Abstract
Dietary patterns are changing severely, especially the consumption of highly processed foods with lots of spices is increasing, carrying an increased risk of immediate hypersensitivity (type I), in sensitised individuals, due to the possible presence of allergens, especially the hidden ones. Paprika is a fruit of the Capsicum genus, which belongs to the Solanaceae family and is commonly consumed fresh or as a spice. Despite recorded cases of anaphylaxis, its allergenicity has yet to be clearly investigated. In this study, we research to identify proteins that could trigger a severe allergic reaction in patients with an equivocal clinical picture. Two types of protein extracts extracted from 3 different paprika spices were immunoblotted with sera from patients with severe allergic symptoms, presumably to paprika. Proteins from the IgE reactive bands obtained were subjected to LC–MS/MS identification and then in silico analysis to assess their possible sensitising capacity and proinflammatory potential using online tools. The spices were shown to contain a number of incompletely investigated highly immunoreactive allergenic proteins, including proteins of foreign origin (contaminants), the presence of which can stimulate inflammatory mechanisms and cross-reactivity with other food allergens, which can threaten life and health and should be investigated in detail.
Similar content being viewed by others
Introduction
Paprika (bell pepper, Capsicum) is a popular plant in various parts of the world. It is cultivated for direct consumption or as a raw material that undergoes technological processing to obtain powdered spices, oleoresin extract or pure capsaicin, which are used in the food, pharmaceutical or cosmetic industries. Its anti-inflammatory, anti-aging, anti-depressive, anti-cancer, and antioxidant properties have been described1. However, more research is needed into its allergenicity, which is still poorly understood.
Food allergens represent a large group of still understudied compounds, often with uncharacterised biological activity, which are additionally subject to change, either naturally or during technological processing2,3,4. Although the knowledge of them is increasing and the most important ones are already listed on food labels (14 in EU countries and 8 in the USA), there are still many under-described allergenic proteins whose accidental ingestion can lead to an adverse immune reaction, especially the hidden ones whose presence is not expected2. The safety of food, both raw materials and products, must be ensured in the global marketplace, involving plant breeding, processing, and goods distribution. So far, the potential microbial hazards of food are well understood, but harmful contaminants can come from trace pests and pesticides, water quality, soil and post-harvest processes, as well as unintentional contaminants that may occur during the production process and may represent, for example, hidden allergens that are difficult to detect. Unintentional cross-contaminants, additives that are not declared on the product label (repackaged products or sold by weight) or whose health risks are unknown, or proteins and protein-based complexes formed during the technological processing are the most difficult to identify and quickly define. They usually pose little threat to the general public but can have serious consequences for sensitive groups of consumers, such as allergy sufferers, whose numbers are growing dramatically.
In the gut, depending on protein solubility and susceptibility to proteolysis, the ingested allergen is internalised and processed in the epithelium, and allergen-derived peptides are presented to lymphocytes, which differentiate into Th2 cells in the presence of activators (such as cytokines, TSLP and DAMP), allowing the activation, differentiation and isotype switching of allergen-specific B cells into IgE-producing plasma cells5,6. Allergen-specific IgE antibodies bind to mast cells and basophils, sensitising them to the allergen. On subsequent contact with the allergen, mast cells and eosinophils degranulate, causing allergic inflammation and associated symptoms. Less obvious and recognised pathways of sensitisation are also suggested7. The main medical marker used to determine allergy status is the level of IgE, but clinical observation does not always correlate with serological test results. A sudden reaction of the body to an allergen can lead to life-threatening anaphylactic shock, which is not always associated with high levels of specific IgE7,8,9. Such cases are sometimes referred to as idiopathic10. Overlapping body reactions and allergic cross-reactions cause additional difficulties in sensitised patient follow-up. Cross-reactions between allergens with highly conserved regions of amino acid sequences and similar three-dimensional structures, as in the case of pan-allergens, are becoming increasingly common and pose a threat to allergists that needs to be thoroughly investigated11. Characterisation of the allergenicity and immunoreactivity of food proteins and peptides provides insight into the level of risk and helps to understand the complex mechanisms that cause food allergy.
Paprika allergy is occasionally diagnosed, so paprika proteins are not among the major food allergens; however accidental ingestion of paprika by an allergic person can cause a severe body reaction, including anaphylactic shock. The scientific literature of the last decade describes cases of anaphylaxis and cross-reactivity of paprika proteins with birch, Prunus representants, profilins and allergenic dyes12,13,14,15. New paprika allergens are also being discovered and verified16. Three allergens, Cap a1, Cap a 2 and Cap a 7 have been already registered in the WHO/IUIS Allergen Nomenclature Database16,17,18. However, the issue remains topical and requires in-depth work, if only because of the widespread use of this raw material. Peppers in raw or processed form can be found in commonly consumed pizzas, goulash soups, meat preparations, vegetable salads, juices, or in other foods where labelling of its presence is not always mandatory. The exception is dried whole chilli peppers, the safety and specification of which is regulated by the United Nations Economic Commission for Europe (UNECE)19. For consumers, spices pose a particular allergy risk because they are difficult to detect in food and are highly processed. Thermal treatments, including dry steam, are commonly used in the decontamination of Capsicum spices, although a combination of non-thermal methods such as infrared, UV and ozonation is also used20. All of these treatments affect the allergenicity of proteins, and as a result of biotic and abiotic environmental stress, defence proteins are produced in the tissues, which may be the hidden allergens11. Apart from clinical case reports, there is very little scientific data on the immunoreactivity of paprika, which may worry given its popularity, especially as a spice. The aim of the study was to detect immunoreactive allergenic proteins of paprika spices, including possible contaminants of environmental origin that may pose an immunological threat to the body. In the spirit of limiting research on living organisms, the extensive in vitro/in silico studies were conducted to identify the potentially most immunoreactive constituents of paprika spices. Such knowledge could be invaluable for further research into paprika allergens and for the development of protective therapies for allergic individuals.
