Abstract
Pearl millet (Pennisetum glaucum [L.] R. Br.) is a nutrient-dense, relatively drought-tolerant cereal crop cultivated in dry regions worldwide. The crop is under-researched, and its grain yield is low (< 0.8 tons ha−1) and stagnant in the major production regions, including Burkina Faso. The low productivity of pearl millet is mainly attributable to a lack of improved varieties, Striga hermonthica [Sh] infestation, downy mildew infection, and recurrent heat and drought stress. Developing high-yielding and Striga-resistant pearl millet varieties that satisfy the farmers’ and market needs requires the identification of yield-promoting genes linked to economic traits to facilitate marker-assisted selection and gene pyramiding. The objective of this study was to undertake genome-wide association analyses of agronomic traits and Sh resistance among 150 pearl millet genotypes to identify genetic markers for marker-assisted breeding and trait introgression. The pearl millet genotypes were phenotyped in Sh hotspot fields and screen house conditions. Twenty-nine million single nucleotide polymorphisms (SNPs) initially generated from 345 pearl millet genotypes were filtered, and 256 K SNPs were selected and used in the present study. Phenotypic data were collected on days to flowering, plant height, number of tillers, panicle length, panicle weight, thousand-grain weight, grain weight, number of emerged Striga and area under the Striga number progress curve (ASNPC). Agronomic and Sh parameters were subjected to combined analysis of variance, while genome-wide association analysis was performed on phenotypic and SNPs data. Significant differences (P < 0.001) were detected among the assessed pearl millet genotypes for Sh parameters and agronomic traits. Further, there were significant genotype by Sh interaction for the number of Sh and ASNPC. Twenty-eight SNPs were significantly associated with a low number of emerged Sh located on chromosomes 1, 2, 3, 4, 6, and 7. Four SNPs were associated with days-to-50%-flowering on chromosomes 3, 5, 6, and 7, while five were associated with panicle length on chromosomes 2, 3, and 4. Seven SNPs were linked to thousand-grain weight on chromosomes 2, 3, and 6. The putative SNP markers associated with a low number of emerged Sh and agronomic traits in the assessed genotypes are valuable genomic resources for accelerated breeding and variety deployment of pearl millet with Sh resistance and farmer- and market-preferred agronomic traits.
Similar content being viewed by others
Introduction
Pearl millet (Pennisetum glaucum [L.] R. Br., 2n = 2x = 14) is a highly nutritious and a key staple food crop in dry regions worldwide. It is the major crop of the Sahel region, including Burkina Faso1. In Africa, pearl millet is cultivated on an estimated area of 13.8 million hectares (ha), with an average yield of 0.7 tons ha−12. In Burkina Faso, an estimated area of 1.2 million ha is devoted to pearl millet production. However, the mean yield of the crop in the country is low (< 0.81 tons ha−1), lesser than the global average of 0.9 tons ha-12. The low grain yield in the farmers’ fields is attributable to various biotic and abiotic constraints, including the use of low-yielding landraces, Striga hermonthica (Sh) infestation, bird damage, insect pests, diseases, heat and drought stresses3,4.
Striga hermonthica (Del.) Benth. is the most significant biotic constraint to pearl millet production and productivity in Burkina Faso and yield losses vary between 40 and 55%5,6. The parasite infests several other major cereal crops, including rice (Oryza glaberrima Steudel and O. Sativa L.), maize (Zea mays L.), sorghum (Sorghum bicolour [L.] Moench), and fonio (Digitaria exilis [Kippist] Stapf)7,8. Farmers often abandon Sh-infested fields and switch from pearl millet to other non-host crops, reducing the crop’s overall production and economic value9. Striga is a highly prolific parasite in Burkina Faso attributed to the host crop being mostly grown in semi-arid parts of the Sahelian and Sudano-Sahelian zones, which are dominated by poor soil fertility, low and erratic rainfall, and high temperatures that favour germination, growth and spread of the weed3.
Striga control is difficult because each parasitic plant can quickly disperse and deposit thousands of seeds into the soil seedbank. Furthermore, Striga seeds can remain viable in the soils for more than 14 years10. Striga hermonthica is a major threat to food security, exacerbating hunger and poverty in many African countries11,12. Monetary losses ranging from 117 to 200 billion US$ is incurred annually due to crop damage, and increases by US $30 million annually13.
