Abstract
Clinical studies have demonstrated an association between high myopia (HM) and neuropsychiatric disorders; however, the underlying mechanism of the association is not clear. We used whole exome sequencing (WES) in combination with the Genetic Variants Classification Criteria and Guidelines published by the American College of Medical Genetics (ACMG) and bioinformatics analysis to clarify the interrelationship between candidate genes. Causative genes for ocular diseases (45.38%) followed by neuropsychiatric disorders (22.69%) accounted for the highest proportion of genes that exhibited high pathogenicity in HM patients were found. Four pathogenic gene mutations were identified according to ACMG guidelines: c.164_165insACAGCA and c.C1760T in POLG, c.G1291A in COL5A1, and c.G10242T in ZNF469. Three causative genes for neuropsychiatric diseases, PTPRN2, PCDH15 and CDH23, were found to fall at the HM locus. The above results suggest that these genes may interact in high myopia and neuropsychiatric diseases.
Similar content being viewed by others

Introduction
High myopia (HM), a common ophthalmic disease, is an important global public health problem, especially in Southeast Asia1,2. HM often leads to a series of complications (such as retinopathy and glaucoma) that can cause blindness. Therefore, development of ocular complications is the key concern during clinical treatment of HM, while the neuropsychiatric disorders that accompany HM tend to be overlooked3.
HM has been shown to be associated with a variety of neuropsychiatric disorders such as anxiety, depression, and cognitive dysfunction4. An estimated one-third of the European population with visual impairment is affected by anxiety or depressive symptoms5. The estimated prevalence of anxiety or depression in patients with HM in East Asia ranges from 22.0% to 25.9%6. Among elderly individuals, the prevalence of cognitive dysfunction in myopic patients was found to be twice as high as that in the individuals with normal vision7.
The development of HM is strongly associated with genetic factors as shown by twin and familial aggregation studies8. As of today, 25 loci are associated with HM (MYP1-MYP3, MYP5-MYP26), based on whole-exome sequencing (WES) and other tests9. Meanwhile, There is a substantial genetic component to most neuropsychiatric disorders, with heritability ranging from 75 to 80%10. In conjunction with clinical manifestations associated with high myopia and neuropsychiatric disorders identification of genes that show interaction with both HM and neuropsychiatric disorders is a potential way to explore the genetic mechanism of causation of neuropsychiatric diseases in HM patients11,12,13. In this study, we explored the interaction between HM and neuropsychiatric genes and identified interacting genes in 83 non-syndromic patients with HM from Northwest China.
Materials and methods
Study population
Eighty-three sporadic patients with HM were recruited in this study. The study was performed in accordance with the tenets of the Declaration of Helsinki. Written informed consent was obtained from the participants or their statutory guardians, with approval from the institutional review board of the People’s Hospital of Ningxia Hui Autonomous Region, Third Clinical Medical College of Ningxia Medical University. Genomic DNA was extracted from peripheral blood samples. All 83 patients included in this study belonged to the Ningxia Hui Autonomous Region and the clinic information of the patients have been show in Supplementary Table 1. Patients were recruited according to the following inclusion criteria: (1) refractive error worse than or equal to − 6 D and/or axial length (AL) > 26.00 mm; and (2) absence of any other known ocular or systemic disorder.
Whole exome sequencing and variant filtering
Exome Enrichment V5 Kit (Agilent Technologies, United States) was used for whole exome sequencing as previously described14. Sequencing of DNA fragments was conducted on Illumina HiSeq 2000, Analyzers (90 cycles per read). In accordance with the manufacturer's instructions, Illumina libraries were prepared and generated using the Hiseq2000 platform from Illumina, Inc. A set of local realignments, quality control text, and variants were called using the Genome Analysis Toolkit (GATK). As described previously, Sequencing Reads were aligned to the human reference sequence (hg19/GRCH37) using Burrows Wheeler Aligner-Maximum Exact Match (BWA-MEM)15. The WES resulted in a mean read depth of 30x. Median coverage of the targeted regions exceeded 95%.
The WES results from patients with HM were filtered as follows: (1) Screening of mutations with minor allele frequency (MAF) below 0.01 in the four major human gene frequency databases (1000G, ExAC, ESP6500, and gnomAD); (2) Retention of mutations shown to be Probably Damaged or Damaged by three or more pathogenic prediction software; (3) Comprehensive analysis of strongly pathogenic mutations in patients based on the American College of Medical Genetics (ACMG) guidelines combined with their clinical phenotypes. The screening process is shown in Fig. 1.