Results
Characteristics of protein isolates
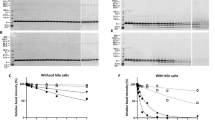
Proteins were isolated from three commercial Capsicum-based spices (mild, chilli and spicy) using two different solvents (based on PBS and TRIS–HCl buffers) and separated by SDS-PAGE (Fig. 1A). Extracts from dried mild and spicy peppers (DMP and DSP, respectively) gave a more diverse profile than that of dried chilli pepper (DCP). For DMP and DSP, in an overall comparison of protein molecular weight profiles, the extraction of 20 kDa molecules in Tris buffer was significantly higher compared to PBS buffer (p < 0.05, Fig. 2A). A similar situation was observed for proteins around 33 kDa, and slightly the opposite for proteins around 50 kDa, where PBS extraction was more efficient for DMP (p < 0.05, Fig. 2B and C, respectively). The Tris-based method yielded the most diverse protein profile from DSP and a well-defined profile of IgE-immunoreactive epitopes compared to the PBS extract (Fig. 1B). However, the 50 kDa PBS/DSP band (extracted from DSP by PBS) gave the strongest reaction with fluorescence-labelled anti human-IgE antibodies (Fig. 1B, arrow). All IgE immunoreactive proteins were detected in the 17–50 kDa molecular mass range.
Electrophoretic separation of pepper protein extracts and their IgE-reactivity with human sera (pooled): (A) Tricine-SDS-PAGE electropherogram of protein isolates (20 µg); (B) IgE-immunoreactive spices protein immunoblot. The underline bands in figure (A) corresponding to IgE-immunoreactive bands in figure (B) were subjected to LC–MS/MS identification. DMP dried mild paprika, DSP dried spicy paprika, DCP dried chili paprika, MW molecular weight marker. Full-length gel and blot are presented in Supplementary Fig. S1.
Relative signal strength for proteins of different molecular weight: (A) 20 kDa; (B) 33 kDa and (C) 50 kDa (ranges characterized as IgE reactive). Significant differences between the products were characterized by the Duncan test. The differences between the bands for different isolation methods were characterized by the student’s t test. Values p < 0.05 were considered significant and were marked with different letters.
Protein identification by LC–MS/MS analysis
The IgE immunoreactive protein bands obtained (Fig. 1) were subjected to LC–MS/MS detection. The raw data were searched by MACOT software against the Green Plant (Viridiplantae) database for all entries, to not exclude possible contaminants, and the results were then compared by BLAST with Capsicum taxid. The identified proteins are shown in Tables 1 and 2 (the raw mass spectrometry data generated and analysed are presented as Supplementary data in the supplementary material). Approximately 85% of the total identified proteins (45 out of 53) were directly assigned to the Solanaceae family by the MASCOT software (Table 1). Of these, approximately 40% (18 out of 45) were identified as being derived from Capsicum annum or C. chinense, and 44% from Solanum lycopersicum, S. tuberosum, or S. peruvianum (20 out of 44), but with high homology (78–98% identity/92–100% query cover) to Capsicum taxIDs. The others were assigned to Nicotiana tomentosiformis, N. tabacum, or N. sylvestris and their identity with Capsicum ranged 68–98% (85–100% query cover). In addition to proteins belonging the Solanaceae family, MASCOT indicated the presence of proteins belonging to other taxa that could be contaminants of the raw material (Table 2). These were proteins from Theobroma cacao, Populus nigra, Triticum aestivum, Arabidopsis thaliana, Malus domestica, Hevea brasiliensis, and Dimocarpus longan. As most of them showed high homology to Capsicum taxa (78–93% identity/71–100% query cover) we did not consider them as contaminants. The exception was rubber elongation factor protein from Hevea brasiliensis which showed low homology (43% identity) to Capsicum and was therefore considered a contaminant of the spices tested.
Allergenicity assessment using online databases
Proteins identified by LC–MS/MS and BLAST software were subjected to in silico analysis to check their allergenic potential using the online allergen databases (AllergenOnline, Allergome and Allermatch). The results for the proteins indicated by MASCOT as belonging to Solanaceae and their Capsicum homologues indicated by BLAST were similar, so we have presented the results obtained for Capsicum. For 22 of the 45 proteins analysed, the FASTA 36 or BLASTP alignment tools used showed greater than 50% identity (79–100% similar) with allergens or putative allergens described in AllergenOnline and/or Allergome databases (Table 3). Of these 22 proteins, 16 showed greater than 70% identity with allergenic sequences, with E < 1e-7. Four of these allergens have been described as allergen or putative allergen of Capsicum, i.e. Cap a Glucanase, (basic beta-1,3-glucanase), Cap a 1.0101 (osmotin-like protein, allergen), the in silico generated Cap ch 17kD (submitted name: major allergen Pru ar 1) and Cap a 4 (pathogenesis-related protein 10), and two others, i.e. Sola l 4.0201 (submitted name: PR10 protein) and Sola l Peroxidase (anionic peroxidase) as putative allergens of Solanum lycopersicum (tomato; the Solanaceae family). Our proteins showed 84–100% and 70–84% sequence identity with Capsicum and Solanum allergens, respectively. In addition, the allergen prediction tools used showed alignments of Capsicum-derived proteins with taxonomically unrelated allergens (Sin a 2, Pers a 1.0101, Cas s 9.0101, Hev b 9 and others) that even exceeded 70% identity at E < 1e-7, indicating a high potential for allergenic cross-reactions (Table 3). A significant number of proteins (23) showed relatively low homology with known allergens and are therefore presented in the supplementary data (Supplementary Table 1S).