Several Striga control strategies are recommended, including hand weeding, mulching crop fields with biomass of the shea tree (Vitellaria paradoxa C.F. Gaertn.) as a bio-control agent, optimal fertilizer application, and soil moisture management7,14. These strategies improve the soil properties, promote crop growth and development, and retard germination and growth of Striga15. Herbicides are less effective in controlling the effect of the parasite after emergence, and they are unaffordable for smallholder farmers.
The use of Striga-resistant pearl millet varieties is the most sustainable and environment-friendly management option for smallholder farmers in semi-arid regions. Resistant cultivars support fewer Striga plants and yield higher16,17. However, with the paucity of locally adapted and Sh-resistant donor sources, breeding for Striga resistance in pearl millet is still challenging compared to other cereals18,19,20,21. In the past decade, intensive research on the interaction of Striga with the host at the molecular level has opened opportunities to develop new management strategies22. For instance, 154 candidate genes associated with Sh resistance traits were identified in maize23. Adewale et al.24 reported 13 associated markers with the Sh resistance trait in early maturing tropical white maize inbred lines.
Genome-wide association studies (GWAS) has been used in pearl millet for the identification of putative genes related to flowering time25, iron, zinc and protein content26, downy mildew resistance27 and Sh resistance28. Also, GWAS has been used in finger millet for the identification of genes associated with Striga resistance29 and grain nutritional contents30 and for genetic diversity analysis31. GWAS is a valuable genomic tool to identify quantitative trait loci (QTLs) linked to Striga resistance for marker-assisted selection. GWAS results depend on the genetic marker and its density, genetic composition and diversity of the test populations.
Genetic markers are landmarks on chromosomes that help pinpoint the location of genes of interest. They can be detected through morphological and molecular markers9. Genetic markers such as GRMZM2G077208, GRMZM2G164502, GRMZM2G018508, and GRMZM2G171986, located on chromosomes 3, 5, 7, and 9 were reportedly significantly associated with Sh count in tropical maize germplasm32. SNP markers are instrumental in the dissection of complex traits such as Striga resistance, and their association with the trait can be revealed through GWAS. Identification of genomic regions linked to Striga resistance in pearl millet breeding would speed up the development of Striga-resistant varieties. Genetic markers improve the efficiency of novel Striga-resistant genes introgression and pyramiding into high-yielding elite varieties through backcross method. Therefore, the objective of this study was to undertake genome-wide association analyses of agronomic traits and Sh resistance among 150 pearl millet genotypes to identify genetic markers for marker-assisted breeding and trait introgression.
Results
Phenotypic variations
Pearl millet genotypes differed significantly (P < 0.001) for days to 50% flowering (DTF), plant height (PH), number of tillers per plant (NT), panicle length (PCL), panicle weight (PWT), thousand-grain weight (TGW), and grain weight (GW) under Sh infestation. Genotypes differed significantly (P < 0.001) for the area under the Striga number progress curve (ASNPC). The genotype by Striga interaction was non-significant for the NT and ASNPC. The genotype by environment interaction differed significantly (P < 0.001) for days to 50% flowering (DTF), plant height (PH), number of tillers per plant (NT), panicle length (PCL), panicle weight (PWT), thousand-grain weight (TGW), and grain weight (GW) under Sh infestation (Table 1). Genotypes with missing data were excluded from the analysis.
Table 2 presented the best linear unbiased prediction means for the response of pearl millet genotypes evaluated under Sh infestation. The DTF and PH ranged from 60.13 to 64.95 days and 132.57 to 159.74 cm in the naturally-infested field and the screen house conditions, respectively. The TGW ranged from 7.60 to 9.03 g under Striga infestation in field and in screen house. Several emerged Striga were recorded during the second counting, particularly in the plastic pots (Fig. 1A) and the hotspot field (Fig. 1B) conditions compared to Striga-free field condition (Fig. 1C). Thousand-grain weight markedly reduced due to high Striga infestations compared to Striga-free field (Table 2).
A low broad-sense heritability value was computed for the number of emerged Sh SN1 (18.17%) and SN2 (28.61%), while high heritability values ranging from 75.07 to 92.42% were recorded for the DTF, PCL, TGW, and PH (Table 2).
Genome-wide association mapping
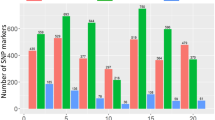
The number of emerged Sh count on the Manhattan plot is presented in Fig. 2. BLINK model analysis for Sh traits led to identifying candidate genetic regions associated with Striga resistance.