Overview of the screening process. A range of strategies were applied to filter the causative genes for high myopia and neuropsychiatric disorders in 83 sporadic patients. Mutation frequency in populations, Mutation pathogenicity prediction, and ACMG are combined to narrow down the candidate genes and mutations.
All the identified variants were assessed using the following tools. The pathogenicity of gene mutations was predicted using 8 bioinformatics tools: SIFT(http://sift.jcvi.Org), Polyphen2(http://genetics.bwh.harvard.edu/pph2/index.Shtml), LRT(genetics.wustl.edu/jfla), Mutation Taster(https://www.mutationtaster.org/), Mutation Assessor(mutationassessor.org/r3), FATHMM(fathmm.biocompute.org.uk), LR(https://doi.org/10.1093/hmg/ddu733), and Radial SVM(https://doi.org/10.1093/hmg/ddu733). Protein interactions were assessed using String (STRING: functional protein association networks (https://string-db.org/)).
Results
Comprehensive analysis of WES results
A total of 109 mutations were predicted to be pathogenic by three or more bioinformatics tools; these included 37 novel mutations that have not been reported and 72 rare mutations (Supplementary Tables 2 and 3) (The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.6084/m9.figshare.19550926.v1). Among these, the strongly pathogenic genes in HM patients were mainly distributed across nine different disease types, with the highest proportion of genes accounting for ocular diseases (45.38%) followed by neuropsychiatric diseases (22.69%) (Fig. 2). After obtaining the proportion of all genes in each disease-related gene, the proportions of various disease-causing genes among the 37 novel mutations and 72 rare mutations were counted. The novel mutations included 18 causative genes for eye diseases (40%), similarly, the proportion of rare mutations in various disease-related genes was 36 (49%) for eye disease genes (Figs. 3, 4). The combined overall data and both partial data indicated that the highest detection rates of mutations in causative genes for ocular and neuropsychiatric disorders were found in patients with HM who had strongly pathogenic genes.
Distribution of strongly pathogenic genes corresponding to various diseases in patients with high myopia. Genes of neuropsychiatric disorders for 22.69%; genes of ocular disease for 45.38%; genes of skeletal-related disease for 5.04%; genes of hormone-related disease for 0.84%; genes of blood disease for 1.68%; genes of mitochondrial disease for 0.84%; genes of deafness for 2.52%; genes of kidney disease for3.36%; genes of others for17.65%.
Distribution of disease-related genes among novel mutations. 18 causative genes were included in novel mutations for eye diseases (40%), 14 causative genes for brain diseases (31%), 4 causative genes for skeletal-related disorders (9%), 2 causative genes for deafness (4%), and only 1 causative gene for both hormone-related genes and blood disease (2%).
Distribution of disease-related genes among rare mutations. The proportion of rare mutations in various disease-related genes was 36 (49%) for eye disease genes, 13 (18%) for brain disease genes, 3 (4%) for kidney disease genes, 2 (3%) for bone-related genes, and 1 (1%) for both deafness and blood-related genes.
Analysis of association between causative genes for high myopia and neuropsychiatric disorders
The genes related to neuropsychiatric disorders and high myopia were among the 37 novel mutations and 72 rare mutations with strong pathogenicity. The results are summarized in Supplementary Tables 4 and 5. Protein interaction analysis of causative genes for high myopia and neuropsychiatric disorders showed that the genes with strong interactions included two gene clusters One of the gene clusters was SOX5, COL9A1, COL4A5, COL5A1, LTBP2, CYP1B1, TGFBI, ZNF469, MYOF, and SLC4A11, with direct interactions between COL9A1, COL4A5, and COL5A1. The other gene cluster was PLCH, INPP5E, ARL13B, NPHP3, BBS2, BBS1, BBS9, NPHP1, CEP78, PLK4, and C5orf42, with the strongest interactions between ARL13B, NPHP3, BBS2, BBS1, and BBS9 (Fig. 5). It indicates that these genes do not act alone in high myopia or neuropsychiatric disorders, but show multiple interactions with each other.
