Abstract
Despite the rising interest in bacteriophages, little is known about their infection cycle and lifestyle in a multicellular host. Even in the model system Streptomyces, only a small number of phages have been sequenced and well characterized so far. Here, we report the complete characterization and genome sequences of Streptomyces phages Vanseggelen and Verabelle isolated using Streptomyces coelicolor as a host. A wide range of Streptomyces strains could be infected by both phages, but neither of the two phages was able to infect members of the closely related sister genus Kitasatospora. The phages Vanseggelen and Verabelle have a double-stranded DNA genome with lengths of 48,720 and 48,126 bp, respectively. Both phage genomes contain 72 putative genes, and the presence of an integrase encoding protein indicates a lysogenic lifestyle. Characterization of the phages revealed their stability over a wide range of temperatures (30–45 °C) and pH values (4–10). In conclusion, Streptomyces phage Vanseggelen and Streptomyces phage Verabelle are newly isolated phages that can be classified as new species in the genus Camvirus, within the subfamily Arquattrovirinae.
Similar content being viewed by others
Introduction
Streptomyces are Gram-positive, spore-forming bacteria, with a relatively high G + C-content1. Their multicellular morphology and life cycle are unique within the bacteria realm. Furthermore, Streptomyces are well-known for their ability to produce natural products with a wide range of biological activities2. These include, for example, metabolites that can act as anti-fungals, anti-virals, anti-tumor agents and many clinically relevant antibiotics3. However, the production of these metabolites from Streptomyces can be negatively influenced by bacteriophage contaminations, which can result in huge economic losses4. Bacteriophages, or phages for short, are viruses that can infect bacteria. Yet, it remains largely unknown how bacteriophages can recognize, attach and infect these multicellular bacteria and more detailed descriptions of new phage species will be beneficial for following reseach5,6. All known Streptomyces phages belong to the class Caudoviricetes, which are tailed bacterial and archaeal viruses with icosahedral capsid heads and double-stranded DNA genomes7. The phages within this class are divided into three morphotypes: long contractile tails (myoviruses), long non-contractile tails (siphoviruses) or short non-contractile tails (podoviruses). Of all characterized Streptomyces phages, 91% have long non-contractile tails and are therefore siphoviruses8. Compared to other genera, relatively few Streptomyces phages have been properly characterized and sequenced. According to The Actinobacteriophage Database9, only 340 Streptomyces phages have been sequenced to date, while there are over 2700 Escherichia coli phage genomes deposited in Genbank10,11.
Here, we describe two newly discovered Actinobacteriophages, which we name Streptomyces phage Vanseggelen and Streptomyces phage Verabelle. These temperate phages belong to the genus Camvirus, within the subfamily Arquattrovirinae. Vanseggelen has an icosahedral capsid head of 53.7 nm ± 7.9 nm and a long non-contractile tail of 228.7 ± 7.5 nm, while Verabelle has an icosahedral capsid head of 63.2 + 1.2 nm and a long non-contractile tail of 200.4 + 1.5 nm. Furthermore, we provide a complete genomic analysis of these newly identified phages. Together, these results will hopefully increase our knowledge on phage-host dynamics in Streptomyces in the future and expanded the list of available, sequenced and characterized Streptomyces phages.
Materials and methods
Bacteriophage isolation
Streptomyces phages Vanseggelen and Verabelle were isolated from a soil sample obtained from the National Park Zuid-Kennemerland in the Netherlands (N52° 23′ 31″, E4° 34′ 49″) using Streptomyces coelicolor as the host. Details on isolation procedures are reported previously in detail12. The collected phage stock solutions were stored at 4 °C.
