Abstract
COVID-19 has resulted in significant morbidity and mortality globally. We develop a model that uses data from thirty days before a fixed time point to forecast the daily number of new COVID-19 cases fourteen days later in the early stages of the pandemic. Various time-dependent factors including the number of daily confirmed cases, reproduction number, policy measures, mobility and flight numbers were collected. A deep-learning model using Bidirectional Long-Short Term Memory (Bi-LSTM) architecture was trained on data from 22nd Jan 2020 to 8 Jan 2021 to forecast the new daily number of COVID-19 cases 14 days in advance across 190 countries, from 9 to 31 Jan 2021. A second model with fewer variables but similar architecture was developed. Results were summarised by mean absolute error (MAE), root mean squared error (RMSE), mean absolute percentage error (MAPE), and total absolute percentage error and compared against results from a classical ARIMA model. Median MAE was 157 daily cases (IQR: 26–666) under the first model, and 150 (IQR: 26–716) under the second. Countries with more accurate forecasts had more daily cases and experienced more waves of COVID-19 infections. Among countries with over 10,000 cases over the prediction period, median total absolute percentage error was 33% (IQR: 18–59%) and 34% (IQR: 16–66%) for the first and second models respectively. Both models had comparable median total absolute percentage errors but lower maximum total absolute percentage errors as compared to the classical ARIMA model. A deep-learning approach using Bi-LSTM architecture and open-source data was validated on 190 countries to forecast the daily number of cases in the early stages of the COVID-19 outbreak. Fewer variables could potentially be used without impacting prediction accuracy.
Similar content being viewed by others
Introduction
Coronavirus disease 2019 (COVID-19) is a global public health crisis declared a pandemic by the World Health Organization. As of March 2021, the virus had infected over 127.6 million people worldwide and the number of deaths had totaled more than 2.7 million 1. Compared to other highly contagious previously identified coronavirus-related diseases, such as Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS), the SARS-CoV-2 virus that resulted in COVID-19 disease appears to be more infectious. It is critical to explore novel approaches to monitor and forecast regional outbreaks in the early phase of the pandemic in order to facilitate better allocation of resources and containment planning 1,2,3 by healthcare providers and policymakers.
A crucial part of planning in this scenario is forecasting the daily confirmed cases of COVID-19. In the short-term, predictions can be performed by time series analysis 2. With the rapid spread of COVID-19, various forecasting, estimation, and modelling approaches are introduced. For instance, to forecast the evolution of confirmed infected cases, both epidemiological models of SIR and SER were used. In the early stages of the epidemic, a single individual can infect several people before isolation but raising public awareness, health, and stringency control, as well as policy controls and movement restrictions, may help control the epidemic. Reproduction Number (Rt), which is characterized by the number of people caused by a single individual at each stage of the outbreak, can also determine the different stages of the infection outbreak. The effective reproduction number of SIR model is used to assess the progress of the epidemic3. Using the forecast for the number of infected (I), recovered (R), and dead (D) individuals, Rt and its temporal evolution are computed. The SIR computes the theoretical number of individuals infected with a contagious illness in a closed population over time with three states: Susceptible people S(t), Infected I(t), and Recovered R(t)4. The susceptible exposed infectious recovered model (SEIR) models population are classified into four categories: S (Susceptible), E (Exposed), I (Infected), and R (Recovered) according to the states of individuals 5,6. In 7, the SIR model outperforms the SEIR model in terms of Akaike Information Criteria (AIC) to forecast and predict the confirmed cases data information.
Some preliminary studies for COVID-19 time series forecasting using Autoregressive Integrated Moving Average (ARIMA) methods have also been done 8,9,10. Many types of research based on traditional time series forecasting models have been explored to forecast future COVID cases 11,12,13. Machine learning and deep learning have developed as promising research 14,15,16 in accurately predicting the number of confirmed COVID-19 cases. In China, a stacked auto-encoder model is designed to fit the epidemic's dynamical propagation and real-time forecasting of confirmed cases 17. For forecasting using time series analysis, deep-learning using recurrent neural networks, or RNN, are proposed as promising methods to predict the risk category trend predictions 18,19. Overall, there have been many developments in the prediction of COVID-19 cases, including the use of LSTM approaches 14,18,20,21 However the analyses are limited to a number of countries (China, India, US, Canada, Australia, and European Countries) and no data on external factors such as containment measures are used in the forecasts. Furthermore, most of the studies use mean squared error (MSE) or mean absolute error (MAE) as a way to evaluate the performance of the models in a single country, which may not be applicable when comparing model performance across multiple countries.