Protein interactions of neuropsychiatric and ocular diseases. One of the gene clusters was SOX5, COL9A1, COL4A5, COL5A1, LTBP2, CYP1B1, TGFBI, ZNF469, MYOF, and SLC4A11, with direct interactions between COL9A1, COL4A5, and COL5A1. The other gene cluster was PLCH, INPP5E, ARL13B, NPHP3, BBS2, BBS1, BBS9, NPHP1, CEP78, PLK4, and C5orf42, with the strongest interactions between ARL13B, NPHP3, BBS2, BBS1, and BBS9.
We performed pooled analysis of the pathogenicity of all neuropsychiatric disorders-related genes and high myopia-related genes using eight pathogenic prediction software. Two software programs that focus on evolutionary conservation, Mutation Assessor and SIFT, yielded consistent scores for all genes, indicating that these genes have remained extremely conserved at the species level over thousands of years of human genetic progression and that, in turn, the mutations in these genes have a significant impact on the structure and function of human proteins. The consistency of scoring the same group of genes across different software programs collectively improves the accuracy of predicting the pathogenicity of these genes (Fig. 6). In addition, the protein interactions of ARL13B, NPHP3, BBS2, BBS1, and BBS9 gene clusters were all predicted to exhibit high pathogenicity.
The mutations were classified according to the ACMG guidelines. The results showed that c.8815G > A in HMCN1 and c.541G > A in SALL4 have both been reported to be benign, combined with the evidence that the allele frequency of the c.8815G > A in HMCN1 is below the gene frequency threshold in normal populations; c.8815G > A in HMCN1 and c.541G > A in SALL4 were determined to be Likely Benign. Another 27 genes were found to be potentially pathogenic, among which four mutations c.164_165insACAGCA, c.1760C > T in POLG, c.1291G > A in COL5A1 and c.10242G > T in ZNF469 were found to qualify the criteria for pathogenic mutations according to ACMG guidelines with PVS1 (pathogenic very strong) and multiple PP1 (probably pathogenic) evidence. Among these, different mutations in the POLG gene were found in two patients, respectively, and the two patients (patients 115 and 116) were not related (Supplementary Table 6).
Neuropsychiatric disorders genes located in regions of high myopia loci
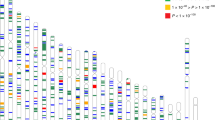
In this study, 62 genes related to ocular diseases and 22 genes related to neuropsychiatric disorders were identified; of these, 15 genes were found to be associated with both ocular and neuropsychiatric diseases (Fig. 7). The causative genes for neuropsychiatric disorders in HM patients were mainly associated with retinopathy, especially retinitis pigmentosa (RP), which involves neuroprotective factors and humoral immunity, and cone optic rod dystrophy, which may develop into RP. The TRMT44 gene is associated with partial epilepsy with pericentric spikes, and the NPHP1 is associated with Joubert's syndrome, which can alter the nuclei of the medulla oblongata in addition to the base of the pons, causing various neuropathological changes. Localization of neuropsychiatric disorders genes revealed that three of them fall in the loci of the HM gene, namely PTPRN2, PCDH15, and CDH23 (Fig. 8). The PCDH15 is located on chromosome 10q21.1, which coincides with the MYP15 region of the HM locus, and has been identified to be associated with the development of Usher syndrome, which is associated with intellectual disability, EEG abnormalities or schizophrenia, in addition to ocular symptoms. CDH23 is located on chromosome 10q21.1 and is in the MYP15 region of the HM locus, like PCDH15, which is also associated with Usher syndrome and retinitis pigmentosa.
In addition to the above-mentioned genes that are located on the HM locus, there are also neuropsychiatric disorders genes that are located very close to the known HM locus regions. PLK4 is located on chromosome 4q28.1, adjacent to the MYP11 (4q22-27) region, POMT2 is on chromosome 14q24.3, similar to the MYP18 (14q22.1–24.2) region, and the TRMT44 is positioned on 4p16.1, which is in the 4p16 region as MYP23 (4p16.3).
Discussion
Studies have shown that patients with high myopia have structural changes in their brain white matter, which are important factors contributing to neuropsychiatric disorders16,17. In this study, causative genes for neuropsychiatric disorders were found to account for 22.69% of strongly pathogenic genes in patients with HM, which represents the highest proportion among disease types other than ocular diseases. From clinical imaging research to genetics, our study deepens the study of high myopia and neuropsychiatric diseases.