Host range analysis
The host range of Vanseggelen and Verabelle was determined by using serial dilutions on double agar overlay plates. The phage stock solutions were serial diluted with Difco Nutrient Broth (DNB) (BD biosciences) supplemented with 4 mM Ca(NO3)2 and 0.5% w/v glucose. Droplets of 3 µl of the phage dilutions were spotted in duplicate on a bacterial lawn of nine different Streptomyces strains and one Kitasatospora strain from the Microbial Biotechnology (MBT) collection at Leiden University13. Streptomyces strains MBT13, MBT61 and MBT86 are new species and have not been taxonomically classified yet. The plates were incubated at 30 °C for at least 24 h before lysis was observed, after which the presence of a lysis zone was scored as a positive result. The efficiency of plating (EOP) for a given strain was calculated relative to the host strain S. coelicolor, only when distinct clear individual plaques were observed.
Morphology analysis
The plaque morphology of Vanseggelen and Verabelle was determined by using serial dilutions on double agar overlay plates on the host S. coelicolor.
Representative images of the phages were made using transmission electron microscopy (TEM). For a single grid preparation, 3 µl of the phage lysates (Vanseggelen = 106 PFU/ml and Verabelle = 107 PFU/ml) was placed on a glow-discharged 200 mesh carbon coated copper grid (EMS) and allowed to set for 30 s before excess sample solution was removed by filter paper. The phages were stained with 2% uranyl acetate for 45 s after which excess liquid was removed and the samples were air-dried for an additional 30 min. The grids were observed using a single tilt specimen holder inside a 120 kV Talos L120C TEM with a Lab6 electron source and Ceta detector at the Netherlands Center for Nanoscopy (NeCEN, Leiden).
Stability analysis
A one-step growth curve analysis was performed in triplicate to determine the latent period and burst size of both phages14. Since Streptomyces phage adsorption was found to be maximal for germlings15, 108 spores ml−1 of S. coelicolor were allowed to germinate in 10 ml DNB medium at 30 °C while shaking at 200 rpm. After approximately 5 h, the culture was infected with Vanseggelen or Verabelle at Multiplicity of Infection (MOI) of 0.01 and 100 µl was sampled at 10 min intervals up to 180 min. The samples were filtered with a 0.20 µm filter, serial diluted with DNB medium and immediately plated on double agar overlay plates. The burst size was calculated using the following formula:
The thermal stability of Vanseggelen and Verabelle was determined by adding 100 µl of phage suspension (Vanseggelen = 105 PFU/ml and Verabelle = 107 PFU/ml) to 900 µl DNB medium and incubating the phages at 25, 30, 37, 45, 55 and 65 °C. The samples were filtered with a 0.20 µm filter after 1 h of incubation. A serial dilution was made with DNB medium, which was immediately spotted on double agar overlay plates to determine the phage titers. To assess viability at common storage temperatures, the phages were incubated in DNB medium without glycerol at -80, -20 and 4 °C for seven days. To determine pH stability, 100 µl of the phage suspensions was added to 900 µl DNB medium that was pH adjusted using 1 M HCl or 1 M NaOH. The samples were incubated at 30 °C for 1 h before filtration. A serial dilution was made with DNB medium, which was immediately spotted on double agar overlay plates to determine the phage titer. All the phage stability measurements were performed in triplicate. All statistical analyses were performed with a Student’s t-test.
DNA extraction and phylogenetic analysis
DNA isolation, whole genome sequencing and de novo assembly of Vanseggelen and Verabelle were performed by the Institute Pasteur using Illumina NovaSeq PE150 sequencing (Paris, France). The complete genome sequences of Vanseggelen and Verabelle were deposited at GenBank under accession number OQ970438 and OQ970439, respectively. Linear maps of the genomes were constructed by Geneious Prime 2022.1.1 (https://www.geneious.com) and genomes were de novo assembled using SPAdes v3.15.5 and aligned starting by the phage terminase16.
A viral proteomic phylogenetic tree and a genomic alignment based on the whole genome sequences of Streptomyces phages Alsaber, Amela, Endor1, Endor2, Hydra, Indigo, Joe, Pablito, phiCAM, Saftant, Sitrop, Verse and Yosif were constructed using ViPtree: the Viral Proteomic Tree version 3.517. The genome sequences of these phages were acquired from the National Center for Biotechnology Information (NCBI). Pairwise genome comparisons of phages within the Camvirus genus were visualized and performed with clinker18. A heatmap to determine intergenomic relatedness was constructed with the use of VIRIDIC19.