Table 2 shows the allergenicity risk of proteins recognised by MASCOT as putative contaminants of the raw material, including proteins finally assigned to Capsicum based on BLAST matching. Three of them showed high homology (79–100% identity, E < 1e-7) to allergenic proteins with partially or fully documented IgE epitopes, including Cas s 9.0101, Hev b 1.0101 and Hev b 3.
Immunomodulatory potential of proteins
Proteins identified in silico as having a high risk of allergenicity, based on high amino acid sequence identity with allergenic proteins, were further screened in silico for the presence of proinflammatory epitopes (PiEs) and antibody-specific B cell epitopes (IgG, IgE and IgA), as well as for the presence of cytokine-inducing sequences (IL-4, IFN-γ and IL-6). The vast majority of them had peptides bearing potential PiEs or capable of inducing proinflammatory cytokines (Supplementary Table 2S). For PiEs, their possible amino acid sequences ranged from 23 to 123 for a window length of 15 and a threshold of 0.9, with the best scores ranging from 1.07 to 1.87. For IL-4 and IFN-γ, with a window length of 15 and a threshold of 0.7, it was 0–10 and 0–24 with best scores of 0.52–1.16 and 0.27–1.39 for IL-4 and IFN-γ, respectively. Possible sequences of IL-6 inducing peptides ranged from 8 to 107 and best scores from 0.18 to 0.61, with a window length of 15 and a threshold of 0.11 (upper value). The in silico analysis revealed that the proteins may differ in their ability to induce antibodies secretion. The number of Abs-inducing peptides was 0–78, 0–17 and 0–20 (best scores 0.92–1.70, 0.92–1.25 and 0.96–1.26), for IgG, IgE and IgA, respectively (Supplementary Table 2S). In turn, IgE epitopes, mapped with the AlgPred 2.0 tool, were present on 10 of all identified proteins (Supplementary Table 2S). The highest number of IgE epitopes was on rubber elongation factor protein, a contaminant of spices.
Immunomodulatory activity of selected peptides
High-risk allergenic proteins and their peptides with pro-inflammatory antigenic regions estimated by ProInflam (those with the highest SVM score and those containing IgE epitopes mapped by AlgPred 2) and peptide sequences with predicted IgE-specific B-cell epitopes predicted by IgPred (those with the highest score) were screened for binding to human major histocompatibility complex class II (MHC II), and the peptides themselves were additionally screened for cytokine induction. The PiE scores of all tested peptides were predominantly highly positive but ranged from − 0.04 (negative) to 1.89 (Table 4). The peptides were able to bind, on average, to 19 of the 24 HLA alleles tested (8.1 per 12 DRB1, 4.8/5 DQ and 6.5/7 DP), and whole proteins were able to bind on average to 23.7 of the 24 alleles tested (11.7/12 DRB1, 4.96/5 DQ and 7/7 DP). With the default settings of the tools used, approximately 24 and 26 of the 45 peptides examined appeared to be potential IL-4 and/or IL-10 inducers, whereas all appeared to be IFN-γ inducers. The inducer scores obtained were 0.22–1.55, 0.32–1.85 and 0.26–1.60 for IL-4, IL-10 and IFN-γ, respectively.
Discussion
Our studies using the immunoblotting technique showed the presence of IgE antibodies to paprika proteins in the serum of patients whose medical tests for these allergens were inconclusive, but the likely cause of the allergic reaction was the ingestion of Capsicum spices, or cross-reactivity with allergens of similar epitope structure. The IgE reactive proteins appeared to be approximately 50, 33 and 20 kDa. A total of 53 proteins were identified in the immunoreactive bonds, five of which showed 100% identity to Capsicum allergens described in the Allergome database: Cap a Glucanase, Cap a 1.0101, the in silico generated Cap ch 17kD and Cap a 4. The IgE reactive protein could also be lichenase, which showed 84% identity with Cap a Glucanase (E < 1e-7). According to the Allergome database, the allergenicity scores for these putative Capsicum allergens were based on IgE immunoblotting tests. Among the paprika allergens registered in the WHO/IUIS database, Cap a 1 (osmotin- thaumatin-like protein) was found in the 20 kDa band. As for the other two allergens, profilin (Cap a 2) and gibberellin-regulated protein (Cap a 7), although they are soluble, our sera did not react positively with proteins with a molecular weight of less than 15 kDa, so they did not react with Cap a 2 and Cap a 7 allergens. Of the remaining proteins identified in the IgE reactive bands, 11S globulin seed storage protein Jug r 4-like, 17.8 kDa and 18.5 kDa class I heat shock proteins, actin-7-like, anionic peroxidase, basic 30 kDa endochitinase precursor, enolase, hypothetical protein BC332_07738, NADPH-dependent aldehyde reductase 1, chloroplastic, and suberization-associated anionic peroxidase 2 showed very high identity (≥ 70; E < 1e-7) to known allergens or putative allergens from other plants used in the food industry. Alignment with such identity scores indicates a potential for allergenic cross-reactions. Cross-reactivity is unlikely for proteins with less than 50% identity to the entire protein sequence and is quite common above 70% identity21. According to the authors of the AllergenOnline database, sequences of two proteins having published evidence of cross-reactivity will align in AllergenOnline.org with a high percent identity (> 50% over nearly full length) and have an E score (statistical expectation score) of less than 1e-7 (0.0000001)22. Our in silico analysis showed that cross-reaction of paprika proteins with latex (Hev b 2, Hev b 9, Hev b 11), tomato (Sola t Glucanase, Sola l Glucanase, Sola I TLP, Sola I Peroxidase, Sola I 4), tobacco (Nic t Osmotin), grapes (Vit v Glucanase), mustard (Sin a 2), kiwi (Act d 2), sesame (Ses i 5, unassigned sesamum seed maturation-like protein group; ACB55491.1), avocado (Pers a 1), wheat (Tri a Endochitinase), maize (Zea a 22), banana (Mus xp 2), chestnut (Cas s 9), hazel (Cor a 13), molds (Asp f gamma Action), meadow plants (Amb a 12, Cyn d 22), cattle (Bos d Enolase), crab (Chi o alpha) and even to mostly in silico generated fish allergens (Sal s alpha Actin; Gas ac 2, Ano fi 2, Sal s 2, Tak ru 2, Ruda ni 2, Ict pu 2, Ory la 