Manhattan plots showing associations between single nucleotide polymorphisms and SN1 and SN2 in naturally Striga-infested fields and greenhouse conditions. Single nucleotide polymorphisms were plotted on the x-axis according to their positions on each chromosome against association with Sh-related traits on the y-axis (−log 10 p-value). The top line indicates genome-wide significant threshold. Note: Blink.Field_SN2 = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of Striga number counted 96 days after sowing in the Striga-infested field; Blink.Pooled_SN1 = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of Striga number counted 70 days after sowing in the Striga-infested field and 116 days after planting in the greenhouse; Blink.Pooled_SN2 = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of Striga number counted 96 days after sowing in the Striga-infested field and 144 days after sowing in the greenhouse; Blink.Greenhouse_SN1 = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of Striga number counted 116 days after planting in the greenhouse, Blink.VarStr_SN1 = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of Striga number counted 116 days after planting in the greenhouse for genotypes with Striga infestation; Blink.VarStr_SN2 = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of Striga number counted 144 days after sowing in the greenhouse for genotypes with Striga infestation.
Twenty-eight SNP markers were significantly (P < 0,001) associated with Sh resistance located on chromosomes 1, 2, 3, 4, 6, and 7. Two SNPs, S3_113184999 and S3_113184999, were located on the same position and were associated with the number of Sh counted (Table 3). Three significant (P < 0,001) SNP markers, S1_75620319, S3_1159738, and S6_231436300, were associated with Striga resistance in the naturally Striga-infested field on chromosomes 1, 3, and 6. In the greenhouse, 10 significant SNPs were associated with Sh-resistance, while 15 SNPs were associated with Sh-resistance on chromosomes 1, 2, 3, 4, 6, and 7 in a pooled analysis.
Agronomic traits
BLINK model analysis of pearl millet agronomic traits under Sh infestation identified candidate genetic regions associated with DTF, PCL, and TGW (Fig. 3). Eleven SNP markers were associated with the assessed pearl millet phenotypic traits. Four SNPs were associated with DTF on chromosomes 3, 5, 6, and 7 in the naturally Striga-infested field; five with PCL on chromosomes 2, 3, and 4 with which two in the naturally Striga-infested field and three in the screen house; and two with TGW on chromosome 6 in the screen house (Table 4).
Manhattan plots showing associations between single nucleotide polymorphisms and panicle length, flowering time and thousand-grain weight under Sh conditions in naturally Striga infested field and greenhouse. Single nucleotide polymorphisms were plotted on the x-axis according to their positions on each chromosome against association with pearl millet-related traits on the y-axis (−log 10 p-value). The top line indicates genome-wide significant threshold. Note: Blink.Greenhouse_PclL = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of panicle length in screen house; Blink.Field_PclL = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of panicle length in the field; Blink.VarStr_PclL = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of panicle length for genotypes with Striga infestation; Blink.Greenhouse_TGW = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of thousand-grain weight in the screen house, Blink.Var_TGW = the Bayesian-information and linkage-disequilibrium iteratively nested keyway of thousand-grain weight for genotypes in the screen house.
Discussion
The assessed genotypes exhibited significant differences in agronomic traits and Sh parameters (Table 1), suggesting substantial genetic variation for selection. The results allowed marker-trait association analysis to be valuable for current and future selection and new variety design and commercialisation. A significant genotype by the Sh interaction effect existed (Table 1), revealing the potential existence of genes controlling Sh resistance among the populations. This concurs with the deductions made by Mrema et al.8 and Shayanowako et al.33, who reported significant variation and differential genotypic responses to Sh infestation among sorghum and maize genotypes, in that order. Thus, the population set evaluated in the current study was suitable for marker-trait association analyses for Sh resistance and economic traits.
Reduced DTF and PH were recorded on pearl millet genotypes under Striga infestation (Table 2), indicating the negative impact of the parasite on the measured traits. These findings concur with reports by Ransom and Odhiambo34. Wilson et al.18 reported a negative correlation between the number of emerged Sh and DTF in pearl millet. Similarly, a reduction of PH by 28% under Sh infestation was reported in pearl millet by Graves et al.35. Badu-Apraku36 reported a negative correlation between Striga damage rating and DTF in maize in Sub-Saharan Africa. Poor crop growth and subsequent low productivity result from the Striga plant attachment and siphoning of nutrients from the host plant's roots.