Results
Morphology and host range
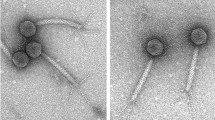
Two novel phages infecting Streptomyces have been isolated from soil samples in the Netherlands. Both phages produced small, clear plaques on the host strain S. coelicolor while using the double agar overlay method (Fig. 1a). TEM images revealed that Vanseggelen has an icosahedral capsid head of 53.7 nm ± 7.9 nm (n = 5) and a long non-contractile tail of 228.7 ± 7.5 nm (n = 4). Verabelle has an icosahedral capsid head of 63.2 ± 1.2 nm (n = 3) and a long non-contractile tail of 200.4 ± 1.5 nm (n = 3) (Fig. 1b).
A host range analysis of Verabelle performed on ten different actinomycetes strains revealed that all nine Streptomyces strains could be infected, and the phage produced clear as well as turbid lysis zone morphologies depending on the host (Table 1). Vanseggelen was not able to infect Streptomyces griseus, Streptomyces lividans and MBT13, but could infect the other six Streptomyces strains tested (Table 2). However, both phages were unable to infect K. virifidaciens. When distinct clear individual plaques were visible in a given strain, the efficiency of plating (EOP) was determined relative to the original host S. coelicolor.
One-step growth curve
The one-step growth curve of Vanseggelen shows a latent period of 140 min, with a burst size of 17 virions per bacterial cell (Fig. 2a). The one-step growth curve of Verabelle also shows a latent period of 140 min, but with a burst size of 8 virions per bacterial cell (Fig. 2b). The latent period of both phages is relatively long compared to other Streptomyces phages12,20.
Phage stability
Vanseggelen and Verabelle were stable up to 45 °C without a significant reduction in viability (Fig. 3). The absence of plaques at 65 °C indicates that Vanseggelen was not viable after incubation at 65 °C for 1 h. In addition, Vanseggelen made significantly less plaques after storing the phage samples at − 80 °C for one week (Fig. 4a), while Verabelle could be incubated at temperatures of − 80, − 20 and 4 °C without loss in viability (Fig. 4b). The optimal pH range for Vanseggelen was between pH 6.0–10.0, from which a decreasing trend in viability was visible at both lower and higher acidity (Fig. 5a). Verabelle remained relatively stable at pH values from 4.0 to 10.0 and no viability was detected at pH values of 3.0 and 11.0 (Fig. 5b).
Temperature stability of Vanseggelen and Verabelle. (a) Vanseggelen was stable from 30 up to 45 °C without a reduction in viability. However, the infectivity significantly decreased when the phage was incubated for one hour at 55 °C or higher. (b) Verabelle remains stable between temperatures ranging from 25 to 45 °C, but the viability significantly decreased at higher temperatures.
Genomic analysis
Whole genome sequencing and de novo assembly of Streptomyces phage Vanseggelen revealed a double-stranded DNA genome of 48,720 base pairs with a G + C content of 65.6% and a 3′-cohesive termini of CGGTACGTGAT. This genome contains 72 potential coding sequences (CDSs), of which 37 encode for hypothetical proteins and the function of 35 CDSs could be predicted (Fig. 6a). Verabelle revealed a slightly smaller double-stranded DNA genome of 48,126 base pairs with G + C content of 65.0% and a 3′-cohesive termini of CGTACCGTCAT. Again, the genome contains 72 potential CDSs, of which 40 are hypothetical proteins and the function of 32 CDSs could be predicted (Fig. 6b). The three additional CDSs that could be predicted in Vanseggelen are an additional minor tail protein, an RNA-polymerase sigma factor and a nucleoid associated Lsr2-like protein. Both genomes possess a putative integrase, indicating the capability of a temperate lifestyle. The Actinobacteriophage Database places both phages in Cluster BD, Subcluster BD39. Although the genomes of both phages are similar, there are some sequence variations that result in differences at the protein level (Fig. 7).