Seitler et al. proposed a hypothesis to explain the higher-than-normal incidence of anxiety and depression in HM patients. He suggested that myopia arises due to tension formed by the tightening of the extraocular muscles around the eye, and that the body develops a resistance mechanism to this tension causing myopia. Further, this tension leads to an interruption of the individual separation process, wherein myopic patients experience individual separation anxiety that leads to feelings of not being able to live a normal life18. In a study of 360 patients with visual impairment in France, Germany, and Italy, one third of the patients with visual impairment were found to be affected by anxiety or depression19. A study by Yokoi et al. indicated that 22.0%–25.9% of patients with HM may experience depression or anxiety20. These findings indicate the high prevalence of anxiety and depression in HM patients in different countries and regions. In this study, we found that neuropsychiatric disorder-related genes accounted for 22.69% of the strongly pathogenic genes. This is consistent with the findings of Yokoi et al20. Our findings support the association of HM with depression and anxiety disorders at the genetic level. Further in-depth studies are required to provide more robust evidence.
In a study of 112 myopic British patients, depression and anxiety in HM patients were found to increase the risk of other diseases, such as myocardial infarction and rheumatoid arthritis21. In our study, HM patient 102 had concomitant symptoms of arthritis. Highly-pathogenic predicted genes B3GALNT2 and C5orf42 were identified in this patient, which are associated with brain abnormalities that can cause a variety of neuropathic alterations, including abnormal cerebellar development (an abnormality that includes both dysplasia and hypoplasia scenarios). In addition, B3GALNT2 and C5orf42 can also cause pathological changes in the nucleus accumbens. Whether the above genes play a role in the development of chronic arthritis deserves further investigation. Moreover, our findings provide genetic evidence for the inference of Rose et al.21.
The prevalence of seasonal affective disorder (SAD) in the healthy population is 1.5% to 9%22. Polish scholars first reported in 2016 that the prevalence of SAD in the visually impaired group was significantly higher than that in the control group23,24. Furthermore, The increase in ocular growth and the occurrence of SAD may be caused by inappropriate exposure to blue light25,26. We identified the PCDH15 in HM patient 103, which is expressed in several tissues and organs of the body. PCDH15 is closely related to electroencephalogram abnormalities in neuropsychiatric diseases, intellectual disability, and the development of schizophrenia27,28,29. The genes CDHR1, INPP5E, BBS1, SLC7A14 and LRP5 which showed strong interactions with high myopia and neuropsychiatric disorders were identified in patients 90, 91, 93 and 96, respectively, all of which have an impact on retinal nutrition and function, thus supporting the hypothesis that retinal dysfunction may play a role in the pathogenesis of some cases of SAD30.
Ong et al. found that in the elderly population, myopic patients were twice as likely to develop cognitive dysfunction compared to normal individuals31. This suggests that myopia may be a major contributing factor in the development of cognitive dysfunction. Some results examining this association suggest a possible correlation between cognitive function and myopia32,33,34. A large cross-sectional study of 1,022,425 Israeli adolescents demonstrated a strong association between cognitive function and myopia35. One hypothesis is based on the biological relationship between myopia and cognitive function, suggesting that myopia arises due to over development of the eye, which itself is correlated with the brain, and thus leads to higher IQ in myopic individuals36,37. Another hypothesis suggests that there is a pleiotropic relationship between cognitive function and myopia. For example, brain disorders and eye disorders are affected by the same gene or set of genes at the same time38,39. Our study identified 62 causative genes for ocular diseases and 22 causative genes for neuropsychiatric disorders among the most pathogenic genes in HM patients. Of these, 15 genes are expressed in both the eye and brain, namely SORBS3, FKRP, SOX5, PLCH2, TRMT44, POMT2, PCDH15, PLK4, ARL13B, BBS1 B3GALNT2, CDH23, BBS9, SALL4, FKRP. This finding provides new evidence to support the second hypothesis that brain and eye diseases may be affected by the same gene or group of genes at the same time. In addition, the association of some neuropsychiatric disorders genes with HM phenotypes has not yet been clarified, but these genes are localized in the locus region of the HM gene and their positioning on the chromosome exhibits absolute positional similarity. Therefore, further studies are required to investigate any potential functional interactions.
