Abstract
Small species with high home fidelity, high ecological specialization or low vagility are particularly prone to suffer from habitat modification and fragmentation. The Lima leaf-toed gecko (Phyllodactylus sentosus) is a critically endangered Peruvian species that shelters mostly in pre-Incan archeological areas called huacas, where the original environmental conditions are maintained. We used genotyping by sequencing to understand the population genomic history of P. sentosus. We found low genetic diversity (He 0.0406–0.134 and nucleotide diversity 0.0812–0.145) and deviations of the observed heterozygosity relative to the expected heterozygosity in some populations (Fis − 0.0202 to 0.0187). In all analyses, a clear population structuring was observed that cannot be explained by isolation by distance alone. Also, low levels of historical gene flow were observed between most populations, which decreased as shown in contemporary migration rate analysis. Demographic inference suggests these populations experienced bottleneck events during the last 5 ka. These results indicate that habitat modification since pre-Incan civilizations severely affected these populations, which currently face even more drastic urbanization threats. Finally, our predictions show that this species could become extinct in a decade without further intervention, which calls for urgent conservation actions being undertaken.
Similar content being viewed by others
Introduction
Habitat loss, degradation and fragmentation stand out among the main anthropogenic threats to biodiversity1,2. One of the primary consequences of these anthropogenic actions is the reduced connectivity of patches of natural habitat, which leads to decreased gene flow among isolated populations that can result in depleting their genetic diversity and increasing genetic drift3,4,5.
Reptiles are a highly diverse taxonomic group that, despite its ecological importance6 and high sensitivity to human landscape modification7,8, has been often overlooked in studies on the effect of anthropogenic habitat alterations9. Although this is a growing field, there is currently a bias toward studies done on temperate countries and on mammals and birds9. Considering the recent assessment that 21% of reptile species are endangered10 and the current expansion of cities and human settlements, the scant studies of the effects of human landscape change on reptile population genetics, especially in the Neotropics, represent a knowledge gap that needs to be addressed in order to develop effective conservation strategies9.
Small species with high home fidelity, high ecological specialization or low vagility are particularly prone to experience adverse effects due to habitat fragmentation5. As a case in point, the Lima leaf-toed gecko (Phyllodactylus sentosus), endemic of the city of Lima (Peru) and classified as critically endangered in the IUCN Red List of Threatened Species11, is constrained to small patches mainly represented by pre-Incan archeological areas called huacas and hilltops12,13,14,15 (Fig. 1), which feature arid soil with scant vegetation and small rocks that resemble the natural habitat previous to human occupancy11. Nowadays, these areas are isolated from each other by completely urbanized areas in Lima, being estimated not to exceed the 500 adult individuals in huacas with the largest census population size16,17,18. Also, unlike other Phyllodactylus species, P. sentosus has low climbing abilities due to their toe structure, which reduces the probability of dispersion and escape from invasive local threats15.
Distribution map of the Lima leaf-toed gecko. (A) Sites where the species had been previously recorded11,13,14,15 in the province of Lima (cross) and sampling localities used in this study (circle). (B) Photograph of a juvenile Lima leaf-toed gecko found in the Pachacamac huaca. Photo by A. Arana. Map created in ArcGIS 10.3. Free vector data from https://www.geogpsperu.com/2014/03/base-de-datos-peru-shapefile-shp-minam.html.
Huacas have been studied as islands of arid habitat in both urban and agricultural landscapes along the coast of Peru, affecting species distributions19,20. However, the effect of this “archeological landscape” on wildlife population genetics has not been analyzed yet. In recent years, the study of the effect of urbanization and human settlements on connectivity and gene flow between populations of wild species has been enhanced by the development of reduced representation sequencing techniques, such as genotyping by sequencing (GBS), which allows the discovery of thousands of genome-wide single nucleotide polymorphisms (SNPs) without the need for a reference genome21,22,23,24. These techniques have also increased the opportunities to explore the demographic history of populations and to perform population viability analysis to inform conservation actions.
In this study, we used GBS data to estimate genetic diversity and assess inbreeding to evaluate the current genetic status of Lima leaf-toed gecko populations. Also, we evaluated the genetic structure, historical and contemporary gene flow and demographic history to gain insights into the population genetics of the species. Finally, we performed a Population Viability Analysis (PVA) to simulate future scenarios of this species in absence of conservation actions.
Results
Data generation
We generated two datasets of GBS data for a total of 50 individuals from 12 localities of the Lima leaf-toed gecko. In the first dataset (i), 147 856 SNPs were obtained using 0.85 as the clustering threshold and 33 as the minimum number of samples that must have information at a locus for it to be retained. From the second dataset (ii), which had no missing data, 10,869 SNPs were obtained.
Genetic diversity and inbreeding
Population-level genetic diversity estimates are given in Table 1. The values observed were quite low for all populations (see “Discussion”). Expected Heterozygosity (He) values ranged from 0.0406 to 0.134 in P. sentosus populations, while values for other four Phyllodactylus species analyzed ranged from 0.2933 to 0.3961 (Fig. 2).
The results of Nucleotide Diversity (π) ranged between 0.0812 and 0.145. The population of Cerro Mulería has the highest value of Nucleotide Diversity (π) despite the fact that only 2 individuals were collected in this population. The deviation of the observed heterozygosity relative to the expected heterozygosity (FIS) ranged between –0.0202 and 0.0187 (Table 1).
Population genetic structure
In all analyses, a clear population structuring was observed, except Huaca San Marcos and Parque de Las Leyendas that were grouped in a single genetic cluster in most analyses (Fig. 3, see Supplementary Figs. S1, S2 online), corresponding to the pre-Hispanic Maranga archeological complex (see “Discussion”).