Objectives
The US CDC has adopted an ensemble forecasting method 22 to generate 4-week forecasts for the number of deaths and confirmed cases and evaluated that accuracy of the model deteriorated at longer prediction horizons of up to four weeks. According to the US CDC 23, the number of deaths and confirmed cases is seen to fall within 30 days after containment measures are taken. We aim to use data from thirty days before a fixed time point to forecast the number of daily cases fourteen days later, which would be a reasonable time frame to facilitate planning dictions (Fig. 1). As various time-dependent factors including the number of daily confirmed cases, reproduction number, containment, and governmental policy measures, mobility and flight data could affect the daily number of cases in the future, these data, where available, were included in the analyses.
Methods
Datasets
Data on daily new cases from the earliest date to 22 Jan 2020 to 31 Jan 2021 were collected from the Johns Hopkins University research databases (https://github.com/CSSEGISandData/COVID-19) 24. Numerical data on twenty-four time-dependent variables of 190 countries were collected from various sources such as the website ourworldindata.com 25. Flight data, where available, were collected from the Official Airline Guide (OAG) 26. Effective Rt is a well-known parameter to evaluate the propagation of the outbreak and is thus used as one of the input variables to predict the daily confirmed cases in this study. The computation of effective Rt is adopted from 3. The details of these variables are listed in Table 1.
The dataset is updated daily with new information. For this experiment, all data from 22 January 2020 to 31 January 2021 were used. Time series points with a missing numbers were replaced with 0.
Overview of approach
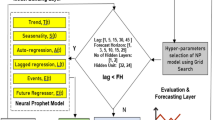
This study proposes a deep-learning framework for COVID-19 time-series prediction. The framework is illustrated in Fig. 2.
Feature engineering
Data pre-processing is one of the crucial steps in machine learning. The time-series data of 190 countries were collected, pre-processed, and analysed for each country. Standard scaling was applied using StandardScaler() in Python 3.7 to scale each of the 24 variables to zero mean and unit variance. As the model requires a sequence of past observations as input and maps it to the output observation, thirty days of time steps up till the current day are used as input, and a one-time step of the target variable fourteen days later is used as output for the one-step prediction that is being learned.
Modelling
Modelling is done individually for each country and has been done in two main stages: the training and testing stage. Data for the training stage comprised data from 22 January 2020 to 8 Jan 2021 for a total of 353 days, and data for the testing stage span the period from 9 Jan 2021 to 31 Jan 2021. The raw data is pre-processed, standardized, and then used to build the deep learning model.
BiLSTM
Time series of daily new confirmed COVID-19 cases were used for generating 14-day forecasts using Bidirectional Long-short Term Memory models (BiLSTM). A BiLSTM is an enhanced version of the LSTM algorithm. LSTMs were designed to process sequences of data and improved upon traditional RNN by using memory cells that can store information in memory for long series and a set of gates to control the flow of this memory information. These innovations allow LSTM to learn longer-term dependencies in sequential data. One of the limitations of LSTM is that the current state can only be reconstructed through the backward context. The BiLSTM algorithm fuses the ideal functions of bidirectional RNN and LSTM. This is done by combining two hidden states, which allow information to come from the backward layer and the forward layer. The BiLSTMs were trained on varying sizes of input sequences—sequence sizes of 128, 64, and 20. Detailed modelling can be seen in Fig. 3.
Hyper-parameter tuning
Hyper-parameter tuning is conducted with trial and error during the training. In the experiments, rmsprop optimizer with a learning rate of 0.1 was used for training the LSTM models, and the mean absolute error was used as the loss function. After that, the models with the selected hyper-parameters were used to forecast the number of COVID cases in the testing stage. The model's accuracy was verified by comparing the measured data with real data via different statistical indicators, including Root Mean Square Error (RMSE), Mean Absolute Error (MAE), Mean Absolute Percentage Error (MAPE) and total absolute percentage error (see evaluation metric).
Features used
Two sets of input features were used for each model as shown in Fig. 4. The first model (Model 1) used all features except for the computed Rt moving average. The second model (Model 2) used all features except the confirmed and recovered cases (as the information was partially captured in daily new cases), estimated Rt (as the information was partially captured in Rt moving average), number of new tests done each day, as well as international travel controls (as the information was partially captured in flight data).