Schematic representation of the double-stranded DNA sequence of Vanseggelen and Verabelle. (a) Overview of the linear genome of Vanseggelen. (b) Overview of the linear genome of Verabelle. (a)/(b) Arrowheads are indicated in the direction of the coding sequence (CDS). Hypothetical proteins are displayed in grey and the predicted proteins are color coded as follows: yellow for head and packaging, red for tail, orange for DNA, RNA and nucleotide metabolism, green for connector, purple for lysis, light blue for transcription regulation, dark blue for integration and excision and pink for other. The blue line represents the GC content, while the green line represents the AT content. The genome maps were generated through Geneious Prime 2022.1.1. (https://www.geneious.com).
Genomic alignment of Vanseggelen and Verabelle. The predicted genes are indicated and the colored vertical blocks between the genomes indicate the level of nucleotide similarity. The genome alignment was generated through ViPTree17.
Phylogenetic analysis (Fig. 8a) based on the whole genome sequences of the closest-related phages, revealed that Vanseggelen is most closest related to Streptomyces phage Endor2 and Verabelle is most closely related to Streptomyces phage Sitrop. They are more distantly related to Streptomyces phages Pablito, Hydra and Yosif, which belong to different genera. These results indicate that both phages belong to the same genus, namely Camvirus. The genomic alignment of all phages within the Camvirus genus (Fig. 8b) show that phage Vanseggelen shares a similar tail fiber protein with phage Endor2, phiCAM, Alsaber, Amela and Endor2, while Verabelle shares a similar tail fiber protein with phage Sitrop, Saftant and Joe. The lack of genome similarity in tail fiber proteins between phage Vanseggelen and Verabelle might explain their difference in host range as seen in Tables 1 and 2. Additionally, phage Vanseggelen and Verabelle have a different integrase protein, indicating that they will likely integrate at different places in their host.
Phylogeny and genome similarity of Vanseggelen and Verabelle based on the whole genome sequences. (a) Streptomyces phages Vanseggelen and Verabelle belong to the genus Camvirus, within the subfamily Arquattrovirinae. The viral proteomic tree was generated through ViPTree17. (b) Genome alligment of all phages within the Camvirus genus starting at the terminase. Vanseggelen and Verabelle don’t share similarity in the tail fiber protein and integrase as indicated by light blue arrowheads in Vanseggelen and dark blue in Verabelle.
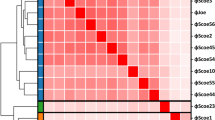
Next, a heatmap of the intergenomic relatedness (Fig. 9) was made. The two phages show a relatedness of 75.0 to each other and Vanseggelen is most related to phage Endor2 with a relatedness of 82.4, while Verabelle is most related to phage Sitrop with 77.8. The Bacterial Viruses Subcommittee of the International Committee on Taxonomy of Viruses has determined that if a phage exhibits ≥ 70% and < 95% nucleotide relatedness, the phage belongs to a new species in an undefined or existing genus. A sequence similarity of ≥ 95% is the cut-off for identifying a phage as a new species. Since the relatedness of Vanseggelen and Verabelle is lower than 95% compared to the top ten most similar Streptomyces phages, these phages are both considered as new species according to the International Committee on Taxonomy of Viruses (ICTV)21. A taxonomic proposal has been submitted to ICTV to officially recognize Vanseggelen and Verabelle as part of the genus Camvirus, as a new species called “Camvirus vanseggelen” and “Camvirus verabelle”.
Intergenomic relatedness. Genomes of the top ten most similar Streptomyces phages Alsaber, Amela, Endor2, Endor1, phiCAM, Joe, Saftant and Sitrop were used to show relatedness. The genomes of Streptomyces phages Pablito, Hydra and Yosif are classified in different genera as shown in green. The heatmap was calculated by VIRIDIC19.