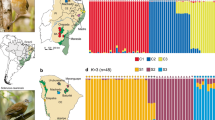
Genetic population structure of Phyllodactylus sentosus in Lima. (a) Discriminant analysis of principal components (DAPC), the axes represent the first two discriminant components. (b) STRUCTURE results from 3737 SNPs, with K = 4. (c) STRUCTURE results from 15,816 SNPs, with K = 6. Group P includes the Puruchuco and Pachacamac samples. Group N2 corresponds to samples from Paraíso, Mulería, UNI and one from SMP. Group N1 includes samples from Tambo Inga, Garagay and one from SMP (San Martin de Porres). (d) Map of the evaluated localities and the average of the coefficients of individual membership to genetic groups in each population from the STRUCTURE analysis with 15,816 SNPs. The size of the pie charts indicates the sample size. Altitudinal relief is represented in gray lines (altitudes higher than 25 m). Map created in ArcGIS 10.3. Free vector data from https://www.geogpsperu.com/2014/03/base-de-datos-peru-shapefile-shp-minam.html.
There are two main rivers that run through the city of Lima: Rímac River and Chillón River (Fig. 3d). In the DAPC analysis, the horizontal axis separates the Tambo Inga population (the only one north Chillón River) from the rest of populations (Fig. 3a). Also, the locations north Rímac River (Cerro Mulería, Garagay, Paraíso, Tambo Inga, San Martín de Porres, and UNI) were separated from the south Rímac River (Mateo Salado, Pachacamac, Parque de Las Leyendas, Pucllana, Puruchuco, and San Marcos) (Fig. 3a).
STRUCTURE analyses revealed high levels of population structuring (Fig. 3d). The dataset which included only SNPs (3373) called in the entire sample set, returned K = 4 as the most likely number of genetic clusters (Fig. 3b). Then we analyzed a larger SNP dataset (15,816) called in 90% of samples in each population, with K = 6 as the most likely number of genetic clusters (Fig. 3c). The main difference between the results of these two analyses occurred in the populations north Rímac River, which showed population structuring (Fig. 3b,c). At K = 4, only one genetic group emerged, while at K = 6 the populations north of the river split into two genetic clusters, called herein as N1 and N2, with the former being composed of populations near the river. In Mateo Salado population, the coefficients of individual genetic group membership (q) obtained from the samples were particularly high (> 0.97).
Gene flow
The divMigrate analysis on the dataset with no missing data yielded values of relative migration rates ranging from 0.03817 (between Mateo Salado and Tambo Inga) to 1 (between San Marcos and Parque de Las Leyendas) (Fig. 4A). The populations at the extremes of the geographical distribution of this species show the lowest migration rates, while those among the four largest census population size16,17,18 (San Marcos, Parque de Las Leyendas, Pucllana and Mateo Salado: Fig. 4, see Supplementary Table S3 online) were slightly higher, but still lower than 0.4, with exception of the populations of San Marcos and Parque de Las Leyendas that showed a high bidirectional gene flow with migration values of 0.99 and 1 (Fig. 4).The results also indicate especially low values of migration to and from Tambo Inga, the only locality north Chillón River (see Supplementary Table S3 online). On average, Tambo Inga had the lowest migration values in both directions, followed by populations located north Rímac River, such as Garagay and El Paraíso, and populations at the southern (Pachacamac) and eastern (Puruchuco), at the extremes of the species distribution (see Supplementary Table S3 online). The Mantel test showed a non-significant positive correlation (r = 0.30; p-value = 0.08), suggesting that gene flow patterns cannot be explained solely by isolation by distance.
On the other hand, contemporary gene flow rates are low across all localities, with the proportion of migrants in most populations being lower than 0.07 (< 7%). Contemporary migration rates between localities range from 0.0149 to 0.166, the latter being the proportion of migrants from Parque de Las Leyendas present in San Marcos. Conversely, the proportion of migrants from San Marcos in Parque de Las Leyendas is 0.0604, showing a lower migration rate than the ones observed between SMP and Parque de Las Leyendas (0.071), and Muleria and San Marcos (0.0712).
Demographic history
The demographic history estimation results for Parque de Las Leyendas, San Marcos, Mateo Salado and Huaca Pucllana revealed the same pattern in all populations, with an accentuated bottleneck and a posterior increase in population size (Fig. 5). The effective population size is estimated to have decreased 3100–5000 years ago and reached its minimum about 2000 years ago in Parque de Las Leyendas, San Marcos, and Mateo Salado. On the other hand, at Pucllana we estimated that the effective population began to decrease more than 10 thousand years ago, and dropped drastically 8000 years ago, reaching its minimum about 5000 years ago (Fig. 5C). The population size started to increase after the bottleneck and remained stable, except for Pucllana, where a decline in the effective population was observed about 200 years ago (Fig. 5C).
Demographic history of P. sentosus. Stairway plot demographic inference of the populations of Parque de Las Leyendas (A), San Marcos (B), Mateo Salado (C), and Huaca Pucllana (D). The red line shows the median estimate of the effective population size (Ne). The pink lines indicate the 75% (dark) and 95% (light) confidence intervals.
Population viability analysis
PVA using Vortex showed that all populations would experience a decrease in their population size resulting in extinction (see Supplementary Fig. S5 online). All simulations resulted in mean extinction times of 9.7 years or less (Table 2).
Discussion
Here we report the first population genomics survey of the critically endangered Lima leaf-toed gecko P. sentosus. According to our results, the populations of this species present low levels of genetic diversity, highly structured populations, and negligible levels of contemporary gene flow between populations. All these results together with the PVA simulations showed an unfavorable scenario for the species.
The locations that host the largest populations of this species are San Marcos, Parque de Las Leyendas, Pucllana and Mateo Salado16,17,18. Large population size is expected to maintain higher levels of genetic diversity. However, even in these localities, very low values relatives of gene diversity were found (He < 0.135). Our analysis of He values for four other Phyllodactylus species revealed values around twice as high as those obtained for P. sentosus when considering all sampled individuals. This reduced genetic diversity could be explained by a recent bottleneck (see below) and by the fragmentation of small populations in restricted areas with limited resources28.