Input features used for prediction. Key: flights—daily number of flights; deaths—cumulative number of COVID-19 deaths, confirmed—cumulative number of confirmed cases; recovery—cumulative number of recovered cases; E0_movil—daily reproduction number, Rt, smoothed; E0_estimated—daily reproduction number, Rt; new_tests_smoothed—daily test numbers; new_tests_smoothed_per_thousand—daily test numbers per thousand population; retail_and_recreation, grocery_and_pharmacy, parks, transit_stations. workplaces, residential—mobility data from Google contact_tracing—level of contact tracing (3 levels); restrictions_internal_movements—restrictions on internal movement during the COVID-19 pandemic (3 levels); containment_index—Containment and Health Index, a composite measure of eleven response metrics; stringency index—Government Stringency Index, a composite measure of nine response metrics; international_travel_controls—government policies on restrictions on international travel controls. (5 levels); facial_coverings—use of face coverings outside-of-the-home; stay_home_requirements—government policies on stay-at-home requirements or household lockdowns; cancel_public_events—government policies on the cancellation of public events; school_closures—government policies on school closures.
Evaluation metric
In addition to RMSE, MAE and MAPE, total absolute percentage error was used for evaluating the performance of the models as shown.
where TotalActual and TotalPredicted refer to the actual and predicted sum of total new cases over the testing period, respectively. A sub-analysis was performed in 84 countries with more than 10,000 cases over the predicted period (or an average of 434 new cases a day) as percentage error could be inflated for models with very few cases and projections would be more useful for capacity planning for countries with a large number of cases. The top five countries with the best and worst performance were analysed in terms of the number of cases per day, phase of infection and number of infections waves experienced.
Comparison with classical ARIMA model
To test the effectiveness of the new method, an Autoregressive Integrated Moving Average (ARIMA) model was used to generate predictions over the same period using only daily new cases as inputs. Results from the classical model were summarised using the same evaluation metrics as the other two models.
Ethics approval
This study is supported by NUS-IRB-2020-812 under National University of Singapore.
Results
Summary by MAE, RMSE, MAPE, percentage error
Median MAE was 157 new daily cases (IQR: 26–666) under the first model, 150 (IQR: 26–716) under the second model, and 130 (IQR: 22–475) under the ARIMA model (Table 2). The countries and their respective performance with Models 1 and 2 are listed in Supplementary Material 1. However, the effectiveness of the model is hard to gauge with MAE and RMSE alone as some countries may report thousands of daily infections, while some others only a handful of cases a day. As seen in Table 3, the worst-performing countries are countries with large numbers reported. For over half of the countries, the percentage error in terms of the total number of cases over the predicted period was at most 51% for the first model, 53% for the second model, and 41% for the ARIMA model However, the maximum error was higher under the ARIMA model as compared to models 1 and 2.
Analysis of the countries with the best and worst results in terms of percentage error shows that countries with better performing results typically had more cases per day and had witnessed more waves of COVID outbreaks over the training period (Table 4). Countries that fared more poorly had fewer daily cases and were usually not in the middle of any covid wave. This is because smaller daily cases would lead to larger percentage errors due to low base effects.
Sub-analysis in countries with more than 10,000 cases
A sub-analysis was performed in 84 countries with more than 10,000 cases over the predicted period (or an average of 434 new cases a day). The median percentage error in terms of the total number of cases over the predicted period was lower when limited to these countries—at 33% for the first model with more variables, and 34% for the second model with fewer variables (Table 5). The percentage error was 16% or less in a quarter of cases in the first model, while the maximum error was 166% for the first model and 191% for the second. While the median percentage error was similar in the ARIMA model (32%, IQR 11–53%), Model 1 (33%, IQR 18–59%), and Model 2 (34%, IQR 16–66%), the maximum error was greater under the classical ARIMA model (462%) than under the other two models (166%, 192%).
Top 5 countries with best and worst model performance
Table 6 summarises the characteristics of the top 5 countries with the best and worst model performance. After limiting the analysis to countries with more than 10,000 cases over the prediction period, both the sets of top-performing and worst-performing models comprised countries from various regions, and most countries in the declining phase of an infection wave. However, the countries which fared better under models 1 and 2 had experienced a greater number of infection waves by the time of prediction and still had a slightly greater number of cases per day. In addition, model 2 appeared to have better performance in countries with a greater number of daily cases per day. As model 2 excluded the use of variables such as the number of tests done, it might appear that such variables are less useful and could introduce noise when the number of cases was relatively high. Model 1 appeared to produce better predictions over a greater variety of trends in daily cases (increasing, out of COVID wave, declining cases), which could suggest better generalizability across country profiles.