Discussion
In this study, the newly isolated Streptomyces phages Vanseggelen and Verabelle have been fully characterized and sequenced. These results increase our insights on phage-host dynamics in Streptomyces and expand available, sequenced and characterized Streptomyces phages. TEM images showed that both phages have a long, flexible non-contractile tail with an icosahedral capsid head typical for siphoviruses. Both phages belong to the class Caudoviricetes, which was confirmed by whole genome sequencing. Since the majority of the sequenced Streptomyces phages have a long non-contractile tail, this tail morphotype is possibly necessary for effectively penetrating the thick peptidoglycan layer of Gram-positive bacteria. The host range analysis showed that both phages can form lysis zones with a turbid morphology with some host strains, indicating that Vanseggelen and Verabelle are likely able to switch from a lytic to a lysogenic lifecycle. Indeed, since all other phages belonging to the genus Camvirus are temperate phages and whole genome sequencing revealed that both genomes contain an integrase protein, we can conclude that Vanseggelen and Verabelle are temperate phages as well22.
Of all sequenced Streptomyces phages available in the Actinobacteriophage database9, we have calculated that the average genome length is 73,556 bp with a G + C content of ~ 61%, which is relatively high, and could be explained by the relatively high G + C-content of their bacterial hosts. Both Vanseggelen and Verabelle have a double-stranded DNA with a genome length of 48,720 bp and 48,126 bp, which is a bit smaller than average. Both phages were able to infect several different Streptomyces strains, but could not infect K. virifidaciens. Kitasatospora is the sister genus of Streptomyces that also belongs to the phylum Actinobacteria. Their morphology and lifecycle are similar to Streptomyces bacteria, but Kitasatospora is usually identified by the fact that they cannot be infected by Streptomyces phages23,24. Possibly, because of the difference in cell wall composition, since the cell wall of Kitasatospora strains contain both LL- and meso-diaminopimelic acid25. It would be interesting to isolate and characterize phages on Kitasatospora species in the future to dissect differences in infectivity between Streptomyces and Kitasatospora phages.
Despite the high degree of nucleotide similarity between the genomes of Vanseggelen and Verabelle, there were sequence variations that resulted in differences at the protein level, which could also account for the differences in host range. Streptomyces phage Verabelle showed a slightly broader host range infectivity than Vanseggelen, which could be explained by potential phage specific defense mechanisms or the difference in tail fiber proteins. Additionally, a different integrase protein was found, which suggest that both the tail fiber protein and integrase are a common source of variation in closely related phages within the Camvirus genus.
The one-step growth curve of Vanseggelen and Verabelle showed a long latent period of 140 min and a relatively low burst size, even when compared to other Streptomyces phages12,20. The low burst size could be the result of the morphological complexity of Streptomyces20. The meaning of MOI loses its significance once spores have germinated and a mycelium is formed as multiple phages are able to attach, but the mycelium would still be counted as one colony forming unit15,26. This notion of MOI in Streptomyces-phage dynamics complicates quantitative experiments and could explain the low burst sizes shown in this study.
Temperature plays a fundamental role in phage attachment, penetration, multiplication, and the length of the latent period, and is therefore a crucial factor for phage viability27. Both phages remain stable at temperatures ranging from 30 to 45 °C while phage viability decreases at higher temperatures. These results are consistent with a previous study that shows that phages most likely lose their DNA from the capsid head between temperatures of 50 and 60 °C due to the tail complex breaking of the capsid head28. This results in the marked loss of infectivity and structural changes in the capsid proteins. It is suspected that denaturation of DNA and proteins within the capsid head only occurs at temperatures above 80 °C29. Another important factor for phage viability is the acidity of the environment in which the phage persists. Streptomyces strains can grow well between a pH range of 5.5–11.528. However, both Vanseggelen and Verabelle remain stable at pH values from 4.0 to 10.0. Although the exact effect of acidity on the capsid head or tail has not been clearly elucidated yet, a previous study showed that extreme pH values, such as highly acidic or highly alkaline conditions, can result in the denaturation or structural changes to phage capsid head proteins30.
The Streptomyces phages described in this study can help to discover new insights in the interactions between Streptomyces bacteria and bacteriophages. More fundamental research is needed to study the attachment, lifecycle and infection of phages targeting multicellular bacteria, like Streptomyces. Since Streptomyces are producing a wide range of clinically relevant antibiotics, such studies become increasingly important in order to keep exploiting Streptomyces’ antimicrobial potential and abolish phage infection in various industries like biotechnology, agriculture and medicine.
