Abstract
Streptomycetes are ubiquitous soil bacteria. Here we report the complete and annotated genome sequence and characterization of Streptomyces phage Pablito, isolated from a soil sample in Haarlem, the Netherlands using Streptomyces lividans as host. This phage was able to infect a diverse range of Streptomyces strains, but none belonging to the sister genus Kitasatospora. Phage Pablito has double-stranded DNA with a genome length of 49,581 base pairs encoding 76 putative proteins, of which 26 could be predicted. The presence of a serine integrase protein indicated the lysogenic nature of phage Pablito. The phage remained stable over a wide range of temperatures (25–45 °C) and at pH ≥ 7.0, but lost infectivity at temperatures above 55 °C or when the pH dropped below 6.0. This newly isolated phage is closely related to Streptomyces phage Janus and Hank144 and considered a new species classified in the genus Janusvirus, within the subfamily Arquattrovirinae.
Similar content being viewed by others
Introduction
Streptomyces spp. are Gram-positive, spore-forming bacteria that thrive in almost all soil environments. They are well-known for their ability to produce a wide array of clinically-relevant antibiotics, such as streptomycin, neomycin, chloramphenicol and many more1,2,3. Most research has been focused on the industrial use of these bacteria and their secretion of secondary metabolites4,5. However, it remains largely unknown how bacteriophages can recognize, attach and infect these multicellular bacteria. Streptomyces have an unusual lifecycle, which starts when spores encounter a favorable environment. This triggers germination and outgrowth into a so-called vegetative mycelium. After a period of vegetative growth, aerial hyphae are formed that can develop into chains of spores. These spores can be dispersed to more favorable environments to establish new colonies. Previous research has shown that Streptomyces phages can only infect young vegetative mycelia, but are unable to infect spores6,7. However, most bacteriophage research has been focused on unicellular bacteria, while the interactions and underlying mechanisms of bacteriophages infecting multicellular bacteria, like Streptomyces, remains largely unexplored. Besides the phage growth limitation system8,9, chemical defense10 and recently discovered cell wall-deficiency11, many more defense mechanisms against phage attack might be present in Streptomyces. In addition, relatively few Streptomyces phages have been properly sequenced and characterized in comparison to other genera. Therefore, it is important to isolate new phages infecting multicellular bacterial species, allowing us to expand our knowledge about the immense viral diversity.
In this paper, we describe a newly discovered Actinobacteriophage, called Streptomyces phage Pablito, which was isolated from a soil sample in the Netherlands. This phage belongs to the genus Janusvirus, within the subfamily Arquattrovirinae. Phage Pablito is a temperate phage with an icosahedral capsid head of 56.1 ± 1.3 nm and a long non-contractile tail of 163.5 ± 0.4 nm. Finally, we provide a whole genome analysis of this newly identified phage. Together, these results shed light on phage taxonomy and can expand the knowledge on phage-Streptomyces interactions in the future.
Materials and methods
Bacteriophage isolation
Phage Pablito was isolated from a soil sample obtained from the National Park Zuid-Kennemerland in the Netherlands (N52°23′31″, E4°34′49″). To this end, 0.5 g soil was mixed with spores from the host S. lividans strain 1326 in 10 ml Difco Nutrient Broth (DNB) (BD Biosciences) supplemented with 0.5% glucose and 4 mM Ca(NO3)2 and incubated overnight at 30 °C, while shaking at 130 RPM. After 24 h, the culture was centrifuged for 10 min at 4000 g and the supernatant containing phages was filtered through a 0.22 µm filter (Millipore). The supernatant was subsequently spread onto a DNB agar plate, followed by applying an overlay (in DNB soft agar) containing 103 spores ml−1 from S. lividans. The plate was incubated at 30 °C for 24–48 h before plaque formation was assessed. Single plaques were purified by streaking them twice to a fresh double agar overlay plate containing the bacterial host. High titre single viral stocks were obtained by picking purified plaques and incubating them with S. lividans in DNB medium for 10 min before plating on double-agar overlay DNB plates. Viral lysates were collected by flooding the plate with 24 ml DNB and gently rocking the plate for 2 h. The supernatant was filtered through a 0.22 µm filter. Collected phages were stored at 4 °C.