Inbreeding in wildlife populations increases homozygosis, reduces genetic diversity, and can represent an important genetic threat if it derives into inbreeding depression, which decreases the overall fitness of individuals endangering the species survival29,30,31,32. The threat of inbreeding depression and reduced genetic diversity in isolated wild populations has been well-documented, such as in wild house sparrows (Passer domesticus) (where inbreeding was associated with shorter telomeres)33, in Bengal tigers (Panthera tigris tigris) (where higher frequency of deleterious mutations were found in isolated populations)34, and even in the last woolly mammoth (Mammuthus primigenius) population35. Three out of four P. sentosus populations herein evaluated showed negative values of Fis which objectively indicate excess of heterozygotes than expected at random mating36, nevertheless this deviation can actually be due to genetic drift as these populations are small and retain low levels of genetic diversity37.
The genetic structure analyses indicated high levels of population structuring, which can be due to the impact of anthropogenic habitat modifications on the P. sentosus populations. High levels of population structuring have been previously observed in different reptile populations, including lizards and geckos of the family Phyllodactylidae38,39. Also, similar effects of urban fragmentation to those observed for P. sentosus have been reported in endemic geckos with restricted distribution such as the Mauritius lowland forest day gecko (Phelsuma guimbeaui) from the island of Mauritius, where its population was fragmented by urban expansion into small and isolated forest patches, resulting into populations with high genetic differentiation even in the absence of natural barriers40.
The historical gene flow results are largely consistent with this scenario, showing low relative migration rates between populations, particularly those at the extremes of the species distribution. This situation got established thousands of generations ago (see below), accelerating the population differentiation by genetic drift. Contemporary gene flow rates are lower than historical ones, showing that currently, the probability that an individual from a huaca could disperse to another is minimal, since the populations are isolated due to the presence of buildings, parks and roads acting as barriers for a species characterized by low vagility and dispersal capability11.
On the other hand, high levels of historical gene flow (0.99–1) were found between San Marcos and Parque de Las Leyendas populations, which is in agreement with population structure analyses where both populations clustered together. Interestingly, both localities were part of the pre-Hispanic Maranga archeological complex and were connected by unpaved areas until the 1950s, when the process of asphalting an avenue was completed and there was an increase in infrastructure in the area including the establishment of a zoo and two of the largest universities in Lima. Contemporary gene flow between these two localities is significantly lower (0.06–0.166), although higher than between most other localities, showing that connectivity between Parque de Las Leyendas and San Marcos stopped more recently. While during pre-hispanic settlement these huacas were connected by the Qhapaq Ñan (the Andean road system that expanded across six Andean countries), these ancient routes have been destroyed during Lima’s urban expansion, with the connection between Parque de Las Leyendas and San Marcos being one of the last remnants until 195041.
In addition, when considering the four largest populations, the observed historical and contemporary migration values to and from Mateo Salado were lower, which is consistent with the high coefficients of genetic group membership (q) previously recorded for this locality (Fig. 3b,c), despite the proximity of the other localities (2.5 km). This marked population divergence exemplifies the power of genetic drift to generate divergent populations. This site was heavily impacted during decades by agriculture and other human activities including automobile repair shops, which could have had an even stronger isolating effect. This locality has since been redeemed in the last decade thanks to efforts to preserve the cultural-historic heritage.
The genetic population structuring, and low gene flow observed among populations could be explained by geographical reasons, since the species distribution is interrupted by the Rímac and Chillón rivers (Fig. 3). However, our results show a more structured scenario than the expected by these landscape features (Fig. 3). In the case of Chillón River, it seems as a natural barrier to gene flow since Tambo Inga (the only population recorded north Chillón River) has the lowest levels of migration compared to other localities. On the other hand, although higher migration values are expectedly observed between populations south of Rímac River than between these and those located north to this barrier, some of the latter populations showed higher migration rates to San Marcos and Parque de Las Leyendas (both south of the river) than to nearby populations. These results may suggest that the rivers may represent an unstable barrier, allowing gene flow in certain periods and acting as a barrier in others, depending on the fluctuation of the water flow. This type of river effect has already been proposed to explain the gene flow observed in the Uruguay marked gecko (Homonota uruguayensis) in the Brazilian and Uruguayan pampas42, in rodent populations43 and lizards44. Another explanation to the population structuring observed here could be an isolation by distance model; however, the Mantel test results indicate that geographic distance does not explain genetic differentiation among populations. Thus, the scenario of population structuring observed among the studied populations of P. sentosus cannot be a product of geographic distance alone. Therefore, the results obtained indicate that anthropogenic habitat modifications may represent an important barrier to the dispersal of this gecko.
The habitat of P. sentosus has been modified constantly by the human occupancy during more than 5000 years. The city of Lima has been inhabited by several human cultures over time. The pre-Inca cultures developed aquifer systems that allowed them to sustain large areas of cultivated lands41,45. This water and soil management modified the landscape of Lima and, therefore, the habitats of local fauna and flora46,47. In the Late Archaic period (~ 4720 to ~ 3520 years ago), the domestication of plants and animals was already recorded in the Rímac valley, as well as the first irrigation canals built in the area47. Subsequently, in the Formative Period (~ 3520 to ~ 1920 years ago), large-scale hydraulic works and an expansion of intensive agriculture were built47 (Fig. 6). With the development of more recent urbanization, the modification of Lima's landscape has been more dramatic, including aspects such as buildings and roads46,48, isolating completely the remnant population of P. sentosus (Figs. 1, 6).