Data availability
The genomes of Streptomyces phages Vanseggelen and Verabelle are available on NCBI (GenBank Accession No. OQ970438 and OQ970439). Both phages are available at the Dutch “Fagenbank” in Delft.
References
Seshadri, R. et al. Expanding the genomic encyclopedia of Actinobacteria with 824 isolate reference genomes. Cell Genom. 2, 100213. https://doi.org/10.1016/j.xgen.2022.100213 (2022).
Bibb, M. J. Understanding and manipulating antibiotic production in actinomycetes. Biochem. Soc. Trans. 41, 1355–1364. https://doi.org/10.1042/BST20130214 (2013).
de Lima Procópio, R. E., da Silva, I. R., Martins, M. K., de Azevedo, J. L. & de Araújo, J. M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16, 466–471 (2012).
Lu, N. et al. Characterization and genome analysis of the temperate bacteriophage ΦSAJS1 from Streptomyces avermitilis. Virus Res. 265, 34–42. https://doi.org/10.1016/j.virusres.2019.03.006 (2019).
Luthe, T., Kever, L., Thormann, K. & Frunzke, J. Bacterial multicellular behavior in antiviral defense. Curr. Opin. Microbiol. 74, 102314 (2023).
Ongenae, V. et al. Reversible bacteriophage resistance by shedding the bacterial cell wall. Open Biol. 12(6), 210379 (2022).
Turner, D. et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 168, 74. https://doi.org/10.1007/s00705-022-05694-2 (2023).
Ackermann, H.-W. Frequency of morphological phage descriptions brief review. 24 (1992).
Russell, D. A. & Hatfull, G. F. PhagesDB: The actinobacteriophage database. Bioinformatics 33, 784–786. https://doi.org/10.1093/bioinformatics/btw711 (2017).
Benson, D. A. et al. GenBank. Nucleic Acids Res. 41, D36–D42. https://doi.org/10.1093/nar/gks1195 (2012).
Olsen, N.S., Kot, W., Junco, L.M.F. & Hansen, L.H. Exploring the Remarkable Diversity of Escherichia Coli Phages in the Danish Wastewater Environment, Including 91 Novel Phage Species (2020). https://doi.org/10.1101/2020.01.19.911818
Ongenae, V. et al. Genome sequence and characterization of Streptomyces phage pablito, representing a new species within the genus Janusvirus. Sci. Rep. 12, 17785. https://doi.org/10.1038/s41598-022-22784-y (2022).
Zhu, H. et al. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 160, 1714–1725. https://doi.org/10.1099/mic.0.078295-0 (2014).
Dowding, J. E. Characterization of a Bacteriophage Virulent for Streptomyces coelicolor A3 (2). J. Gen. Microbiol. 76, 163–176. https://doi.org/10.1099/00221287-76-1-163 (1973).
Rosner, A. & Gutstein, R. Adsorption of actinophage Pal 6 to developing mycelium of Streptomyces albus. Can. J. Microbiol. 27, 254–257. https://doi.org/10.1139/m81-039 (1981).
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A. & Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 70(1), e102 (2020).
Nishimura, Y. et al. ViPTree: The viral proteomic tree server. Bioinformatics 33, 2379–2380. https://doi.org/10.1093/bioinformatics/btx157 (2017).
van den Belt, M. et al. CAGECAT: The CompArative GEne cluster analysis toolbox for rapid search and visualisation of homologous gene clusters. BMC Bioinform. 24, 181. https://doi.org/10.1186/s12859-023-05311-2 (2023).
Moraru, C., Varsani, A. & Kropinski, A. M. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12, 1268. https://doi.org/10.3390/v12111268 (2020).
Hardy, A., Sharma, V., Kever, L. & Frunzke, J. Genome sequence and characterization of five bacteriophages infecting Streptomyces coelicolor and Streptomyces venezuelae: Alderaan, Coruscant, Dagobah, Endor1 and Endor2. Viruses 12, 1065. https://doi.org/10.3390/v12101065 (2020).