The recorded TGW of 7.60 to 9.03 g in the present study (Table 2) aligns with the 6.9 to 12.9 g reported by Kanatti et al.37. The relatively high ranges of values in the current results are probably attributed to the large grain size, which increased the yield of pearl millet37,38. The broad-sense heritability (H2) for agronomic traits ranged from 75.07 to 92.42% and 18.17 to 28.61% for emerged Sh count (Table 2). The high broad-sense heritability estimates of 75.07 to 92.42% computed for DTF, PH, PCL, and TGW indicated that the traits are mainly governed by genes with limited influence by the test environment39. Traits with high heritability are easy to select and improve using marker-assisted selections and pyramiding in a desirable genetic background. The lower broad-sense heritability estimates of 18.17 and 28.61% for the number of emerged Striga (SN1 and SN2) (Table 2) suggests that the genetic variation was small and genetic gain for those traits will be slow because both genetic and phenotypic constituents of the genotypes are affected by Striga infestation stress39. Robert40 and Kaewchumnong and Price41 reported a low heritability estimate for Striga resistance-related traits in sorghum and rice, respectively.
The seven pearl millet chromosomes harbour several genes (Table 4) conditioning Striga resistance and agronomic traits. Each chromosome had at least two significant marker-trait associations in the present study. After successful validation, the 28 significant SNP markers associated with Sh emergence on chromosomes 1, 2, 3, 4, 6, and 7 (Table 3) can be used for marker-assisted selection and trait introgression to improve Sh resistance in pearl millet. Dawud28 identified 16 SNP markers associated with the area under the Striga number progress curve on chromosomes 2, 3, 4, 5, and 7 in pearl millet. The findings confirm that the chromosomes harbour some beneficial alleles influencing Sh resistance. Dawud28 reported significant gene markers related to Striga resistance on chromosomes 1, 2, 3, and 5 in pearl millet. Markers associated with Striga resistance traits have also been reported in sorghum and maize44,45. The two SNP makers located adjacent to each other and associated with the low number of Sh count could be tightly linked and co-segregating. Hence, the respective candidate genes can be selected and introgressed simultaneously24. Identifying genetic markers associated with agronomic traits will facilitate marker-assisted breeding in pearl millet (Fig. 3). Using SSR markers, Kannan et al.46 detected significant markers associated with pearl millet panicle length and thousand-grain weight in Striga-free conditions. In maize, some quantitative trait loci associated with grain yield and ear aspect have been reported by Stanley et al.47 and Badu-Apraku et al.23. Dawud28 also reported Significant SNP markers related to the number of tillers in pearl millet. This study identified 28 SNP markers associated with low Sh emergence on chromosomes 1, 2, 3, 4, 6, and 7, involving genetic analysis of 150 genetically diverse pearl millet genotypes in Burkina Faso (Fig. 3). The candidate markers and genotypes are novel genomic and genetic resources for Striga resistance breeding programs in the country and elsewhere.
Materials and methods
Study sites
A field experiment was conducted in the 2019/20 main growing season in a naturally infested Striga hotspot field at the Didri site in Burkina Faso, and a screen house evaluation was conducted at the main station of the Institute of Environment and Agricultural Research (INERA) in the offseason of 2020/21. The Didri site is located at 12° 12′ 15" N and 1° 14′ 13" W and is a hotspot site for Sh affecting pearl millet, maize and sorghum crops. The site received an annual rainfall of 748.5 mm for 46 days during the 2019/20 rainy season and has sandy soils. The INERA site is located at 12°28/27 N and 1°33/31W.
Plant materials
The study used 148 pearl millet genotypes collected from the International Crop Research Institute for the Semi-arid Tropics (ICRISAT) in Niger and two elite breeding lines from INERA/Burkina Faso. The descriptions of the test genotypes are summarised in Table 5. The pearl millet genotypes acquired from ICRISAT are part of the pearl millet germplasm association panel (PMiGAP) comprising 250 inbred lines representing cultivated germplasm from Africa and Asia. They are included in the present study to identify unique genetic resources with unique agronomic and farmers’ preferred traits, and because of their wide genetic diversity.