Host range analysis
The host range of phage Pablito was determined using serial dilutions on double agar overlay plates. In short, 3 µl of the phage dilution was spotted in duplicate on a bacterial lawn of different Streptomyces and Kitasatospora strains from the Microbial biotechnology (MBT) collection at Leiden University12, which represents an in-house collection of isolated actinomycetes. The plates were incubated at 30 °C for at least 24 h before lysis was observed. The presence of a lysis zone was scored as a positive result. The efficiency of plating (EOP) for a given strain was calculated relative to the host strain S. lividans when plaques were observed.
Stability of phage Pablito
A one step-growth curve analysis was performed in triplicate to calculate the latent period and phage burst size13. In short, 105 spores ml−1 of S. lividans were allowed to germinate in 10 ml DNB medium at 30 °C for 5 h. At T = 0, the culture was infected with phage Pablito at Multiplicity of Infection (MOI) = 0.01 and 100 µl was sampled at 10 min intervals up to 180 min. The samples were filtered, diluted and immediately plated on double agar plates. Burst size was calculated using the following formula:
The thermal stability of phage Pablito was determined by incubating 1 ml of lysate (106 PFU ml−1) at 25, 30, 37, 45, 55 and 65 °C. The samples were filtered after 1 h of incubation and immediately plated on double agar plate to determine the phage titre. For pH stability, 10 µl of a phage Pablito suspension (106 phages ml−1) was added to 990 µl DNB medium adjusted to different pH values (2–9) using 6 M HCl or 1 M NaOH. The tubes were then incubated at 30 °C for 1 h before the samples were filtered through a 0.22 µm filter and plated using the double agar overlay method. All measurements were performed in triplicate.
DNA extraction and phylogenetic analysis
DNA of phage Pablito was isolated using the Phage DNA Isolation Kit from Norgen (Thorold, Canada) according to the manufacturers protocol. After isolation, multiple samples were pooled and DNA was concentrated using the Concentrator Plus (Eppendorf) for 20 min. Whole-genome sequencing and de novo assembly of phage Pablito was performed by BaseClear using Illumina NovaSeq PE150 sequencing with a mean coverage of 118.21 (Leiden, the Netherlands). The complete genome sequence of Streptomyces phage Pablito was deposited at GenBank under accession number OK412919. A linear map of the genome was constructed by Geneious Prime 2020.1.2 and PhageTerm was used to determine genome ends14.
Phylogenetic analysis was performed using the putative protein sequence of the major capsid head protein. The protein sequences of Streptomyces phage Janus, Hank144, Pablito and Joe were acquired from NCBI and aligned using MUSCLE15. A Maximum Likelihood phylogenetic tree was constructed through Molecular Evolutionary Genetics Analysis Version X (MEGAX) using 100 bootstrap replicates to estimate the genus of Phage Pablito.
Electron microscopy
Two liquid cultures of S. lividans infected with phage Pablito were filtered, combined and precipitated overnight with 5 × PEG solution (20% PEG8000 in 2.5 M NaCl) at 4 °C and centrifuged for 70 min at 3500 g after which the supernatant was discarded. Of the remaining pellet, 3 µl was placed on a glow discharged 200 mesh carbon coated copper grid (EMS) and allowed to set for 30 s before excess sample solution was removed by filter paper. The phages were stained with 2% uranyl acetate for 45 s and air-dried for an additional 30 min. The grids were observed using a single tilt specimen holder inside a 120 kV Talos L120C TEM with a Lab6 electron source and Ceta detector at the Netherlands Center for Nanoscopy (NeCEN, Leiden).