Lefkowitz, E. J. et al. Virus taxonomy: The database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 46, D708–D717. https://doi.org/10.1093/nar/gkx932 (2018).
Groth, A. C. & Calos, M. P. Phage integrases: Biology and applications. J. Mol. Biol. 335, 667–678. https://doi.org/10.1016/j.jmb.2003.09.082 (2004).
Wellington, E. M. H., Stackebrandt, E., Sanders, D., Wolstrup, J. & Jorgensen, N. O. G. Taxonomic status of kitasatosporia, and proposed unification with Streptomyces on the basis of phenotypic and 16S RRNA analysis and emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int. J. Syst. Bacteriol. 42, 156–160. https://doi.org/10.1099/00207713-42-1-156 (1992).
Groth, I. et al. Five novel Kitasatospora species from soil: Kitasatospora arboriphila Sp. Nov., K. gansuensis Sp. Nov., K. nipponensis Sp. Nov., K. paranensis Sp. Nov. and K. terrestris Sp. Nov. Int. J. Syst. Evol. Microbiol. 54, 2121–2129. https://doi.org/10.1099/ijs.0.63070-0 (2004).
Takahashi, Y. Genus Kitasatospora, taxonomic features and diversity of secondary metabolites. J. Antibiot. 70, 506–513. https://doi.org/10.1038/ja.2017.8 (2017).
Gilmour, C. M., Noller, E. C. & Watkins, B. Studies on Streptomyces phage. J Bacteriol 78, 186–192 (1959).
Jończyk, E., Kłak, M., Międzybrodzki, R. & Górski, A. The influence of external factors on bacteriophages—Review. Folia Microbiol. 56, 191–200. https://doi.org/10.1007/s12223-011-0039-8 (2011).
Vörös, Z., Csík, G., Herényi, L. & Kellermayer, M. Temperature-dependent nanomechanics and topography of bacteriophage T7. J. Virol. https://doi.org/10.1128/JVI.01236-18 (2018).
Olson, M. R., Axler, R. P. & Hicks, R. E. Effects of freezing and storage temperature on MS2 viability. J. Virol. Methods 122, 147–152. https://doi.org/10.1016/j.jviromet.2004.08.010 (2004).
Sharma, M., Kumar, D. & Poluri, K. M. Elucidating the PH-dependent structural transition of T7 bacteriophage endolysin. Biochemistry 55, 4614–4625. https://doi.org/10.1021/acs.biochem.6b00240 (2016).
Acknowledgements
We would like to thank ICTV for their help in characterizing Streptomyces phage Vanseggelen and Verabelle and setting up the proposal to make both phages officially new species in the genus Camvirus, within the subfamily Arquattrovirinae. In addition, we thank Laurence Ma, from Biomics Platform, C2RT, Institut Pasteur, Paris, France, supported by France Génomique (ANR-10-INBS-09) and IBISA.
Funding
This research was funded by a VICI grant, grant number VI.C.192.002 and an NWO XS grant, grant number OCENW.XS.041. Microscope access was funded by the Netherlands Electron Microscopy Infrastructure Grant 84.034.014.
Author information
Authors and Affiliations
Contributions
Conceptualization, V.O., D.R., A.B. and D.C.; methodology, all; validation, all; formal analysis, all; investigation, A.K., V.N., V.O., H.S. and F.T.; resources, A.B. and D.C.; data curation, V.O., H.S. and F.T.; writing—original draft preparation, A.K., V.N. and V.O.; writing—review and editing, all; visualization, A.K., V.N. and V.O.; supervision, A.B., D.R. and D.C.; project administration, A.B., D.R. and D.C.; funding acquisition, A.B. and D.C. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ongenae, V., Kempff, A., van Neer, V. et al. Genome sequence and characterization of Streptomyces phages Vanseggelen and Verabelle, representing two new species within the genus Camvirus. Sci Rep 13, 20153 (2023). https://doi.org/10.1038/s41598-023-47634-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47634-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.