Experimental design and trial management
The field and greenhouse experiments were laid out using a 10 × 15 alpha lattice design with two replicates. In the greenhouse, 5L plastic pots were used and filled with a soil medium composed of clay, sand, and organic manure at a ratio of 2:1:1 respectively. Two weeks before planting, each pot was infested with a scoop of sand mixed with 0.05 g of 1-year-old Sh seed collected from farmers' fields in Burkina Faso. Pearl millet seeds in the naturally Striga-infested field (hereafter designated as GenInf), were sown during the main crop growing season from June to October 2019. Genotypes were established in 4.2 m long rows spaced at an inter-row spacing of 160 cm and intra-row spacing of 60 cm, providing a total plot size of 6.72 m2 per genotype. Four seeds were initially sown per hill and later thinned to one plant two weeks after planting. A total of three plants was selected randomly from the middle of the experimental unit and tagged for agronomic data collection. In the greenhouse, one healthy and vigorous plant was grown per pot for the test genotypes with Sh (hereafter denoted as GenStr), and genotypes without Sh (hereafter referred to as control) treatments. Standard agronomic practices recommended for pearl millet production were followed. Experimental units were fertilized using nitrogen, phosphorus and potassium (NPK: 14:23:14) and applied as a microdose of 3 g per hill 15 days after planting. Hand weeding was routinely done after the first hoeing to remove all other weeds except Striga.
Data collection
Phenotypic data
The following agronomic parameters were collected from pearl millet: days-to-50%-flowering (DTF) were recorded as the days when 50% of the plants in each plot had intruded stigma. Plant height (PH) was measured in cm from the base to the top of the panicle of the main tiller. The number of tillers per plant (NT) was recorded by counting the number of tillers with panicles for the tagged plants. Panicle length (PCL) was measured in cm from the base to the top of the main tiller panicle. Panicle weight (PWT) was recorded in grams by weighing the harvested panicles for each entry after 14 days of sun-dry, and thousand-grain weight (TGW) was determined in grams by weighing one thousand-grain for each of the entries. Grain weight per plant (GW) was determined in grams by weighing the grain after threshing and dividing it by the number of harvested plants for each plot.
For Striga parameters, the number of emerged Sh plants per plot were recorded at 70 and 96 days after sowing in the naturally infested field for each row, excluding the borders. The number of emerged Sh plants were counted per pot 116 and 144 days after sowing in the greenhouse. The area under the Striga Number Progress Curve (ASNPC) was computed using the successive Striga counts according to Haussmann et al.48 as follows:
where n is the number of Striga assessment dates, Yi is the Striga count at the ith assessment date, Y(i+1) is the Striga count at the ith plus 1 assessment date, ti is the number of days after planting (DAP) at the ith assessment date, t(i+1) is DAP at the ith plus 1 assessment date.
Phenotypic data analysis
Both the crop and Striga data were subjected to analysis of variance using GenStat 19th Edition (http://www.genstat.co.uk). Homogeneity of variance test was done for each site using the Bartlett49 procedure before combined analyses. The treatment, genotype, and genotype × treatment interaction significance tests were computed using GenStat. The Best Linear Unbiased Prediction (BLUP) was calculated according to Haslett and Puntanen50 to predict the accuracy and to aid selection. The area under the Striga number progress curve was drawn using R. ASReml-R Version 4 was used to fit the linear mixed models using Residual Maximum Likelihood (REML) in R51.