Figures 5 and 6 show the scatter plot of ranking in terms of percentage error, and absolute percentage error respectively. There were some countries in which Model 1 performed better (Moldova, Jordan, Croatia, Switzerland), and which Model 2 performed better (Czechia, UK, Turkey, Germany). The charts showing the predictions and actual cases for these seven countries are in Supplementary Material 2. In both cases, the poorer-performing model tended to overpredict the number of cases. However, Models 1 and 2 performed more poorly under different scenarios. Model 1 wrongly predicted that the rise in cases would continue when cases were at the peak and about to decline (Czechia, UK), while Model 2 provided higher estimates during a declining phase of infections (Moldovia, Jordan, Croatia, Switzerland).
Limitations and strengths
The models' limitations are their restricted applicability only on some outbreak stages and with the availability of enough data. During model development, it was assumed that the intensity and coverage of surveillance and testing were consistent throughout the whole period as well as across the different countries, which realistically may not be possible due to a potential shortage of resources. With the roll-out of vaccination programmes, daily number of new cases is expected to decrease with the same input variables. Therefore, the validation was only performed using data collected before 31 January 2021, when the vaccination campaign has just started globally. On the other hand, the emergence of new virus variants with different transmissibility could also impact the performance of the models. The Omicron variant had become the dominant strain of the virus and is known to be more transmissible but less deadly. Nevertheless, the availability of open-source data and previous training of the model developed may make it useful in forecasting for outbreaks of a similar nature, especially during the early stages of an outbreak. The model with fewer inputs performed reasonably well compared to the model with more inputs, suggesting that in the case of fewer data available, a reasonable forecast could still be obtained. Nevertheless, given the varying results, it is recommended that individual models with individual sets of variables be trained specifically in those countries, using all variables as a starting point.
The strength of the models is that they draw upon readily available data on a country’s national, healthcare, social and economic status to generate predictions, and have been validated on 84 countries with more than 10,000 total cases over the prediction period that are different geographically, politically, and culturally. By running the model on 84 countries, an estimate of the maximum possible error is obtained, allowing for planning of best and worst case scenarios. An additional strength of the models is the ability to generate predictions fourteen days in advance, without knowledge of the number of cases or changes in the upcoming thirteen days. As such, these predictions would be useful for facilitating the better allocation of resources and containment planning by healthcare providers and policymakers over a longer time horizon.
Discussion
Previous work has been done on COVID-19 forecasting using both classical and machine learning methods. Miralles-Pechuán et al. compared the performance of state-of-the-art machine learning algorithms, such as long-short-term memory networks, against that of online incremental machine learning algorithms to predict the coronavirus cases for the 50 countries with the most cases during 2020 27. Kasilingam et al. used exponential growth modelling studies to understand the spreading patterns of SARS-CoV-2 and identify countries that showed early signs of containment until March 26, 2020 28. Saba et al. applied time-series and machine learning models to forecast daily confirmed infected cases and deaths due to COVID-19 for countries under various types of lockdown (partial, herd, complete) 29. While these studies used machine learning and acknowledged the impact of containment measures on the daily case numbers, our study is novel in terms of the variates used and approach.
To our best knowledge, this study is the first that leverages open-source data including flight data to perform COVID-19 time series forecasting on 190 countries using a machine learning approach. Only data up to 31 January 2021 could be accessed at the time the analysis was conducted. The model was able to predict the total number of cases across a period of 14 days in advance from 9 Jan 2021 to 31 Jan 2021 with a median of 35% error amongst countries with more than 10,000 cases over the predicted period, or an average of 434 new cases per day. When tested on countries with more than 10,000 cases over the predicted period, maximum error was much smaller for the Bi-LSTM model than a classical ARIMA model, suggesting that using more variables and machine learning methods could help to minimise the maximum error.
The model was developed in 190 countries but validated over 84 countries with more daily cases. Further fine-tuning of the models to create a country-specific model is warranted given the varying results across different countries.
Upon analysis of the key characteristics of the top five countries with best and worst performance found that countries with the best performance in terms of percentage error had experienced more waves of COVID-19 infections prior, such that the prediction method would be more suitable for countries who had more historical data for training on. Cases were on the decline for most countries, and thus the model might be better in prediction when the trend in daily cases is stable and less accurate in predicting sudden surges 14 days in advance.