Habitat modification by human activity in the Rimac Valley, Lima (Peru). Landscape modification in the area has been done mainly by agricultural activities (A,C) and urbanization (B,D). (A) Photograph of the agricultural area surrounding the Mateo Salado huaca taken in 1944 by the National Aerophotographic Service of the Peruvian Air Force (FAP). (B) Current photograph of the Mateo Salado huaca surrounded by the Lima urban landscape. Photo by A. Arana. (C) The area around the Rímac River has been modified by pre-Incan human activity by the development of water canals for the spread of agricultural land since 1500 BCE (maps modified from ANA, 2016). Agricultural land established by pre-Incan cultures is depicted in green. (D) Further modification arose in the Rimac Valley by the establishment and growth of Lima city since 1920 (based on Sáez Giraldez et al., 2010). Modern urban environment is depicted in orange. Sampling localities of the Lima leaf-toed gecko are represented by red dots. Altitudinal relief is represented in gray lines (altitudes higher than 25 m). Maps created in ArcGIS 10.3. Free vector data from https://www.geogpsperu.com/2014/03/base-de-datos-peru-shapefile-shp-minam.html.
The genetic effect of these changes in the landscape can be observed in the demographic history of P. sentosus populations (Fig. 5). With the development of irrigation canals and agriculture in the area, this gecko, a specialist species adapted to arid areas, lost most of its habitat, but found refuge in the huacas, where population fragmentation, decrease in effective population size and loss of genetic diversity started. Bottleneck effects were observed in all populations analyzed during the last 5000 and 2000 years ago, except for Pucllana that experience a bottleneck between 10,000 and 5000 years ago. After the bottleneck all populations recovered and reached a stable size, showing a period of adaptations to the new environmental conditions. Based on global assessments5,8, it is expected that the new pressures brought from the urbanization and city growth (Fig. 6) will negatively affect the remnant populations. This process can be observed in Pucllana where a drop in the effective population size was estimated to occur about 200 years ago (Fig. 5). However, estimating with accuracy very recent demographic changes that are complex and could include several bottlenecks events is not possible with the method used in this study, which is more appropriate to detect demographic change at the scale of thousands of years49. Methods that can discern very recent demographic changes use linkage disequilibrium analysis and therefore require a reference genome49,50,51. This would be an important next step in the study of this endangered species.
Another factor that could have affected the landscape of the area, during this period is the El Niño Southern Oscillation (ENSO) event, which can produce flooding, variations in temperature and precipitation. However, climate simulations during the Holocene in South America show that in the central coast of Peru there was no major variation in precipitation and vegetation in the last 8000 years, unlike what happened in northern and southern Peru52, allowing an increase in human populations in the sub-continent53. Conversely, it has been estimated that floods occurred more frequently on the coast of Peru between 7000 to 8000 years ago54,55. The Pucllana population may have been affected by a flood caused by an ENSO event in years prior to the settlement of pre-Inca communities. Nonetheless, an event affecting only this locality is unlikely. Therefore, it is possible that the variation in the estimated dates of the bottleneck at Pucllana is due to bias effects related to recent migrations or admixture (see Fig. 2). This type of bias due to demographic processes has been detected in different types of demographic history analyses56.
The whole scenario for P. sentosus conservation does not seems to be favorable. Unlike the previous PVA57, in which no initial genetic diversity value was included, we included a starting initial genetic diversity value of 0.15 based on our genetic diversity results. PVA results showed a mean extinction time of less than 10 years, in contrast with the previous one, in which the mean extinction time was longer than 33 years (if extinct at all). The addition of an initial genetic diversity value causes the effects of this low diversity to be evident from the beginning and as the simulations progress58 these effects include inbreeding depression and the increased probability of lethal alleles occurrences59. This implies that, in a period of 10 years, if no changes are made in favor of its conservation and considering that there are no populations with more than 1000 individuals, all populations of P. sentosus would become extinct.
We must mention that these results are based on simulations performed by the Vortex program based on the information that is available about the natural history of P. sentosus11,16,57 and that they are not definitive results but are a prediction that may vary as more is known about this species and actions are taken to preserve it. The drastic decline in the following years is plausible because the species could enter an extinction vortex in which both a deterioration of population and genetic characteristics occur gradually60,61. In addition, the rate of population decline increases the closer the population gets to extinction62. These predictions represent a red flag for a species that need urgent conservation measures.
Nowadays, P. sentosus is only found in a few huacas and hills that maintain sandy substrate with rocks11. Most of these sites are legally protected for cultural reasons63, but still face several threats in practice such as illegal land appropriation and informal garbage dumps. Furthermore, protection measures may not necessarily align with the goal of conserving wildlife inhabiting the huacas and can either not address threats relevant to the Lima leaf-toed gecko (like the presence of invasive predators, e.g., cats and rats) or directly endanger them with the removal of the gecko’s retreat sites during archeological restoration.
Based on the results obtained, we recommend a management plan for the conservation of P. sentosus that incorporates the recovery of gene flow between populations through the translocation of individuals between localities. Management actions should be coordinated with national biodiversity authorities as with local archeological institutions, considering that most populations of P. sentosus live in archeological areas that are protected by the national government authorities. Also, our attention should be drawn to the populations in areas that do not have protection and that are at risk of disappearing. Another important future step should be producing a high-quality genome assembly, as this would provide more information about the genomic health of these populations (for example by analyzing the population frequency of runs of homozygosity and deleterious alleles to assess the effects of inbreeding), and about genomic signatures of local adaptation64. Finally, a management plan should be integrated with the ongoing efforts in captive breeding that have already produced protocols for artificial incubation for this species65 and future ex-situ conservation programs that include zoo-kept gecko populations to build insurance colonies for this endangered taxon66. This would benefit from implementing more monitoring, ecological and reproductive studies of P. sentosus. These studies are crucial for incorporating more precise information into population genetic analyses and population viability simulations, to gain a deeper understanding of the natural history of this species.