Resting-state functional magnetic resonance imaging analysis shows that the brains of HM patients exhibit low-frequency fluctuations and amplitude changes of the default mode network (DMN) that are different from healthy people, demonstrating the correlation between HM and cognitive changes40. In this study, a total of 22 genes in HM patients were identified as strongly pathogenic and expressed in the brain, 11 of which were adjacent to the MYP locus, PTPRN2 on MYP4, PCDH15 and CDH23 on MYP15, PLK4 immediately adjacent to the MYP11 region, POMT2 near the MYP18 region, TRMT44 is in the 4p16 interval as well as MYP23, and the PLCH2, B3GALNT2, NPHP3, C5orf42 and SORBS3 have corresponding high proximity loci nearby. Our finding may provide biological evidence for the above imaging scan results. According to Huang et al. structural brain dysfunction may result from HM accompanied by damage to the visual cortex41. Liu J et al. reveals a positive correlation between mGMV and c-fos and NeuN expression in visual cortex of form-deprivation myopia (FDM) rats, also suggesting the relationship between cortical activity and the structural plasticity in visual cortex42. Our study identified 15 genes expressed both in the eye and neuropsychiatric disorders, among which protein interactions between PLCH2, BBS9, ARL13B, and BBS1 were evident. Further studies are required to determine whether these genes are associated with altered gray matter volume in the brain.
WES is an important tool for the detection of genetic disorders. It enables in-depth characterization of the relationship between specific genetic mutations and different diseases. However, due to the widespread genetic heterogeneity and clinical phenotypic heterogeneity, the understanding of both phenomena is still evolving. In this study, WES of patients with HM phenotype revealed a large proportion of neuropsychiatric disorders gene clusters in addition to ocular disease genes, which provides a new perspective to better understand the pathogenesis in patients with complex clinical phenotypes. Most previous studies have entailed biased analysis of WES data after identification of clinical phenotypes; however, comprehensive analysis of WES data can also facilitate the discovery of relevant complex clinical phenotypes. In particular, comprehensive analysis of WES data in a patient population with well-defined clinical phenotypes may reveal associations between different clinical phenotypes while exploring the disease itself. The two complement each other, ultimately forming a closed loop for accurate diagnosis of genetic diseases.
The study had some limitations. For one thing, this study lack of comprehensive clinic data from patients with HM, as an eye hospital without neuropsychiatric testing facilities, we cannot refer patients to other hospitals for neuropsychiatric testing if the patients report no relevant signs or symptoms. For another, the study is limited by the small sample size, although our synthesis-driven analysis of WES data has improved the accuracy of genetic pathogenicity in HM patients. Whether the genes we discovered act on both high myopia and neuropsychiatric diseases needs to be verified with a larger HM sample size. Coincidentally, 10 of 15 genes that act on both high myopia and neuropsychiatric diseases included FKRP, POMT2, PCDH15, PLK4, ARL13B, BBS1, CDH23, BBS9, SALL4, FKRP were also discovered in the WES data of high myopia population genetics study by Wan L et al.43. Additionally, PCDH15, CDH23, BBS1, and BBS9 were simultaneously verified in the WES sequencing results of 325 high myopia patients in Guangdong, China, and BBS1, BBS9 were verified again in the WES data of 6215 school-aged children with high myopia44,45.
In summary, we found Causative genes for neuropsychiatric disorders accounted for the high proportion of genes that exhibited high pathogenicity in HM patients, to our knowledge, this is the first study of genes interacting with HM and Neuropsychiatric Disorders in Northwest China. The findings presented three causative genes for neuropsychiatric diseases falled at the HM locus. These findings expand our knowledge of the WES data, and provide clues for further genetic study of complex diseases.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Change history
21 January 2024
The original online version of this Article was revised: In the original version of this Article the email addresses for authors Wei-Gang Xu and Wen-Juan Zhuang were interchanged. Correspondence and requests for materials should be addressed to Wei-Gang Xu (dr.xuwg@163.com) or Wen-Juan Zhuang (zh_wenj@163.com).
References
Zhao, Q. et al. Quantitative analysis of peripheral retinal defocus checked by multispectral refraction topography in myopia among youth. Chin. Med. J. 136(4), 476–478 (2023).
Deng, J. et al. Myopic maculopathy among Chinese children with high myopia and its association with choroidal and retinal changes: the SCALE-HM study. Br. J. Ophthalmol. https://doi.org/10.1136/bjo-2022-321839 (2023).