Results and discussion
Morphology and host range determination of Pablito
Bacteriophage Pablito produced clear small plaques on the S. lividans host strain using the double agar overlay method (Fig. 1a). Transmission electron microscopy (TEM) was used to reveal the morphology of phage Pablito. This demonstrated that phage Pablito has an icosahedral capsid head of 56.1 ± 1.3 nm (n = 3) and a long non-contractile tail of 163.5 ± 0.4 (n = 3) (Fig. 1b). The long, flexible tail together with the icosahedral capsid head were the first indication that this phage is a siphovirus belonging to the class Caudoviricites, which was further confirmed by whole-genome sequencing (see below).
A host range assay on 10 actinomycetes revealed that all eight Streptomyces strains tested could be infected by phage Pablito (Table 1). Depending on the host strain, some lysis zones showed a turbid morphology, which is common for several Streptomyces phages16, and was the first indication that phage Pablito might be a temperate phage. The EOP for a strain relative to the host strain S. lividans from which the phage was isolated, is given when plaques were countable. On the contrary, the two species that remained uninfected (i.e. MBT66 and K. viridifaciens) belong to the sister genus Kitasatospora, members of actinomycetes which are morphologically similar to Streptomyces but generally classified by their resistance against Streptomyces phages17,18.
Genomic analysis
Whole genome sequencing and de novo assembly of phage Pablito revealed a double-stranded DNA genome of 49,581 base pairs. While its host S. lividans has relatively high G + C content of 72%, Streptomyces phage Pablito has a G + C content of 66.4% and a 3′-cohesive termini of CGCCGTGTCTT (Fig. 2). The genome contains 76 potential coding sequences (CDSs), of which 50 encode for hypothetical proteins, while the function of 26 CDSs could be predicted. The latter were categorized as morphogenesis proteins, nucleotide metabolism and DNA replication proteins, lysogenic module and a bacterial lysis protein as indicated in Fig. 2.
Schematic representation of the dsDNA genome of Streptomyces phage Pablito. Arrowheads are indicated in the direction of the CDS. Hypothetical proteins are displayed in yellow and the color code for predicted proteins is as follows: green for nucleotide metabolism and DNA replication, orange for morphogenesis, red for lysogeny and dark blue for bacterial lysis. The green line represents AT content, while the blue line represents GC content. The genome map was generated through Geneious Prime 2020 1.2.
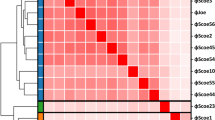
Phylogenetic analysis (Fig. 3a) based on the putative major capsid head protein revealed that Streptomyces phage Pablito is similar to Streptomyces phage Janus and Streptomyces phage Hank144, while Streptomyces phage Joe is more distantly related. Based on the criteria from the Bacterial Viruses Subcommittee, 70% nucleotide identity is established as the cut-off for genera, while 95% identity assigns phages to the same species19. As calculated by the intergenomic distance calculator VIRIDIC20, Streptomyces phage Pablito shows 70.0 and 72.5% relatedness to Streptomyces phages Janus and Hank144, respectively (Fig. 3b). Since the relatedness of Streptomyces phage Pablito is lower than 95%, this phage is considered a new species according to the International Committee on Taxonomy of Viruses (ICTV)20. Because Streptomyces phage Janus and Streptomyces phage Hank144 are both temperate phages and whole genome sequencing revealed the presence of a serine integrase protein in Streptomyces phage Pablito, we conclude that phage Pablito is a temperate phage as well21. Together, these data indicate that Streptomyces phage Pablito is a newly discovered species that can be included in the genus Janusvirus. A taxonomic proposal has been submitted to ICTV to officially recognize phage Pablito as part of the genus Janusvirus as a new species called “Janusvirus Pablito”. The Actinobacteriophage Database (https://phagesdb.org/) places phage Pablito in Cluster BD, Subcluster BD2. In addition, phage Pablito is peripherally related to two prophages found in Streptomyces finlayi strain NBSH44 and Streptomyces venezuelae strain ATCC 14583.