2, Dan re 2, Gil mi 2, Ruda m 2, Pan h 2, Tet ni 2) are particularly like. Unfortunately, apart from negative results for tomato and potato, we have no confirmed information on whether our patients were hypersensitive to these proteins, so we cannot indicate what cross-reactions, if any, occurred. Reactivity of plant proteins with animal allergens seems unlikely at present, as it has not yet been clearly described. However, cross-reactivity of pollen with food proteins is increasingly observed. A dangerous allergen in peppers seems to be Cap a Glucanase, which has a high identity with the latex allergen Hev b 2. The same is true for enolase, which shows 90% identity with Hev b 9. MS analysis showed that spices were contaminated with latex. This could have exacerbated the allergic reaction through cross-reactivity between allergens. Unfortunately, we have no information whether our patients were allergic to latex. Latex has many recognised IgE epitopes, that can cross-react with many food proteins. Cases of paprika allergy associated with a latex-fruit syndrome have been reported, and seems to be quite common23,24. Nevertheless, Estrada-Rodriguez et al.25 described a case of paprika allergy in which they excluded latex and rubber allergy. Although the specific IgE was low (0.34 kU/L), they, like us, confirmed the presence of IgE-reactive proteins by immunoblotting technique. Palomares et al.26 detected reactive IgG and IgE peptide epitopes common to 1,3-beta-glucanase (Ole e 9) in extracts of ash and birch pollen, tomato, potato, banana, latex and paprika. However, the described latex food allergy syndrome is most commonly recognized in patients with hypersensitivity to latex, banana, kiwi, avocado, tomato, potato, chestnut, and peach27.
Allergic diseases can lead to eating disorders, psychosocial disadvantages, and inflammatory autoimmune diseases28,29. We tested the proinflammatory potential of proteins with high allergenic risk by in silico mapping some inflammatory and IL-4, IFN-γ and IL-6 inducing peptides, as well as those with IgG-, IgE- and IgA-specific B cell epitopes. The storage and defence proteins seem to stand out in terms of bioactivity tested. The storage protein prunin 1 Pru du 6.0101 showed the highest scores for the proinflammatory markers tested. Although it does not appear to have IgE-specific B cell epitopes, it does have short IgE epitopes, identified using AlgPred 2.0 software. This protein and the 11S globulin seed storage protein Ana o 2.0101 and 11S globulin seed storage protein Jug r 4-like showed the highest ability to induce IL-6, a proinflammatory cytokine that stimulates acute phase responses, haematopoiesis, and specific immune responses. In terms of IgE-specific B cell epitopes, 36% of the proteins with a high risk of allergenicity showed their presence, but the most remarkable results we observed for the hypothetical protein BC332_07738, serpin-ZX-like, 11S globulin seed storage protein Jug r 4-like, 11S globulin seed storage protein Ana o 2.0101 and rubber elongation factor protein. They have not yet been reported as potential paprika allergens, although they are likely to influence allergic reactions in sensitised individuals, which should be investigated.
The affinity of dietary peptides for MHC II is crucial in the development of allergy. The most common HLA (human MHC) alleles corresponding to MHC class II are HLA-DRB1 (12), HLA-DQ (5), and HLA-PD (7). HLA molecules act as receptors that bind lysosomal processed antigens and present them to T lymphocytes. This initiates an immune response; the production of cytokines, antigen-specific antibodies by B lymphocytes, and the formation of cytotoxic lymphocytes. Depending on the cytokines secreted, CD4+ T (T helper) cells polarise into Th1, Th2, Th17 or iTregs populations30. The paprika proteins studied carry peptides capable of inducing antibodies and proinflammatory cytokines, such as IL-4 or IL-6. IL-4 plays a key role in antibody isotype switching, stimulating IgE production, haematopoiesis and inflammation, and the development of appropriate effector T cell responses31. Its secretion is a characteristic Th2 cell response, resulting from the maturation of Th0 lymphocytes in the presence of IL-4 produced by previously activated Th2 cells, mast cells, basophils and NKT cells (Natural Killer T cells)31. Proteochemometric analysis using the EpiTOP3 tool revealed a high capacity of the tested proteins to bind to HLA alleles. We found the presence of numerous peptides that can be bound by the HLA-DRB1,—DQ and -DP alleles found on antigen-presenting cells. Twenty three of the 45 peptides examined appeared to be IL-4 inducers, including 11 with SVM-motif-based scores above 1. Six of them (derived from: actin-7-like, basic beta-1,3-glucanase, osmotin-like protein (Cap a 1 allergen), pathogenesis-related protein 10, prunin 1 Pru du 6.0101 and stress related protein) showed strong proinflammatory features, indicating a high probability of allergic reaction to their parent proteins, especially since, except for two derived from prunin 1 Pru du 6.0101 and osmotin-like protein, the peptides did not induce IL-10. IL-10 plays a crucial role in the development of tolerance by suppressing inflammation, altering the profile of activated effector cells, increasing the expression of tight junction proteins in the mucosa, and increasing the number of goblet cells32. Stress related protein significantly induced IFN-γ. The generation of IFN-γ by MHC class II activated CD4+ Th cells is important in the context of the accompanying proinflammatory response, but also plays an important role, depending on the allergen dose, in immune suppression and the induction of tolerance to allergenic proteins31,33. Only a reduction of the secretion of proinflammatory cytokines, e.g. IL-4 and IFN-γ, while increasing the levels of regulatory cytokines, such as IL-10, in the context of peptide potential discrimination, offers hope for a more targeted immunotherapy34,35. Of the above-mentioned highly immunoreactive proteins capable of inducing IgE, only basic beta-1,3-glucanase and pathogenesis-related protein 10 have been reported as potential paprika allergens (Cap ch 17kD and Cap a 4).