Tariq, F. et al. Advances in myopia prevention strategies for school-aged children: A comprehensive review. Front. Public Health https://doi.org/10.3389/fpubh.2023.1226438 (2023).
Zhu, X. et al. CCL2-mediated inflammatory pathogenesis underlies high myopia-related anxiety. Cell Discov. 9(1), 94 (2023).
Binder, K. W., Wrzesińska, M. A. & Kocur, J. Anxiety in persons with visual impairment. Psychiatr Pol 54(2), 279–288 (2020).
Sankaridurg, P. et al. IMI impact of myopia. Invest. Ophthalmol. Vis. Sci. 62(5), 2 (2021).
Ali, S. G. et al. A systematic review: Virtual-reality-based techniques for human exercises and health improvement. Front. Public Health 11, 1143947 (2023).
Martínez-Albert, N., Bueno-Gimeno, I. & Gené-Sampedro, A. Risk factors for myopia: A review. J. Clin. Med. 12(18), 6062 (2023).
Tian, Q. et al. GLRA2 gene mutations cause high myopia in humans and mice. J. Med. Genet. 60(2), 193–203 (2023).
Bray, N. J. & O’Donovan, M. C. The genetics of neuropsychiatric disorders. Brain Neurosci. Adv. 2, 2398212818799271 (2019).
Li, K., Wang, Q., Wang, L. & Huang, Y. Cognitive dysfunctions in high myopia: An overview of potential neural morpho-functional mechanisms. Front. Neurol. 13, 1022944 (2022).
Vilboux, T. et al. Cystic cerebellar dysplasia and biallelic LAMA1 mutations: A lamininopathy associated with tics, obsessive compulsive traits and myopia due to cell adhesion and migration defects. J. Med. Genet. 53(5), 318–329 (2016).
von Scheibler, E. N. M. M. et al. Ocular findings in 22q11.2 deletion syndrome: A systematic literature review and results of a Dutch multicenter study. Am. J. Med. Genet. A 188(2), 569–578 (2022).
Liu, Y. et al. Whole-exome sequencing in a cohort of high myopia patients in northwest China. Front. Cell Dev. Biol. 9, 645501 (2021).
Craven, K. E. et al. Optimizing insertion and deletion detection using next-generation sequencing in the clinical laboratory. J. Mol. Diagn. 24(12), 1217–1231 (2022).
Wang, H. et al. Disrupted topological organization of white matter structural networks in high myopia patients revealed by diffusion kurtosis imaging and tractography. Front. Neurosci. 17, 1158928 (2023).
Ou, Y. N. et al. The genetic architecture of fornix white matter microstructure and their involvement in neuropsychiatric disorders. Transl. Psychiatry 13(1), 180 (2023).
Seitler, B. N. Separation-individuation issues and castration anxiety: their curious influence on the epigenesis of myopia. Am. J. Psychoanal. 69(3), 221–237 (2009).
Augustin, A. et al. Anxiety and depression prevalence rates in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 48(4), 1498–1503 (2007).
Yokoi, T. et al. Predictive factors for comorbid psychiatric disorders and their impact on vision-related quality of life in patients with high myopia. Int. Ophthalmol. 34, 171–183 (2014).
Rose, K. E. & Tullo, A. B. Myopia. Br. J. Ophthalmol. 82(10), 1220–1220 (1998).
Thaler, M., Biedermann, R., Lair, J., Krismer, M. & Landauer, F. Cost-effectiveness of universal ultrasound screening compared with clinical examination alone in the diagnosis and treatment of neonatal hip dysplasia in Austria. J. Bone Jt. Surg. Br. 93(8), 1126–1130 (2011).
Łazarczyk, J. B. et al. The differences in level of trait anxiety among girls and boys aged 13–17 years with myopia and emmetropia. BMC Ophthalmol. 16, 1–7 (2016).
Furman, D. J., Hamilton, J. P., Joormann, J. & Gotlib, I. H. Altered timing of amygdala activation during sad mood elaboration as a function of 5-HTTLPR. Soc. Cogn. Affect. Neurosci. 6(3), 270–276 (2011).
Cougnard-Gregoire, A. et al. Blue light exposure: Ocular hazards and prevention-a narrative review. Ophthalmol. Ther. 12(2), 755–788 (2023).