Genotyping and GWAS analysis
To assemble the pearl millet genome, whole genome shotgun (WGS) and bacterial artificial chromosome (BAC) sequencing were used. Ten small inserts (of ~ 170, 250, 500 and 800 bp) and 13 large inserts (of ~ 2, 5, 10, 20 and 40 kb) WGS libraries were constructed using Tift 23D2B1-P1-P510 genotype. These libraries were sequenced on the Illumina HiSeq 2000, and 520 Gb of sequence data, representing 296 × genome coverage. Two BAC libraries, with an average insert size of ~ 120 kb, were constructed from Tift 23D2B1-P1-P5 using EcoRI and HindIII. Nine hundred seventy-two Gb of sequence data were generated from 100,608 BAC clones at ~ 80 × genome coverage. In brief, 1.49 Tb of sequence data, after stringent filtering and correction steps, were assembled into 1.58 Gb of contigs and 1.82 Gb of scaffolds (https://doi.org/10.1038/nbt.3943). A raw marker set consisting of 29 million SNPs generated from 345 pearl millet genotypes from which 148 genotypes used in the study was sourced from the Nature Paper Pearl Millet (https://doi.org/10.1038/nbt.3943) and filtered using Tassel v4.2 for site coverage of 90%, minor allele frequency of 0.1, taxa coverage of 30% and maximum heterozygosity of 50%. The resulting final set of variants contained 256 K SNPs was used for the current analysis. Phenotypic data collected from 150 genotypes were available for marker-trait association analysis. Principal component analysis (PCA) was calculated, and the resulting eigenvalue (7) was used for genome-wide association analysis (GWAS) following multiple methods procedures generated from GAPIT v3.052. The Bayesian-information and linkage-disequilibrium iteratively nested keyway (BLINK) software was used to determine the significant variations among pearl millet and Sh traits owing to its ability to produce fewer false positives and more true positives than the GWAS method, FarmCPU42. Liu et al.43 reported the power of BLINK to outperform FarmCPU relative to statistical capabilities vs False Discovery Rate (FDR) and statistical power vs type I error.
Conclusion
The current study detected significant genetic variability and markers for Sh resistance and agronomic traits through GWAS involving 150 pearl millet genotypes in Burkina Faso. There were significant genotypes by Sh interaction for assessed agronomic traits, the number of Sh and ASNPC. Twenty-eight SNPs were associated with Sh traits on chromosomes 1, 2, 3, 4, 6, and 7. SNPs markers associated with DTF, PCL, and TGW were located on chromosomes 2, 5, 6, and 7; chromosomes 2, 3, and 4; and chromosome 6, respectively. After successful validation, the new markers would be deployed for marker-assisted breeding emphasising the above agronomic traits and Sh resistance in pearl millet in Burkina Faso or related agro-ecologies.
Data availability
The datasets generated and/or analysed during the current study are available in the [OAR@ICRISAT] repository, [https://cegresources.icrisat.org/data_public/PM_SNPs/SNP_calls/].
Change history
25 October 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41598-023-45365-z
References
Bationo, A. & Ntare, B. Rotation and nitrogen fertilizer effects on pearl millet, cowpea and groundnut yield and soil chemical properties in a sandy soil in the semi-arid tropics, West Africa. J. Agric. Sci. 134(3), 277–284 (2000).
FAOSTAT, FAOSTAT online database 2022: Rome Italy, 2022. (2022).
Drabo, I. et al. Identifying farmers’preferences and constraints to pearl millet production in the sahel and north-sudan zones of burkina faso. Exp. Agricult. 55(5), 765–775 (2018).
Rouamba, A. et al. Constraints to pearl millet (Pennisetum glaucum) production and farmers’ approaches to Striga hermonthica management in burkina faso. Sustainability 13(15), 8460 (2021).
Zombré, P. & Nikiéma, S. Importance et effet de Striga hermonthica (Del.) Benth sur la production du sorgho en zone Nord Soudanienne du Burkina Faso: Cas de Linonghin. Revue du réseau pour l’amélioration de la productivité agricole en milieu aride 4, 103–112 (1992).
Traoré, H. & Yonli, D. Striga et autres adventices: perception paysanne et inventaire des méthodes endogènes de lutte dans l’Est du Burkina Faso. Sci. Tech., Sci. Nat. Agron. 25(1), 46–59 (2001).
Boussim, I. et al. Etat d’infestation, connaissance endogène et approche systématique des espèces du genre Striga au Burkina Faso. Int. J. Biol. Chem. Sci. 5(4), 1374–1386 (2011).
Mrema, E. et al. Screening of sorghum genotypes for resistance to Striga hermonthica and S. asiatica and compatibility with Fusarium oxysporum f. sp. strigae. Acta Agricult. Scandinavica Sect. B Soil Plant Sci. 67(5), 395–404 (2017).
Acquaah, G., Principles of Plant Genetics and Breeding. Second ed., 9600 Garsington Road, Oxford, OX4 2DQ, UK. The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK 111 River Street, Hoboken, NJ 07030–5774, USA: John Wiley & Sons, Ltd. 740 (2012).
Emechebe, A. et al. Farmers’perception of the Striga problem and its control in Northern Nigeria. Exp. Agricult. 40(2), 215–232 (2004).
Pennisi, E. Armed and dangerous: Sciences. New Series 327, 804–805 (2010).