Conclusions
The study showed that Capsicum spices possess many highly immunoreactive allergenic proteins/peptides, the presence of which can stimulate potent inflammatory mechanisms. Basic beta-1,3-glucanase (Cap a Glucanase), osmotin-like protein (Cap a 1.0101), pathogenesis related proteins 10 (Cap a 4, Cap ch 17kD) and putative pathogenesis related proteins (Cap ch 17kD) have already been reported as allergens or putative paprika allergens. However, other proteins may also be highly allergenic , such as 11S globulin seed storage protein Ana o 2.0101, 11S globulin seed storage protein Jug r 4-like, actin-7-like, hypothetical protein BC322_07738, lichenase, prunin 1 Pru du 6.0101, serpin-ZX-like, stress related protein or vicilin Jug r 2.0101 showing strong proinflammatory features. In addition, cross-reactivity of paprika proteins with latex (possible paprika contaminant), tomato, tobacco, grapes, mustard, kiwi, sesame, avocado, wheat, maize, banana, chestnut, hazel, molds, meadow plants, and even cattle, crab and fishes is possible and should be taken into account in allergy diagnosis, especially in the cases of idiopathic and non-IgE-mediated anaphylaxis, without exceeding norms of specific IgE antibodies.
Materials and methods
Protein extracts
Commercially available peppers spices (mild, spicy and chili) were used in the study. Proteins were extracted in (a) 10 mmol/L PBS (pH 7.0) containing 2% (w/v) polyvinylpolypyrrolidone (PVPP), 2 mmol/L ethylenediaminetetraacetic acid (EDTA), 10 mmol/L sodium diethyldithiocarbamate (DIECA) and 3 mmol/L sodium azide36, and in (b) 20 mmol/L Tris/HCL buffer (pH 7.4) containing 150 mM NaCl, 0.05% Tween 20, 1% sodium dodecyl sulfate (SDS) and 7% 2-mercaptoethanol (ME)37 by overnight shaking at 4 °C. After centrifugation at 12000×g for 60 min at 4 °C, the supernatants were collected and further centrifuged in Amicon Ultra centrifugal 3K devices (Merck Millipore Ltd., Cork, IRL) at 5000×g for 20 min. The concentrated extracts were collected, and aliquots were stored at − 20 °C for analysis. Protein content was determined by the Bradford method.
Serum
Human sera were selected from the bank of sera collected at the IAR&FR PAS in Olsztyn between 2010 and 201438. All procedures were approved by the Bioethics Committee of the Faculty of Medical Sciences of the University of Warmia and Mazury in Olsztyn (decision No. 2/2010 and 2/2016) and were performed in accordance with the standards of the Helsinki Declaration. Written informed consent was obtained from all subjects. The sera tested (3) were from patients (aged 32–57 years, female) with severe allergic reactions, presumably to paprika, including one episode of anaphylaxis (see Supplementary Table 3S). The EUROLINE Atopy Screen Panel normally used to diagnose the sera (EUROIMMUN AG, Lübeck, Germany) did not include paprika, so the sera were analysed using the Allercoat™ 6-ELISA and the Allergy Profile Pollen-Food Cross Reactions test (EUROIMMUN AG, Lübeck, Germany). Paprika-specific serum IgE levels were < 0.35 kU/L. Potato- and tomato-specific IgE antibodies were also not elevated. Sera were pooled due to similar clinical findings and the intended immunoblotting analysis.
SDS-PAGE analysis
Extracted proteins (20 μg) were separated in the 12.5% polyacrylamide gel in the presence of the Tris–glycine buffer (192 mmol/L glycine, 25 mmol/L Tris and 0.1% SDS, pH 8.3; according to Laemmli39, using 4μL of Odyssey® Protein Molecular Weight Marker (10–250 kDa) (Li-COR Biotechnology, Lincoln, NE, USA). Electrophoresis was performed in a Mini PROTEAN 3 Cell apparatus (Bio-Rad Laboratories, Hercules, CA, USA) at 140V for 75 min. Gels were stained with a 0.1% solution of Coomassie Brilliant Blue R-250. Bands were detected on the ChemiDoc Imaging System (Bio-Rad Laboratories) and analysed using Image Lab software (Bio-Rad Laboratories) including densitometric analysis.