Methods
Sample collection and genomic DNA extraction
We collected 50 tissue samples from 12 P. sentosus localities in Lima, Peru (Fig. 1) under the research and sampling permit (SERFOR N° AUT-IFS-2018-40). We conducted opportunistic visual encounter surveys between 18:00 and 24:00 h by taking identification photos upon capture. We obtained more samples from localities where larger census population sizes were previously registered: San Marcos, Parque de Las Leyendas, Pucllana and Mateo Salado16,17,18. In contrast, much smaller census population sizes have been recorded in other localities (Pérez pers. commun.). We used sterilized surgical steel scissors to cut the tail tip up to 15 mm from the tip. In a few cases autotomized tail parts were collected. Geckos were all safely released after processing. Tissue samples were stored in 96% ethanol at − 18 °C prior to DNA extraction. We then extracted genomic DNA using an innuPREP DNA Mini Kit (Analytik Jena, Jena, Germany) and quantified the extracted DNA with NanoDrop Lite (Thermo Scientific, Waltham, USA).
Library preparation
GBS libraries were generated via labor contract with University of Minnesota Genomics Center (https://genomics.umn.edu/services/gbs). Here we briefly describe the protocol, which is based on the one proposed by Elshire et al.24. A total of 100 ng of DNA was digested with 10 units of the restriction enzymes PstI-HF and MspI (New England Biolabs, Inc. MA, USA); and incubated at 37 °C for 2 h. DNA samples were ligated with 200 units of T4 ligase (New England Biolabs, Inc. MA, USA) and phase adaptors with -TGCA and CG overhangs at 22 °C for 1 h and heat killed. Ligated libraries were purified with SPRI beads and then amplified for 18 cycles with 2 × Taq Master Mix (New England Biolabs, Inc. MA, USA) to add the barcodes. GBS libraries were then purified, quantified using a NanoDrop™ Lite Spectrophotometer, and pooled. Fragments with the 300–744 bp size region were selected and diluted to 2 nM for sequencing on the Illumina NovaSeq 550 (Illumina, CA, USA) using 1 × 150 single-end reads.
SNP genotyping and filtering
First, cutadapt67 and fastp68 were used to remove adapters. Then sequencing reads were processed using ipyrad v. 0.7.1369 with the following parameters: minimum depth for statistical base calling = 6; maximum heterozygous sites per locus = 0.5; clustering threshold = 0.85. We generated two data sets with ipyrad from 50 individuals from 12 localities, one dataset (i) with no missing data (loci shared by all individuals) and a second one (ii) with loci shared by at least 33 individuals (66% of the total number of individuals collected).
Genetic diversity and inbreeding coefficient
Two measures of genetic diversity were applied at the population level using the population tool from Stacks70: (1) nucleotide diversity (π) defined as the mean number of differences per nucleotide site between two sequences from a pair of individuals randomly selected from a population71,72; and (2) expected heterozygosity (He). To facilitate a meaningful comparison with our results, we also calculated expected heterozygosity for four other Phyllodactylus species using data from a study conducted by Ramírez-Reyes et al. in 202073. The inbreeding coefficient (FIS) indicating the deviation of the observed heterozygosity relative to the expected heterozygosity36, was also obtained using the Stacks tool set. Fis was calculated only for the four largest populations (largest census size16,17,18 and sample size of 10 individuals: Table 1).
Population genetic structure
To analyze the genetic structure, a principal component analysis (PCA) was performed using the glPCA function of the R package adegenet74. We used the ipyrad-analysis toolkit for a t-SNE analysis, which is a big data visualization method developed by machine learning (Automatic Learning) within the field of artificial intelligence and has been proposed as a better method for visualizing SNPs than PCA75,76. Also, a discriminant analysis of principal components (DAPC) was conducted using the function dapc of the R package adegenet74, and the number of principal components that were retained for the DAPC was chosen with the function xvalDapc using the cross-validation method as recommended by Jombart and Collins77. Finally, a STRUCTURE78 analysis was performed using the ipyrad packages and the admixture model with a correlated allele frequency. Also, we used two different mincov options, filtering all missing data or maintaining only SNPs with 0.9 of sample coverage. The number of genetic groups (K) was determined by the largest ΔK79.
Gene flow
We created a file in GENEPOP80 format from the vcf file using unlinked_snps_to_genepop from ipyrad tools (github.com/brunoasm/ipyrad_tools). We then examined the pairwise historical migration patterns of local populations using the divMigrate method25 implemented in the R package diveRsity81. This method estimates relative directional migration based on asymmetric distributions of allele frequencies25. Results are presented in relative migration rates with values between 0 and 125. We made a modification in the code to allow the incorporation of populations that had a single individual as a sample (see Supplementary Material S6 online). GST was chosen among the migration measures offered by divMigrate25.
We used the Mantel test to assess if there was a correlation between geographical and genetic distances between populations, following the isolation by distance model (IBD)82,83 while using the R package hierfstat84 to obtain Nei's FST, which was used as a measure of genetic distance. Geographic distances were obtained using the Geographic Distance Matrix Generator v. 1.2.385. The Mantel test with the pairwise genetic distances (FST/(1 − FST)) and the pairwise geographic distances (log transformed) was performed using the Mantel function of the R package vegan86 with 10,000 permutations.
Contemporary gene flow was estimated using a modification of BayesAss 3.0427, called BA3-SNPs v. 3.0.426, which accepts large SNP datasets and estimates migration rates as proportion of migrants using a Bayesian framework and Markov chain Monte Carlo (MCMC). We used BA3-SNP-autotune v 3.0.426 to determine the optimal mixing parameters for our dataset for migration rates (deltaM = 0.9991), allele frequencies (deltaA = 0.7750), and inbreeding coefficients (deltaF = 0.0250). We ran BA3-SNPs for 100 million iterations, with a burn-in of 10 million. Chain convergence was assessed in Tracer v 1.7.187.
Demographic history
To infer the demographic history of the Lima leaf-toed gecko we first generated a folded SFS file with easySFS (https://github.com/isaacovercast/easySFS). We then used Stairway plot v. 2.188 to estimate the historical effective population size changes of the largest gecko populations in Lima16,17,18: Huaca San Marcos, Parque de Las Leyendas, Mateo Salado and Huaca Pucllana. We did not analyze the gecko population as a whole, since high levels of population structure are considered factors that artificially modify the results56.