The models in our study were trained and tested in isolation on each country, that is, model weights obtained from training on one country were not used for prediction on another country. Given that the models performed relatively well on countries which had experienced earlier of outbreaks, one area for future work would be to investigate if pre-training the model on countries with more cases and fine-tuning it on another country with fewer would produce better results. Model 1 appeared to generate more reliable estimates across a variety of stages of COVID infections, suggesting that generally Model 1 should be used as a default model for all countries first. However, Model 2 seemed to generate better predictions for countries with higher daily cases, suggesting a more parsimonious model could be used instead to achieve better accuracy.
In addition, given the tendency of Model 1 to predict sharp increases when cases were on the decline, any sharp increases predicted by Model 1 should be further substantiated with information on the current situation in the country. As Model 2 tended to overpredict the number of cases during a decline, predictions from model 2 may be taken as an upper bound prediction, rather than the actual number when cases are starting to decline.
As discussed, there are some limitations of this study due to limitations in data availability. Underlying factors may have been missed when data were obtained, adding a degree of uncertainty in the predictions. An example of this is that daily case counts may be drastically high during the forecast phase with the ease of restrictive measures, and these pieces of information may not be present in historical data. It is also unrealistic to fully account for these potential uncertainties, which directly affect the performance of predictive models and cause inaccurate predictions of future cases.
Forecasts can provide potentially useful information to facilitate better allocation of resources and containment planning by healthcare providers and help policymakers manage the consequences of COVID-19 over a longer time horizon. For future work, an ensembling approach to combine both models and potentially other time-series candidate models can be explored. This stems from our observation that a single model might not be able to capture and predict the complex nature of the virus transmission, and a combination of different models will be able to account for the inherent weaknesses of each candidate model. Additionally, for future pandemics involving new variants or viruses, there is potential to apply transfer learning to model them, utilizing the pre-trained bidirectional LSTM developed in this work. This may be able to speed up prediction efforts in a bid to curb the viral spread effectively.
The approach of our study is both model-driven and data driven. While input variables had been selected after literature review on the factors affecting COVID-19 transmission, the data-driven aspect came from the daily data that were provided for each country that was used to fine-tune the model. We believe the data science approach presented in this paper can be generalised for other time-series forecasting applications which use multivariate data.
Conclusion
A deep-learning approach using Bi-LSTM architecture and open-source data was developed to forecast the new daily number of COVID-19 cases 14 days in advance across 190 countries during the early phase of a pandemic and evaluated using absolute percentage error. The model with fewer variables performed reasonably well compared to the model with more inputs. A deep-learning approach using Bi-LSTM architecture and open-source data can be used as a starting point for forecasting the new daily number of COVID-19 cases 14 days in advance and fewer variables could potentially be used without impacting prediction accuracy.
Data availability
The datasets generated during and/or analysed during the current study are available from the Johns Hopkins University research database (github.com/CSSEGISandData/COVID-19), Our World in Data (ourworldindata.com) and OAG (oag.com).
Code availability
Software application or custom code: The code that support the findings of this study are available from the corresponding author upon reasonable request.
References
Worldometers info. https://www.worldometers.info/coronavirus/. Accessed 2021.
Ceylan, Z. Estimation of COVID-19 prevalence in Italy, Spain, and France. Sci. Total Environ. 729, 138817 (2020).
Medina-Ortiz, D., Contreras, S., Barrera-Saavedra, Y., Cabas-Mora, G. & Olivera-Nappa, Á. Country-wise forecast model for the effective reproduction number RT of coronavirus disease. Front. Phys. 8, 304 (2020).
Liu, Z., Magal, P., Seydi, O., & Webb, G. Predicting the cumulative number of cases for the COVID-19 epidemic in China from early data. arXiv preprint arXiv:2002.12298 (2020).
Peng, L., Yang, W., Zhang, D., Zhuge, C., & Hong, L. Epidemic analysis of COVID-19 in China by dynamical modeling. arXiv preprint arXiv:2002.06563 (2020).
Jia, L., Li, K., Jiang, Y., & Guo, X. Prediction and analysis of coronavirus disease 2019. arXiv preprint arXiv:2003.05447 (2020).
Roosa, K. et al. Real-time forecasts of the COVID-19 epidemic in China from February 5th to February 24th, 2020. Infect. Disease Model. 5, 256–263 (2020).
Bayyurt, L., & Bayyurt, B. Forecasting of COVID-19 cases and deaths using ARIMA models. medrxiv (2020).