Streptomyces phage Pablito belongs to the subfamily Arquattrovirinae, genus Janusvirus. (a) Evolutionary history of phage Pablito based on the major capsid head protein. Total of 100 bootstraps replicates were used to calculate the Maximum-Likelihood tree in MEGA.X. (b) Heatmap calculated by VIRIDIC20 to show pairwise intergenomic distance/relatedness amongst phage genomes. Genomes of Streptomyces phage Janus (NC_054660.1) and Streptomyces phage Hank144 (NC_054661.1) were used to show relatedness, while the genome of Streptomyces phage Joe (NC_054674.1) was used as an outlier.
One-step growth curve
Streptomyces phage adsorption was found to be maximal for germlings16. We therefore incubated S. lividans spores first for 5 h in DNB medium before infecting the culture with bacteriophages. The one-step growth curve of phage Pablito showed a relatively long latent period of 80 min, with a burst size of 22 virions per cell (Fig. 4). However, another rise of viral titer was observed around 150 min, which can happen when multiple phages bind to one germinated spore, as a sign of delayed adsorption or as an indication of a lysogenic infection cycle22. The highest titre obtained was around 106 PFU ml−1 after 24 h. A different range of MOI, longer incubation periods and even flooding of a confluent plaque plate did not result in an increase of the total number of phages ml−1.
Phage stability
We also assessed the effect of temperature and pH on phage viability. Phage Pablito was stable up to 45 °C without a reduction in viability (Fig. 5a). However, the infectivity significantly decreased when the phage was incubated for an hour at 55 °C or higher. Phage Pablito was relatively stable at pH 7.0–9.0, while infectivity steeply declined when the pH was lower than 6.0 (Fig. 5b). No plaques were found when the pH was below 4.0, while only a small amount of activity was retained at pH 5.0 and pH 6.0.
Conclusion
This study presents Streptomyces phage Pablito as a temperate siphophage being able to infect diverse Streptomyces strains. Genomic analysis and TEM images showed that this phage could be classified as a new species in the genus Janusvirus within the subfamily Arquattrovirinae. Members of this subfamily/genus have long non-contractile tails and icosahedral capsid heads. Phage Pablito showed a burst size of 22 virions per cell and was stable at temperatures ranging from 25 to 45 °C and a pH between 7.0 and 9.0. This phage can be used in future studies to shed new light on bacteriophage-Streptomyces interactions and disclose how this phage affects soil community.
Data availability
The genome of Streptomyces phage Pablito is available on NCBI (GenBank Accession No. OK412919).
References
Fernández-Martínez, L. T. et al. New insights into chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712. Antimicrob. Agents Chemother. 58, 7441–7450 (2014).
de Pocópio, R. E. L. et al. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16, 466–471 (2012).
Zheng, J., Li, Y., Guan, H., Zhang, J. & Tan, H. Enhancement of neomycin production by engineering the entire biosynthetic gene cluster and feeding key precursors in Streptomyces fradiae CGMCC 4.576. Appl. Microbiol. Biotechnol. 103, 2263–2275 (2019).
Hwang, K.-S., Kim, H. U., Charusanti, P., Palsson, B. Ø. & Lee, S. Y. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 32, 255–268 (2014).
van Wezel, G. P. & McDowall, K. J. The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat. Prod. Rep. 28, 1311–1333 (2011).
Alstyne, M. H. V., Otto, R. H. & McCoy, E. Characteristics of Streptomyces griseus strains resistant to phage. J. Bacteriol. 70, 113–119 (1955).
Jones, L. A. & Bradley, S. The life-cycle of an actinophage for Streptomyces venezuelae. Microbiology 40, 191–198 (1965).
Hoskisson, P. A., Sumby, P. & Smith, M. C. The phage growth limitation system in Streptomyces coelicolor A (3) 2 is a toxin/antitoxin system, comprising enzymes with DNA methyltransferase, protein kinase and ATPase activity. Virology 477, 100–109 (2015).