Mure, L. S. Intrinsically photosensitive retinal ganglion cells of the human retina. Front. Neurol. 12, 636330 (2021).
Albaradie, R., Uzair, M. & Bashir, S. Sensorineural hearing loss due to a novel mutation in the PCDH15 gene: A case study. Brain Disord. 9, 100064 (2023).
Bruno, L. P. et al. New candidates for autism/intellectual disability identified by whole-exome sequencing. Int. J. Mol. Sci. 22(24), 13439 (2021).
Ishii, T. et al. In vitro modeling of the bipolar disorder and schizophrenia using patient-derived induced pluripotent stem cells with copy number variations of PCDH15 and RELN. Eneuro https://doi.org/10.1523/ENEURO.0403-18.2019 (2019).
Dam, H. & Hageman, I. High prevalence of seasonal affective disorder among persons with severe visual impairment. Br. J. Psychiatry 208(1), 56–61 (2016).
Ong, S. Y. et al. Myopia and cognitive dysfunction: The Singapore Malay eye study. Invest. Ophthalmol. Vis. Sci. 54(1), 799–803 (2013).
Morgan, I. G. et al. IMI risk factors for myopia. Invest. Ophthalmol. Vis. Sci. 62(5), 3 (2021).
Peled, A. et al. Myopia and BMI: A nationwide study of 1.3 million adolescents. Obesity 30(8), 1691–1698 (2022).
Dudley, S. E. & Xie, Z. Designing a choice architecture for regulators. Public Admin. Rev. 80(1), 151–156 (2020).
Megreli, J., Barak, A., Bez, M., Bez, D. & Levine, H. Association of Myopia with cognitive function among one million adolescents. BMC Public Health 20(1), 1–9 (2020).
Hirsch, M. J. The relationship between refractive state of the eye and intelligence test scores. Optometry Vision Sci. 36(1), 12–21 (1959).
Miller, E. M. On the correlation of myopia and intelligence. Genet. Soc. Gen. Psychol. Monogr. 118(4), 361–383 (1992).
Karlsson, J. L. Influence of the myopia gene on brain development. Clin. Genet. 8(5), 314–318 (1975).
Mak, W. et al. Myopia as a latent phenotype of a pleiotropic gene positively selected for facilitating neurocognitive development, and the effects of environmental factors in its expression. Med. Hypotheses 66(6), 1209–1215 (2006).
Zhang, X. W., Dai, R. P., Cheng, G. W., Zhang, W. H. & Long, Q. Altered amplitude of low-frequency fluctuations and default mode network connectivity in high myopia: A resting-state fMRI study. Int. J. Ophthalmol. 13(10), 1629 (2020).
Huang, X. et al. Altered whole-brain gray matter volume in high myopia patients: A voxel-based morphometry study. Neuroreport 29(9), 760 (2018).
Liu, J. et al. Altered whole-brain gray matter volume in form-deprivation myopia rats based on voxel-based morphometry: A pilot study. Front. Cell Neurosci. 17, 1113578 (2023).
Wan, L., Deng, B., Wu, Z. & Chen, X. Exome sequencing study of 20 patients with high myopia. PeerJ 6, e5552 (2018).
Zhou, L., Xiao, X., Li, S., Jia, X. & Zhang, Q. Frequent mutations of RetNet genes in eoHM: Further confirmation in 325 probands and comparison with late-onset high myopia based on exome sequencing. Exp. Eye Res. 171, 76–91 (2018).
Yu, X. et al. Whole-exome sequencing on 6215 school-aged children reveals the importance of genetic testing in high myopia. medRxiv 60, 2023–06 (2023).
Acknowledgements
National Natural Science Foundation of China (82060182), National Natural Science Foundation of China project pre-experiment in People's Hospital of Ningxia Hui Autonomous Region (2021GZRYSY034) and The seventh batch of young scientific and technological talents in the Ningxia autonomous region Provide financial support.
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.L.(1); Sample Collection: Y.L.(2), W.Z.; Bioinformatics Analysis: Z.-Q.X., F.-X.Z.; Writing—Preparing the manuscript: Y.L.(1); Writing—Reviewing and Editing: W.-G.X, W.-J.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, Y., Liu, Y., Zhang, W. et al. Screening of genes interacting with high myopia and neuropsychiatric disorders. Sci Rep 13, 18347 (2023). https://doi.org/10.1038/s41598-023-45463-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-45463-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.