Khan, Z. R. et al. Achieving food security for one million sub-Saharan African poor through push–pull innovation by 2020. Philos. Trans. R. Soc. B: Biol. Sci. 369(1639), 20120284 (2014).
Rodenburg, J. et al. Parasitic weed incidence and related economic losses in rice in Africa. Agricult. Ecosyst. Environ. 235, 306–317. https://doi.org/10.1016/j.agee.2016.10.020 (2016).
Tesso, T. T. & Ejeta, G. Integrating multiple control options enhances Striga management and sorghum yield on heavily infested soils. Agronomy Journal 103(5), 1464–1471 (2011).
Reda, F. & Verkleij, J. The biology and control of Striga: a review. Pest Manag. J. Ethiopia 8, 1–13 (2004).
Doggett, H., Sorghum 2nd Edition Tropical Agriculture. Series Longman Scientific & Technical, Essex, England, (1988).
Gebisa, E., Butler, L., Babiker, A. New approaches to the control of Striga: Striga research at Purdue University. Purdue University Agricultural Experiment Station Research Bulletin RB-991: 27 (1991)
Wilson, J., Hess, D. & Hanna, W. Resistance to Striga hermonthica in wild accessions of the primary gene pool of Pennisetum glaucum. Phytopathology 90(10), 1169–1172 (2000).
Wilson, J. et al. Pennisetum glaucum subsp. monodii accessions with Striga resistance in West Africa. Crop Protect. 23(9), 865–870 (2004).
Kountche, B. A. et al. Development of a pearl millet Striga-resistant genepool: Response to five cycles of recurrent selection under Striga-infested field conditions in West Africa. Field Crops Res. 154, 82–90 (2013).
Sattler, F. et al. Characterization of West and Central African accessions from a pearl millet reference collection for agro-morphological traits and Striga resistance. Plant Genetic Resour. 16(3), 260–272 (2018).
Jamil, M., Kountche, B. A. & Al-Babili, S. Current progress in Striga management. Plant Physiol. 185(4), 1339–1352 (2021).
Badu-Apraku, B. et al. Identification of QTLs for grain yield and other traits in tropical maize under Striga infestation. PloS one 15(9), e0239205 (2020).
Adewale, S. A. et al. Genome-wide association study of Striga resistance in early maturing white tropical maize inbred lines. BMC Plant Biol. 20(1), 1–16 (2020).
Diack, O. et al. GWAS unveils features between early-and late-flowering pearl millets. BMC Genom. 21(1), 1–11 (2020).
Pujar, M. et al. Genome-wide association study uncovers genomic regions associated with grain iron, zinc and protein content in pearl millet. Sci. Rep. 10(1), 1–15 (2020).
Drabo, I. Breeding Pearl Millet (Pennisetum glaucum (L) R. BR.) for Downy Mildew Resistance and Improved Yield in Burkina Faso. Ph.D. thesis, West Africa Centre for Crop Improvement, College of Basic and Applied Sciences University of Ghana (2016).
Dawud, M.A., Genetic Studies of Pearl Millet [Pennisetum Glaucum (L.) R. Br.] For Resistance to Striga Hermonthica. Ph.D. thesis, West Africa Centre for Crop Improvement, College of Basic and Applied Sciences University of Ghana (2018).
Nyongesa, S.P., Field and molecular screening for Striga resistance in selected finger millet (Eleusine coracana, l. gaertn) germplasm in western Kenya. University of Eldoret (2017).
Puranik, S. et al. Genome-wide association mapping and comparative genomics identifies genomic regions governing grain nutritional traits in finger millet (Eleusine coracana L .Gaertn). Plants, People, Planet 2(6), 649–662 (2020).
Backiyalakshmi, C. et al. Genome-wide assessment of population structure and genetic diversity of the global finger millet germplasm panel conserved at the ICRISAT Genebank. Fron. Plant Sci. 12, 692463 (2021).
Gowda, M. et al. Genetic dissection of Striga hermonthica (Del.) Benth. Resistance via genome-wide association and genomic prediction in tropical maize germplasm. Theor. Appl. Gene. 134(3), 941–958 (2021).
Shayanowako, A. I. et al. Striga resistance and compatibility of maize genotypes to a biocontrol agent, Fusarium oxysporum f. sp. Strigae. J. Crop Improve. 34(4), 437–454 (2020).