The parameters for the Stairway Plot analysis were as follows: 67% for the training sites; random breakpoints at 4, 9, 14, and 18; and one year was assumed as generation time (J Pérez, pers. commun.). The mutation rate was assumed to be 2.4 × 10–9, since this value is frequently used in studies on species of the order Squamata89,90.
Population viability analysis
To carry out the Population Viability Analysis (PVA) we used Vortex (v. 10.2.13.0)58 which performs simulations, based on given conditions, for different populations. Five populations were simulated, each one with a different initial census population size (N0 = 50, 100, 200, 500, 1000) and a carrying capacity (K) that corresponds to 125% of the initial census population size. The maximum age was 4 years old. These conditions were chosen based on previous ecological population studies16,18 and previous PVAs57. Simulation parameters are summarized in Supplementary Table S7 (see online). These parameters were carefully selected based on an extensive review of the existing literature on P. sentosus11,16,57, and through in-depth consultations with experts actively engaged in the study of this species.
Ethical statement
Field protocols for the capture, handling, release of Lima leaf-toed geckos, and tail tips sampling were approved by the competent authority: the National Forestry and Wildlife Service (SERFOR) of the Ministry of Agriculture and Irrigation of Peru (permit: SERFOR N° AUT-IFS-2018-40). All methods were performed in accordance with the relevant SERFOR guidelines and regulations, as well as in compliance with the IUCN Policy Statement on Research Involving Species at Risk of Extinction.
Data availability
Genotyping-By-Sequencing fastq data were deposited to GenBank under BioProject PRJNA939276. Other data generated and analyzed are presented in the figures or tables of Supplementary Material.
References
Harrison, S. & Bruna, E. Habitat fragmentation and large-scale conservation: What do we know for sure?. Ecography 22, 225–232 (1999).
Rogan, J. E. & Lacher, T. E. Impacts of habitat loss and fragmentation on terrestrial biodiversity. In Reference Module in Earth Systems and Environmental Sciences 1–18 (Elsevier Inc., 2018). https://doi.org/10.1016/b978-0-12-409548-9.10913-3.
Miles, L. S., Rivkin, L. R., Johnson, M. T. J., Munshi-South, J. & Verrelli, B. C. Gene flow and genetic drift in urban environments. Mol. Ecol. 28, 4138–4151 (2019).
Hoehn, M., Sarre, S. D. & Henle, K. The tales of two geckos: Does dispersal prevent extinction in recently fragmented populations?. Mol. Ecol. 16, 3299–3312 (2007).
French, S. S., Webb, A. C., Hudson, S. B. & Virgin, E. E. Town and country reptiles: A review of reptilian responses to urbanization. Integr. Comp. Biol. 58, 948–966 (2018).
Cortés-Gomez, A. M., Ruiz-Agudelo, C. A., Valencia-Aguilar, A. & Ladle, R. J. Ecological functions of neotropical amphibians and reptiles: A review. Universitas Scientiarum 20, 229–245. https://doi.org/10.11144/Javeriana.SC20-2.efna (2015).
Böhm, M. et al. The conservation status of the world’s reptiles. Biol. Conserv. 157, 372–385 (2013).
Cordier, J. M. et al. A global assessment of amphibian and reptile responses to land-use changes. Biol. Conserv. 253, 108863 (2021).
Brum, P. H. R., Gonçalves, S. R. A., Strüssmann, C. & Teixido, A. L. A global assessment of research on urban ecology of reptiles: Patterns, gaps and future directions. Anim. Conserv. 26, 1–13. https://doi.org/10.1111/acv.12799 (2023).
Cox, N. et al. A global reptile assessment highlights shared conservation needs of tetrapods. Nature 605, 285–290 (2022).
Perez, J. & Balta, K. Phyllodactylus sentosus. The IUCN Red List of Threatened Species.https://doi.org/10.2305/IUCN.UK.2016-1.RLTS.T48442971A48442982.en
Cossíos, E. D. & Icochea, J. Nuevos registros del gecko de Lima, Phyllodactylus sentosus (Reptilia, Geckonidae). Ecología Aplicada 5, 182–184 (2006).
Olivera, D., Castillo, L. & Gutiérrez, G. Primer registro de Phyllodactylus sentosus (Squamata: Phyllodactylidae) para el valle del río Chillón, Lima, Perú. Rev. Peru Biol. 23, 321–324 (2016).
Pérez, J., Ramírez, C. & Balta, K. A new record of Phyllodactylus sentosus (Dixon & Huey, 1970) (Squamata: Phyllodactylidae) for the coastal desert of Peru. Cuadernos de Herpetología 27, 2013 (2013).
Dixon, J. R. & Huey, R. B. Systematics of the lizards of the gekkonid genus Phyllodactylus of mainland South America. Contrib. Sci. 192, 48–50 (1970).
Valdez, F., Iannacone, J., Luna, A. & Cossios, E. D. Population size and dynamics of the Lima Leaf-Toed Gecko, Phyllodactylus sentosus, in one of its last refuges. J. Herpetol. 54, 155–160 (2020).
Milla Villegas, J. Ecología térmica de Phyllodactylus sentosus (Squamata: Phyllodactilidae) en la Huaca San Marcos, Lima, Perú (Honors thesis). (Universidad Nacional Mayor de San Marcos, 2022).
Fernández Rodríguez, S. Evaluación de la dinámica poblacional del gecko de Lima Phyllodactylus sentosus (Reptilia: Phyllodactylidae) (Honors Thesis). (Universidad Peruana Cayetano Heredia, 2019).
Leiva González, S. Flora y fauna de la Huaca del Rosario, Magdalena de Cao, Ascope, región La Libertad, Perú. Arnaldoa https://doi.org/10.22497/arnaldoa.253.25316 (2018).
Salinas, L., Arana, C., Gavancho, L. & Garrido, A. Distribución actual del Huerequeque (Burhinus superciliaris) en Lima. Revista Científica Dilloniana 3, 97 (2003).