Immunoblotting for IgE binding assay
Proteins were transferred onto nitrocellulose membranes (Sigma-Aldrich, St. Louis, MO, USA) by wet electrotransfer in a buffer of Tris–glycine (pH 8.3) with methanol (192 mmol/L glycine, 25 mmol/L Tris and 20% (v/v) methanol) according to Towbin et al.40 at 25 mA for 20 h. Membranes were washed in PBS (pH 7.4) for 5 min at room temperature (RT), then blocked in the Odyssey® blocking buffer (Li-COR Biotechnology), pH 7.2–7.6, for 2 h at RT according to Markiewicz et al.41 and incubated overnight at 4 °C in a solution of human sera diluted twice in blocking buffer containing 0.1% Tween 20. The membranes were then rinsed four times in the PBS-T buffer (PBS, pH 7.4, containing 20% Tween 20). Detection of human IgE reactive proteins was performed by incubating the membranes for 90 min at RT in a solution containing mouse monoclonal anti-human IgE antibodies (Sigma-Aldrich) labelled with IRDye® 800CW (Li-COR Biotechnology). Anti-human IgE secondary antibodies were diluted 1:500 with Odyssey® blocking buffer (pH 7.2–7.6) containing 0.1% Tween 20 and 0.01% SDS. Signal detection was performed using the ChemiDoc Imaging System (Bio-Rad Laboratories) and analysed using Image Lab software (Bio-Rad Laboratories).
Identification of proteins by LC–MS/MS analysis
Bands identified as IgE reactive were excised from the gel, destained in 50 mM NH4HCO3 solution in 50% ACN, reduced with 10 mM DTT in 100 mM NH4HCO3 and alkylated with 50 mM iodoacetamide solution in 100 mM NH4HCO3. Proteins were then identified by mass spectrometry (MS) after in-gel digestion with 10 ng/mL trypsin (Promega, Madison, WI, USA) overnight at 37 °C. Trifluoroacetic acid was added to a final concentration of 0.1% to stop digestion. MS analysis was performed by LC–MS/MS technique in the Laboratory of Mass Spectrometry (IBB PAS, Warsaw) using a nanoACQUITY UPLC system (Waters Corporation, Milford, MA, USA) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The sample was applied to the nanoACQUITY UPLC trapping column (Waters Corporation, Milford, MA, USA) using water containing 0.1% formic acid as the mobile phase. The peptide mixture was then transferred to the nanoACQUITY UPLC BEH C18 column (Waters Corporation, 75 µm inner diameter, 250 mm long) and a CAN gradient (5–35% over 180 min) was applied in the presence of 0.1% formic acid at a flow rate of 250 nL/min. Eluted peptides were electrosprayed directly into the mass spectrometer operating in positive ion mode at a voltage of 2 kV. Spectra were recorded in full MS mode in profile mode at 60,000 resolution with a scan range of 400–2000 m/z. Each sample was washed three times prior to measurement to avoid cross-contamination and the final MS wash was checked for cleanliness. Raw data were searched using MASCOT (Matrix Science Ltd., London, UK) against the SwissProt database—taxa Green Plants (Viridiplantae), but also against all entries. Search parameters were: enzyme, trypsin; peptide mass tolerance, 20 ppm; fragment ion tolerance, 0.1 Da; fixed modifications, carbamidomethyl (C); variable modifications, oxidation (M). For each identified protein, the significance threshold of p < 0.05, the ions score or expected cut-off-43 and the highest emPAI value were considered significant. Finally, the identification results were checked using the Basic Local Alignment Search Tool (BLAST) against Capsicum taxid (https://www.ncbi.nlm.nih.gov). Proteins indicated by BLAST with the peptides on which the identification by MASCOT was based were found to be derived from Capsicum.
In silico analysis of proteins and peptides
Protein allergenicity and proinflammatory activity were investigated using online tools. The in silico protein sequence analysis used was partially described by Ogrodowczyk et al.42.
Recognition of protein allergenicity
Sequences of proteins identified by MS analysis and BLAST were retrieved from NCBI and used for in silico allergenicity analyses. Proteins with sensitising potential were selected based on sequences deposited in the Allergome (https://www.allergome.org) and AllergenOnline v. 21 (FARRP; http://www.allergenonline.org) databases. Questionable results were further checked using the Allermatch database (http://allermatch.org). The allergenic potential of the protein was estimated using the full-length alignment and, in the absence of positive results, using 8- or 6-amino acid exact match methods. Prediction of IgE epitopes was performed using the AlgPred 2.0 server (https://webs.iiitd.edu.in/raghava/algpred2/)43.
Screening for proinflammatory activity of proteins with high risk of allergenicity
The ProInflam web server (http://metagenomics.iiserb.ac.in/proinflam) was used to predict antigenic regions that induce a proinflammatory response, the IL4pred tool (https://webs.iiitd.edu.in/raghava/il4pred/) to map IL-4 inducing peptides, while IFNepitope (https://webs.iiitd.edu.in/raghava/ifnepitope/) and IL-6Pred (https://webs.iiitd.edu.in/raghava/il6pred/) were used to map INF-γ and IL-6 inducing peptides, respectively44,45,46. The IgPred web server (https://webs.iiitd.edu.in/raghava/igpred/) was used to predict protein IgG, IgE and IgA specific B cell epitopes47.
Prediction of peptide-MHC II binding
The high-risk allergenic proteins and their peptides with proinflammatory antigenic regions estimated by ProInflam (the one with the highest SVM score and those containing IgE epitopes mapped by AlgPred 2) and peptide sequences with predicted IgE-specific B-cell epitopes predicted by IgPred (those with the highest score) were screened for binding to human major histocompatibility complex class II (MHC II). Protein sequences and peptides were uploaded to the EpiTOP3 server (http://www.ddg-pharmfac.net/EpiTOP3/), which is designated to predict binding to human leukocyte antigen (HLA) alleles corresponding to MHC class II using proteochemometric models. An IC50 threshold of 6.3 for peptide/HLA complexes was used for analysis48,49.
Prediction of peptide/MHC II complexes inducing IL-4, IL-10 and IFN-γ
Peptide sequences analysed as above were further screened for their ability to induce IL-4, IL-10 and IFN-γ. The IL4pred (http://crdd.osdd.net/raghava/il4pred/), IL-10pred (http://crdd.osdd.net/raghava/IL-10pred/) and IFNepitope (https://webs.iiitd.edu.in/raghava/ifnepitope/) tools were used for analysis with default settings31,44,50.