Sumby, P. & Smith, M. C. Genetics of the phage growth limitation (Pgl) system of Streptomyces coelicolor A3 (2). Mol. Microbiol. 44, 489–500 (2002).
Kronheim, S. et al. A chemical defence against phage infection. Nature 564, 283–286. https://doi.org/10.1038/s41586-018-0767-x (2018).
Ongenae, V. et al. Reversible bacteriophage resistance by shedding the bacterial cell wall. Open Biol. https://doi.org/10.1098/rsob.210379 (2022).
Zhu, H. et al. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 160, 1714–1726. https://doi.org/10.1099/mic.0.078295-0 (2014).
Dowding, J. Characterization of a bacteriophage virulent for Streptomyces coelicolor A3 (2). Microbiology 76, 163–176 (1973).
Garneau, J. R., Depardieu, F., Fortier, L.-C., Bikard, D. & Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 7, 1–10 (2017).
Edgar, R. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Hardy, A., Sharma, V., Kever, L. & Frunzke, J. Genome sequence and characterization of five bacteriophages infecting Streptomyces coelicolor and Streptomyces venezuelae: Alderaan, Coruscant, Dagobah, Endor1 and Endor2. Viruses 12, 1065 (2020).
Groth, I. et al. Five novel Kitasatospora species from soil: Kitasatospora arboriphila sp. nov., K. gansuensis sp. nov., K. nipponensis sp. nov., K. paranensis sp., nov. and K. terrestris sp. nov. Int. J. Syst. Evolut. Microbiol. 54, 2121–2129. https://doi.org/10.1099/ijs.0.63070-0 (2004).
Wellington, E. M. H., Stackebrandt, E., Sanders, D., Wolstrup, J. & Jorgensen, N. O. G. Taxonomic status of Kitasatosporia, and proposed unification with Streptomyces on the basis of phenotypic and 16S rRNA analysis and emendation of Streptomyces Waksman and Henrici 1943, 339(AL). Int. J. Syst. Bacteriol. 42, 156–160. https://doi.org/10.1099/00207713-42-1-156 (1992).
Turner, D., Adriaenssens, E. M., Tolstoy, I. & Kropinski, A. M. Phage annotation guide: Guidelines for assembly and high-quality annotation. PHAGE 2, 170–182 (2021).
Moraru, C., Varsani, A. & Kropinski, A. M. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12, 1268 (2020).
Groth, A. C. & Calos, M. P. Phage integrases: Biology and applications. J. Mol. Biol. 335, 667–678 (2004).
Dennehy, J. J. & Abedon, S. T. Phage infection and lysis. Bacteriophages Biol., Technol. Ther., 341–383. https://doi.org/10.1007/978-3-319-41986-2_53 (2021).
Acknowledgements
We would like to thank ICTV for their help in characterizing phage Pablito and setting up the proposal to make phage Pablito officially a new species in the genus Janusvirus, subfamily Arquattrovirinae.
Funding
This work was funded by a VICI grant from the Dutch Research Council (NWO) to DC (Grant Number VI.C.192.002) and an NWO XS grant to AB (Grant Number OCENW.XS.041). Microscope access was supported by the Netherlands Center for Electron Nanoscopy and partially funded by the Netherlands Electron Microscopy Infrastructure Grant 84.034.014.
Author information
Authors and Affiliations
Contributions
The experimental plan was designed by V.O., A.B. and D.C.. The phage genetics were analyzed by V.O. and A.M.K.. The paper was written by V.O. and the final manuscript was shaped, edited and reviewed by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ongenae, V., Azeredo, J., Kropinski, A.M. et al. Genome sequence and characterization of Streptomyces phage Pablito, representing a new species within the genus Janusvirus. Sci Rep 12, 17785 (2022). https://doi.org/10.1038/s41598-022-22784-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22784-y
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.