Ekblom, R. & Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107, 1–15 (2011).
Morin, P. A., Martien, K. K. & Taylor, B. L. Assessing statistical power of SNPs for population structure and conservation studies. Mol. Ecol. Resour. 9, 66–73 (2009).
Wallace, J. G. & Mitchell, S. E. Genotyping-by-sequencing. Curr. Protoc. Plant Biol. 2, 64–77 (2017).
Elshire, R. J. et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6, e19379 (2011).
Sundqvist, L., Keenan, K., Zackrisson, M., Prodöhl, P. & Kleinhans, D. Directional genetic differentiation and relative migration. Ecol. Evol. 6, 3461–3475 (2016).
Mussmann, S. M., Douglas, M. R., Chafin, T. K. & Douglas, M. E. BA3-SNPs: Contemporary migration reconfigured in BayesAss for next-generation sequence data. Methods Ecol. Evol. 10, 1808–1813 (2019).
Wilson, G. A. & Rannala, B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191 (2003).
Frankham, R., Ballou, J. & Briscoe, D. Introduction to Conservation Genetics (Cambridge University Press, 2010). https://doi.org/10.1071/pc010217.
Madsen, T., Shine, R., Olsson, M. & Wittzell, H. Restoration of an inbred adder population. Nature 402, 34–35 (1999).
Charlesworth, D. Effects of inbreeding on the genetic diversity of populations. Philos. Trans. R. Soc. B Biol. Sci. 358, 1051–1070 (2003).
Keller, L. F., Reid, J. M. & Arcese, P. Testing evolutionary models of senescence in a natural population: Age and inbreeding effects on fitness components in song sparrows. Proc. R. Soc. B Biol. Sci. 275, 597–604 (2008).
Powell, J. R. & Evans, B. R. How much does inbreeding reduce heterozygosity? Empirical results from Aedes aegypti. Am. J. Trop. Med. Hyg. 96, 157–158 (2017).
Pepke, M. L. et al. Inbreeding is associated with shorter early-life telomere length in a wild passerine. Conserv. Genet. 23, 639–651 (2022).
Khan, A. et al. Genomic evidence for inbreeding depression and purging of deleterious genetic variation in Indian tigers. PNAS 118, e2023018118 (2021).
Palkopoulou, E. et al. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol. 25, 1395–1400 (2015).
Keller, L. F. & Waller, D. M. Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 (2002).
Kerr, W. E. & Wright, S. Experimental studies of the distribution of gene frequencies in very small populations of Drosophila melanogaster: I. Forked. Evolution 8, 172–177 (1954).
Delaney, K. S., Riley, S. P. D. & Fisher, R. N. A rapid, strong, and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS ONE 5, 1–11 (2010).
Blair, C., Jiménez Arcos, V. H., Mendez de la Cruz, F. R. & Murphy, R. W. Landscape genetics of leaf-toed geckos in the tropical dry forest of northern Mexico. PLoS ONE 8, e57433 (2013).
Buckland, S. et al. High risks of losing genetic diversity in an endemic Mauritian Gecko: Implications for conservation. PLoS ONE 9, e93387 (2014).
Bonilla, E. et al. Guía de arquitectura y paisaje Lima y El Callao (Universidad Ricardo Palma, 2009).
Felappi, J. F., Vieira, R. C., Fagundes, N. J. R. & Verrastro, L. V. So far away, yet so close: Strong genetic structure in Homonota uruguayensis (Squamata, Phyllodactylidae), a species with restricted geographic distribution in the Brazilian and Uruguayan Pampas. PLoS ONE 10, 1–19 (2015).
Nascimento, F. F. et al. The role of historical barriers in the diversification processes in open vegetation formations during the Miocene/Pliocene using an ancient rodent lineage as a model. PLoS ONE 8, e61924 (2013).
Siedchlag, A. C., Benozzati, M. L., Passoni, J. C. & Rodrigues, M. T. Genetic structure, phylogeny, and biogeography of Brazilian eyelid-less lizards of genera Calyptommatus and Nothobachia (Squamata, Gymnophthalmidae) as inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 56, 622–630 (2010).
Stotz, D. F., Saboya Del Castillo, P. & Laverde-R, O. Aves. In Perú: Medio Putumayo-Algodón. Rapid Biological and Social Inventories Report 28 (2016).
del Castillo, J. M. Lima biotopo: ecosistemas de montaña, patrimonio arqueológico indígena y activismo en los intersticios urbanos de la megalópolis andina. in VII Seminario Internacional de Investigación en Urbanismo, Barcelona-Montevideo, junio 2015 (Facultad de Arquitectura. Universidad de la República, 2015). https://doi.org/10.5821/siiu.6178.
Autoridad Nacional del Agua. Rímac: Historia del Río Hablador (ANA, 2016).
Sáez Giraldez, E., García Calderón, J. & La Roch Peña, F. Ciudad Desde La Casa: Ciudades Espontáneas En Lima. Revista INVI 25, 77–116 (2010).
Nadachowska-Brzyska, K., Konczal, M. & Babik, W. Navigating the temporal continuum of effective population size. Methods Ecol. Evol. 13, 22–41 (2022).
Santiago, E. et al. Recent demographic history inferred by high-resolution analysis of linkage disequilibrium. Mol. Biol. Evol. 37, 3642–3653 (2020).
Browning, S. R. & Browning, B. L. Accurate Non-parametric estimation of recent effective population size from segments of identity by descent. Am. J. Hum. Genet. 97, 404–418 (2015).
Maksic, J. et al. Simulation of the Holocene climate over South America and impacts on the vegetation. Holocene 29, 287–299 (2019).
Riris, P. & Arroyo-Kalin, M. Widespread population decline in South America correlates with mid-Holocene climate change. Sci. Rep. 9, 1–10 (2019).
Carré, M. et al. Holocene history of ENSO variance and asymmetry in the eastern tropical Pacific. Science 1979(345), 1045–1048 (2014).