Benvenuto, D., Giovanetti, M., Vassallo, L., Angeletti, S. & Ciccozzi, M. Application of the ARIMA model on the COVID-2019 epidemic dataset. Data Brief 29, 105340 (2020).
Chintalapudi, N., Battineni, G. & Amenta, F. COVID-19 virus outbreak forecasting of registered and recovered cases after sixty day lockdown in Italy: A data driven model approach. J. Microbiol. Immunol. Infect. 53(3), 396–403 (2020).
Wu, J. T., Leung, K. & Leung, G. M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 395(10225), 689–697 (2020).
Zhuang, Z. et al. Preliminary estimation of the novel coronavirus disease (COVID-19) cases in Iran: A modelling analysis based on overseas cases and air travel data. Int. J. Infect. Dis. 94, 29–31 (2020).
Kucharski, A. J. et al. Early dynamics of transmission and control of COVID-19: A mathematical modelling study. Lancet Infect. Dis. 20(5), 553–558 (2020).
Kırbaş, İ, Sözen, A., Tuncer, A. D. & Kazancıoğlu, F. Ş. Comparative analysis and forecasting of COVID-19 cases in various European countries with ARIMA, NARNN and LSTM approaches. Chaos Solitons Fractals 138, 110015 (2020).
Rustam, F. et al. COVID-19 future forecasting using supervised machine learning models. IEEE Access 8, 101489–101499 (2020).
Tuli, S., Tuli, S., Tuli, R. & Gill, S. S. Predicting the growth and trend of COVID-19 pandemic using machine learning and cloud computing. Internet Things 11, 100222 (2020).
Hu, Z., Ge, Q., Li, S., Jin, L., & Xiong, M. Artificial intelligence forecasting of covid-19 in China. arXiv preprint arXiv:2002.07112 (2020).
Chimmula, V. K. R. & Zhang, L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos Solitons Fractals 135, 109864 (2020).
Swapnarekha, H., Behera, H. S., Nayak, J. & Naik, B. Role of intelligent computing in COVID-19 prognosis: A state-of-the-art review. Chaos Solitons Fractals 138, 109947 (2020).
Zeroual, A., Harrou, F., Dairi, A. & Sun, Y. Deep learning methods for forecasting COVID-19 time-Series data: A comparative study. Chaos Solitons Fractals 140, 110121 (2020).
Shastri, S., Singh, K., Kumar, S., Kour, P. & Mansotra, V. Time series forecasting of Covid-19 using deep learning models: India-USA comparative case study. Chaos Solitons Fractals 140, 110227 (2020).
Ray, E. L. et al. Ensemble Forecasts of Coronavirus Disease 2019 (COVID-19) in the US. MedRXiv (2020).
Deb, P., Furceri, D., Ostry, J. D., & Tawk, N. The effect of containment measures on the COVID-19 pandemic (2020).
Dong, E., Du, H. & Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Diseases 20(5), 533–534 (2020).
O. W. i. Data. https://ourworldindata.org/. Accessed 2021.
O. f. data. https://www.oag.com/flight-status-data. Accessed 2021.
Miralles-Pechuán, L., Kumar, A. & Suárez-Cetrulo, A. L. Forecasting COVID-19 cases using dynamic time warping and ncremental machine learning methods. Expert Syst. 40, e13237 (2023).
Kasilingam, D. et al. Exploring the growth of COVID-19 cases using exponential modelling across 42 countries and predicting signs of early containment using machine learning. Transbound. Emerg. Dis. 68, 1001–1018 (2021).
Saba, T., Abunadi, I., Shahzad, M. N. & Khan, A. R. Machine learning techniques to detect and forecast the daily total COVID-19 infected and deaths cases under different lockdown types. Microsc. Res. Tech. 84, 1462–1474 (2021).
Acknowledgements
The authors would like to thank Á.Olivera-Nappa and D. Medina-Ortiz for providing the methodology for estimating estimating the daily Rt values in each country as mentioned in the paper "Country-Wise Forecast Model for the Effective Reproduction Number Rt of Coronavirus Disease".
Author information
Authors and Affiliations
Contributions
N.N.A. and H.X.T. designed the study, acquired the data and analysed and interpreted the results. J.P. and M.C.H.C. were involved in drafting the paper and revising it critically for important intellectual content. All authors give final approval of the version to be submitted.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aung, N.N., Pang, J., Chua, M.C.H. et al. A novel bidirectional LSTM deep learning approach for COVID-19 forecasting. Sci Rep 13, 17953 (2023). https://doi.org/10.1038/s41598-023-44924-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-44924-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.