Ransom, J. K. & Odhiambo, G. D. Effect of corn (Zea mays) genotypes which vary in maturity length on Striga hermonthica parasitism. Weed Technol. 9(1), 63–67 (1995).
Graves, J. et al. Growth and carbon allocation in Pennisetum typhoides infected with the parasitic angiosperm Striga hermonthica. Plant, Cell Environ. 13(4), 367–373 (1990).
Badu-Apraku, B. Genetic variances and correlations in an early tropical white maize population after three cycles of recurrent selection for Striga resistance. Maydica 52(2), 205 (2007).
Kanatti, A. et al. Grain iron and zinc density in pearl millet: combining ability, heterosis and association with grain yield and grain size. SpringerPlus 3(1), 1–12 (2014).
Jukanti, A. et al. Crops that feed the world 11. Pearl Millet (Pennisetum glaucum L.): an important source of food security, nutrition and health in the arid and semi-arid tropics. Food Secur. 8(2), 307–329 (2016).
Afolayan, G. et al. Genetic variation for Striga hermonthica resistance and yield among sorghum accessions in Nigeria. J. Agricult. Sci. 12(7), 192–202 (2020).
Robert, O.J., Genetic analysis of Striga hermonthica resistance in Sorghum (Sorghum bicolor) genotypes in Eastern Uganda. Doctoral dissertation (2011).
Kaewchumnong, K. & Price, A. H. A study on the susceptibility of rice cultivars to Striga hermonthica and mapping of Striga tolerance quantitative trait loci in rice. New Phytol. 180(1), 206–216 (2008).
Huang, M. et al. BLINK: a package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 8(2), giy154 (2019).
Liu, X. et al. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Gene. 12(2), e1005767 (2016).
Khangura, M.E., A Genome-wide Association Study of the Quantitative Resistance to Striga hermonthica and Plant Architecture of Sorghum bicolor in Northwestern Ethiopia. Purdue University Graduate School (2019).
Pfunye, A., et al. Genome-Wide Association Studies for Striga asiatica Resistance in Tropical Maize. Int. J. Genom. (2021).
Kannan, B., et al. Association analysis of SSR markers with phenology, grain, and stover-yield related traits in pearl millet (Pennisetum glaucum (L.) R. Br.). Sci. World J. (2014).
Stanley, A. et al. Association analysis for resistance to Striga hermonthica in diverse tropical maize inbred lines. Sci. Rep. 11(1), 1–14 (2021).
Haussmann, B. I., Hess, D. E., Welz, H.-G. & Geiger, H. H. Improved methodologies for breeding Striga-resistant sorghums. Field Crops Res. 66, 195–211 (2000).
Bartlett, M. S. Properties of sufficiency and statistical tests. Proc. R. Soc. London. Ser. A Math. Phys. Sci. 160(901), 268–282 (1937).
Haslett, S.J. and S. Puntanen, Best linear unbiased prediction (BLUP). Wiley StatsRef: Statistics Reference Online, p. 1–6 (2014).
Butler, D., et al. ASReml-R reference manual version 4. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK, (2017).
Lipka, A. E. et al. GAPIT: genome association and prediction integrated tool. Bioinformatics 28(18), 2397–2399 (2012).
Acknowledgements
The research was funded by the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) through ‘Harnessing Opportunities for Productivity Enhancement (HOPE II) for Sorghum and Millets in sub-Saharan Africa and CIMMYT through the Accelerated Varietal Improvement and Seed delivery of legumes and cereals in Africa (AVISA) (grant number OPP1198373), funded by the Bill & Melinda Gates Foundation (BMGF).
Author information
Authors and Affiliations
Contributions
A.R.: Methodology, data collection, data analysis, and writing of the manuscript draft. H.S.: supervision, writing conceptualization, review, and editing. I.D.: experiment designing, writing, reviewing and editing. E.M.: reviewing and editing. C.O.O.: reviewing and editing. L.M.: reviewing and editing. A.R.: GWAS analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The Acknowledgements section in the original version of this Article was incomplete. Full information regarding the corrections made can be found in the correction notice for this Article.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rouamba, A., Shimelis, H., Drabo, I. et al. Genome-wide association analyses of agronomic traits and Striga hermonthica resistance in pearl millet. Sci Rep 13, 17152 (2023). https://doi.org/10.1038/s41598-023-44046-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-44046-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.