Statistical analysis
Statistical parameters used in analyses requiring specialised software linked to an instrument/tool are described in the analytical method/online tool used and were briefly summarized by Ogrodowczyk et al.42. Densitometric data were expressed as mean ± SD from three independent assays. Student’s t test was used to compare isolation methods, while one-way ANOVA followed by post hoc Duncan or Kruskal–Wallis tests were used to compare protein isolates in the tested spices. Calculations were performed using Statistica v. 13 (Statsoft, Kraków, Poland). Differences were considered significant at p < 0.05.
All procedures and methods were performed in accordance with relevant guidelines and regulations.
Data availability
The mass spectrometry proteomics data were deposited at the ProteomeXchange Consortium via the PRIDE partner repository51 under the accession numbers PXD039651 and https://doi.org/10.6019/PXD039651.
References
Mendes, N. S. & Gonçalves, E. C. B. A. The role of bioactive components found in peppers. Trends Food Sci. Technol. 99, 229–243 (2020).
Anibarro, B., Seoane, F. J. & Mugica, M. V. Involvement of hidden allergens in food allergic reactions. J. Investig. Allergol. Clin. Immunol. 17, 168–172 (2007).
Chudzik-Kozłowska, J., Wasilewska, E. & Złotkowska, D. Evaluation of immunoreactivity of pea (Pisum sativum) albumins in BALB/c and C57BL/6 mice. J. Agric. Food Chem. 68, 3891–3902 (2020).
Xiong, Y. L. & Guo, A. Animal and plant protein oxidation: Chemical and functional property significance. Foods 10, 40 (2021).
Schülke, S. & Albrecht, M. Mouse models for food allergies: Where do we stand?. Cells 8, 546 (2019).
Nguyen, S. M. T. et al. Mechanisms governing anaphylaxis: Inflammatory cells, mediators, endothelial gap junctions and beyond. Int. J. Mol. Sci. 22, 7785 (2021).
Cianferoni, A. Non-IgE-mediated anaphylaxis. J. Allergy Clin. Immunol. 147, 1123–1131 (2021).
Simons, F. E. R. et al. Risk assessment in anaphylaxis: Current and future approaches. J. Allergy Clin. Immunol. 120(Suppl), S2-24 (2007).
Reber, L. L., Hernandez, J. D. & Galli, S. J. The pathophysiology of anaphylaxis. J. Allergy Clin. Immunol. 140, 335–348 (2017).
Gulen, T. & Akin, C. Idiopathic anaphylaxis: a perplexing diagnostic challenge for allergists. Cur. Allergy Asthma Rep. 21, 11 (2021).
Arora, R., Kumar, A., Kumar Singh, I. & Singh, A. Pathogenesis related proteins: A defensin for plants but an allergen for humans. Int. J. Biol. Macromol. 157, 659–672 (2020).
Yamada, Y. et al. High-yield production of the major birch pollen allergen Bet v 1 with allergen immunogenicity in Nicotiana benthamiana. Front. Plant Sci. 11, 1–10 (2020).
Walker, M. J. et al. Almond or mahaleb? Orthogonal allergen analysis during a live incident investigation by ELISA, molecular biology, and protein mass spectrometry. J. AOAC Int. 101, 162–169 (2018).
Alessandri, C. et al. Molecular approach to a patient’s tailored diagnosis of the oral allergy syndrome. Clin. Transl. Allergy 10, 1–18 (2020).
Trentanni Hansen, G. J. et al. NIR-based sudan I to IV and para-red food adulterants screening. Food Addit. Contam. A https://doi.org/10.1080/19440049.2019.1619940 (2019).
Takei, M. et al. Capsicum allergy: Involvement of Cap a 7, a new clinically relevant gibberellin-regulated protein cross-reactive with Cry j 7, the gibberellin-regulated protein from Japanese cedar pollen. Allergy Asthma Immunol. Res. 14, 328–338 (2022).
Leitner, A. et al. Allergens in pepper and paprika. Immunologic investigation of the celery-birch-mugwort-spice syndrome. Allergy 53, 36–41 (1998).
Jensen-Jarolim, E. et al. Bell peppers (Capsicum annuum) express allergens (profilin, pathogenesis-related protein P23 and Bet v 1) depending on the horticultural strain. Int. Arch. Allergy Immunol. 116, 103–109 (1998).
DDP-24 Whole Dried Chilli Peppers (UNICE Standards). https://unece.org/fileadmin/DAM/trade/agr/standard/dry/dry_e/DDP24_WholeDriedChilliPepper_2013_e.pdf (2013).
Watson, I., Kamblea, P., Shanksa, C., Khana, Z. & El Darrac, N. Decontamination of chilli flakes in a fluidized bed using combined technologies: Infrared, UV and ozone. Innov. Food Sci. Emerg. Technol. 59, 102248 (2020).
Aalberse, R. C. Structural biology of allergens. J. Allergy Clin. Immunol. 106, 228–238 (2000).
Goodman, R. E. Practical and predictive bioinformatics methods for the identification of potentially cross reactive protein matches. Mol. Nutr. Food Res. 50, 655–660 (2006).
Wagner, et al. Characterization of cross-reactive bell pepper allergens involved in the latex-fruit syndrome. Clin. Exp. Allergy 34, 1739–1746 (2004).
Callero, et al. Bell pepper allergy: Different sensitization profiles. J. Investig. Allergol. Clin. Immunol. 28, 340–342 (2018).
Estrada-Rodriguez, et al. A clinical case of paprika allergy. Curr. Allergy Clin. Immunol. 33, 183–184 (2020).
