 457–462. https://doi.org/10.3343/alm.2016.36.5.457 (2016).
Acknowledgements
This study was supported by Nano Material Technology Development Program (Nanoconnect Program) through the National Research Foundation of Korea (NRF) funded by Ministry of Science and ICT (No. 2022M3H4A4097528), the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) (No. RS-2020-KD000004) and Future Medicine 2030 Project of the Samsung Medical Center (SMX1230761). We thank SD BIOSENSOR for providing the STANDARD M SARS-CoV-2 Real-Time Detection Kit used in this study. SD BIOSENSOR had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: T.Y.K, M.K and H.J.H; Data curation: T.Y.K and M.K; Methodology: T.Y.K, J-Y.K, S.A.Y, E.J, M.K and H.J.H; Validation: E.J and M.K; Writing—original draft: M.K and E.J; Writing—review and editing: M.K, E.J, J-H.J, M-A.J, T.Y.K, H.J.H, and N.Y.L. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kang, M., Jeong, E., Kim, JY. et al. Optimization of extraction-free protocols for SARS-CoV-2 detection using a commercial rRT-PCR assay. Sci Rep 13, 20364 (2023). https://doi.org/10.1038/s41598-023-47645-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47645-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.