Carré, M. et al. High-resolution marine data and transient simulations support orbital forcing of ENSO amplitude since the mid-Holocene. Quat. Sci. Rev. 268, 107125 (2021).
Gattepaille, L. M., Jakobsson, M. & Blum, M. G. B. Inferring population size changes with sequence and SNP data: Lessons from human bottlenecks. Heredity 110, 409–419 (2013).
Perez, J., Elías, R., Balta, K., Rodríguez, J. & Matamoros, Y. Análisis de Viabilidad de la Población y hábitat (PHVA) del gecko de Lima (Phyllodactylus sentosus). Universidad Peruana Cayetano Heredia-IUCN Specialists Group (2018).
Lacy, R. C. Structure of the VORTEX simulation model for population viability analysis. Ecol. Bull. 48, 191–203 (2000).
Ralls, K., Ballou, J. D. & Templeton, A. Estimates of lethal equivalents and the cost of inbreeding in mammals. Conserv. Biol. 2, 185–193 (1988).
Soulé, M., Gilpin, M., Conway, W. & Foose, T. The millenium ark: How long a voyage, how many staterooms, how many passengers?. Zoo Biol. 5, 101–113 (1986).
Fagan, W. F. & Holmes, E. E. Quantifying the extinction vortex. Ecol. Lett. 9, 51–60 (2005).
Williams, N., McRae, L., Freeman, R., Capdevila, P. & Clements, C. Scaling the extinction vortex: Body size as a predictor of population dynamics close to extinction events. Ecol. Evol. 11, 7069–7079 (2021).
Meijers, S. C. Archeological sites in Lima and their inheritors: Community engagement in the management of huacas in Lima. Inter-Section 1, 6–13 (2015).
Paez, S., Kraus, R. H. S., Shapiro, B., Gilbert, M. T. P. & Jarvis, E. D. Reference genomes for conservation. Science 1979(377), 364–366 (2022).
Sánchez, M. V., Manrique, Z. M. & Elías, P. R. Artificial incubation of eggs of Lima gecko (Phyllodactylus sentosus) (Reptilia, Geckonidae) kept in captivity. Revista de Investigaciones Veterinarias del Peru 33, e22896 (2022).
Rech, I., Ginal, P., Rauhaus, A., Ziegler, T. & Rödder, D. Geckos in zoos: A global approach on distribution patterns of threatened geckos (Gekkota) in zoological institutions. J. Nat. Conserv. 75, 126467 (2023).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Eaton, D. A. R. & Overcast, I. ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 2–4 (2020).
Catchen, J., Hohenlohe, P. A., Bassham, S., Amores, A. & Cresko, W. A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140 (2013).
Innan, H., Terauchi, R., Kahl, G. & Tajima, F. A method for estimating nucleotide diversity from AFLP data. Genetics 151, 1157–1164 (1999).
Hughes, A. R., Inouye, B. D., Johnson, M. T. J., Underwood, N. & Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623 (2008).
Ramírez-Reyes, T. et al. Phylogenomics and molecular species delimitation reveals great cryptic diversity of leaf-toed geckos (Phyllodactylidae: Phyllodactylus), ancient origins, and diversification in Mexico. Mol. Phylogenet. Evol. 150, 106880 (2020).
Jombart, T. & Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011).
Li, W., Cerise, J. E., Yang, Y. & Han, H. Application of t-SNE to human genetic data. J. Bioinform. Comput. Biol. 15(4), 1750017 (2017).
Platzer, A. Visualization of SNPs with t-SNE. PLoS ONE 8, e56883 (2013).
Jombart, T. & Collins, C. A tutorial for discriminant analysis of principal components (DAPC) using adegenet 2.1.3. (Imperial College London, MRC Centre for Outbreak Analysis and Modelling, 2021).
Pritchard, J., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Rousset, F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106 (2008).
Keenan, K., McGinnity, P., Cross, T. F., Crozier, W. W. & Prodöhl, P. A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 4, 782–788 (2013).
Wright, S. Isolation by distance. Genetics 28, 114–138 (1943).
Slatkin, M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47, 264–279 (1993).
Goudet, J. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186 (2005).
Ersts, P. J. Geographic Distance Matrix Generator (version 1.2.3). American Museum of Natural History, Center for Biodiversity and Conservation (2022).
Oksanen, J. et al. vegan: Community ecology package. (2013).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Liu, X. & Fu, Y. X. Exploring population size changes using SNP frequency spectra. Nat. Genet. 47, 555–559 (2015).
Pasquesi, G. I. M. et al. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat. Commun. 9, 2774 (2018).
Green, R. E. et al. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 1979(346), 1254449–1254449 (2014).
Acknowledgements
This work was funded by CONCYTEC-PROCIENCIA within the frame of the call for Proyectos de Investigación Básica 2019-01 [Contrato N° 443-2019-FONDECYT] and VRIP-UNMSM PCONFIG-B19100931. The authors are thankful to Sol Fernández for the valuable samples from Mateo Salado. PGJ acknowledges CNPq/Brazil (303524/2019-7). We thank the reviewers for their valuable suggestions, insightful comments, and careful editing, which have significantly improved the quality and clarity of our manuscript.
Author information
Authors and Affiliations
Contributions
A.A., J.E., J.P., and J.R.: conceived and designed the experiments; A.A., J.E., and J.R.: performed all experiments; R.R., J.P., P.G., and J.R.: provided scientific advice and resources. A.A., J.E., R.R., P.G., J.P., and J.R.: wrote the manuscript and prepared figures. All authors reviewed and read the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Arana, A., Esteves, J., Ramírez, R. et al. Population genomics reveals how 5 ka of human occupancy led the Lima leaf-toed gecko (Phyllodactylus sentosus) to the brink of extinction. Sci Rep 13, 18465 (2023). https://doi.org/10.1038/s41598-023-45715-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-45715-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.