Palomares, O., Villalba, M., Quiraltew, J., Polozand, F. & Rodrıguez, R. 1,3-beta-glucanases as candidates in latex–pollen–vegetable food cross-reactivity. Clin. Exp. Allergy 35, 345–351 (2005).
De Martinis, M., Sirufo, M. M., Suppa, M. & Ginaldi, L. New perspectives in food allergy. Int. J. Mol. Sci. 21, 1474–1495 (2020).
Kreiner, E. et al. Shared genetic variants suggest common pathways in allergy and autoimmune diseases. J. Allergy Clin. Immunol. 140, 771–781 (2017).
Wróblewska, B., Szyc, A. M., Markiewicz, L. H., Zakrzewska, M. & Romaszko, E. Increased prevalence of eating disorders as a biopsychosocial implication of food allergy. PLoS One 13, e0198607 (2018).
Gutcher, I. & Becher, B. APC-derived cytokines and T cell polarization in autoimmune inflammation. J. Clin. Investig. 117, 1119–1127 (2007).
Dhanda, S. K., Gupta, S., Vir, P. & Raghava, G. P. S. Prediction of IL4 inducing peptides. Clin. Dev. Immunol. 2013, 263952 (2013).
Nguyen, H. D., Aljamaei, H. M. & Stadnyk, A. W. The production and function of endogenous interleukin-10 in intestinal epithelial cells and gut homeostasis. Cell. Mol. Gastroenterol. Hepatol. 12, 1343–1352 (2021).
Wijdeven, R. H. et al. Chemical and genetic control of IFNγ-induced MHCII expression. EMBO Rep. 19, e45553 (2018).
Ng, T. H. S. et al. Regulation of adaptive immunity; the role of interleukin-10. Front. Immunol. 4, 129 (2013).
Wróblewska, B. et al. Effect of low-immunogenic yogurt drinks and probiotic bacteria on immunoreactivity of cow’s milk proteins and tolerance induction - in vitro and in vivo studies. Nutrients 12, 3390 (2020).
Jensen-Jarolim, E. et al. Characterization of allergens in Apiaceae spices: Anise, fennel, coriander and cumin. Clin. Exp. Allergy 27, 1299–1306 (1997).
Watanabe, Y. et al. Novel ELISA for the detection of raw and processed egg using extraction buffer containing a surfactant and a reducing agent. J. Immunol. Methods 300, 115–123 (2005).
Ogrodowczyk, A. M., Zakrzewska, M., Romaszko, E. & Wróblewska, B. Gestational dysfunction-driven diets and probiotic supplementation correlate with the profile of allergen-specific antibodies in the serum of allergy sufferers. Nutrients 12, 2381 (2020).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Towbin, H., Staehelin, T. & Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Biochemistry 76, 4350–4357 (1979).
Markiewicz, L. H., Szymkiewicz, A., Szyc, A. & Wróblewska, B. A simultaneous two-color detection method of human IgG- and IgE-reactive proteins from lactic acid bacteria. J. Microbiol. Methods 126, 72–75 (2016).
Ogrodowczyk, A. M., Dimitrov, I. & Wróblewska, B. Two faces of milk proteins peptides with both allergenic and multidimensional health beneficial impact - integrated in vitro/in silico approach. Foods 10, 163 (2021).
Sharma, N. et al. AlgPred 2.0: An improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief. Bioinform. 22, bbaa294 (2021).
Dhanda, S. K., Vir, P. & Raghava, G. P. S. Designing of interferon-gamma inducing MHC class-II binders. Biol. Dir. 8, 30 (2013).
Gupta, S., Madhu, M. K., Sharma, A. K. & Sharma, V. K. ProInflam: A webserver for the prediction of proinflammatory antigenicity of peptides and proteins. J. Transl. Med. 14, 178 (2016).
Dhall, A., Patiyal, S., Sharma, N., Usmani, S. S. & Raghava, G. P. S. Computer-aided prediction and design of IL-6 inducing peptides: IL-6 plays a crucial role in COVID-19. Brief. Bioinform. 22, 936–945 (2020).
Saha, S. & Raghava, G. P. S. Prediction methods for B-cell epitopes. In Methods in Molecular Biology, Immunoinformatics: Predicting Immunogenicity in Silico Vol. 409 (ed. Flower, D. R.) 387–394 (Humana Press Inc, 2007).
Dimitrov, I., Garnev, P., Flower, D. R. & Doytchinova, I. EpiTOP-a proteochemometric tool for MHC class II binding prediction. Bioinformatics 26, 2066–2068 (2010).
Dimitrov, I. & Doytchinova, I. Associations between milk and egg allergens and the HLA-DRB1/DQ polymorphism: A bioinformatics approach. Int. Arch. Allergy Immunol. 169, 33–39 (2016).
Nagpal, G. et al. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci. Rep. 7, 42851 (2017).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Acknowledgements
This project was supported by IAR&FR PAS.
Author information
Authors and Affiliations
Contributions
Conceived the idea and secured funding: B.W. Conceptualization and design of the study: E.W. and A.O. Acquisition of data: E.W., A.O. and B.W. Analysis and/or interpretation of data: E.W. and A.O. Drafting the manuscript: E.W. and B.W. Revising the manuscript critically for important intellectual content: E.W., A.O. and B.W. Approval of the version to be published: E.W., A.O. and B.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wróblewska, B., Ogrodowczyk, A. & Wasilewska, E. Immunoreactive proteins of Capsicum-based spices as a threat to human health: mass spectrometry analysis and in silico mapping. Sci Rep 13, 17723 (2023). https://doi.org/10.1038/s41598-023-44775-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-44775-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.