Abstract
Systemic lupus erythematosus (SLE) is a chronic rheumatic disorder. Endothelin-1, a vasoconstrictor, belongs to the endothelin family and is associated with vascular-related damages. To date, association between ET-1 and pathogenesis of SLE remains unclear. This case–control study was carried out by 314 SLE, 252 non-SLE diseases patients and 500 healthy controls. Serum ET-1, CCN3, IL-28B levels were detected by ELISA, and ET-1 gene polymorphisms (rs5369, rs5370, rs1476046, rs2070699, rs2071942, rs2071943, rs3087459, rs4145451, rs6458155, rs9369217) were genotyped with Kompetitive Allele-Specific PCR. SLE patients had high levels of ET-1, which were correlated with some clinical, laboratory features. Serum CCN3, IL-28B levels were higher in SLE patients, and ET-1 levels were positively correlated with the two cytokines. Rs5370, rs1476046, rs2070699, rs2071942, rs2071943, rs3087459, rs6458155 and rs2070699 were associated with SLE risk. Rs2070699 (T, TT) was related to SLE patients with alopecia. Rs5370 (T, TT, TG), rs1476046 (G,GA), rs2071942 (G,GA) and rs2071943 (G,GA) were associated with SLE patients with pericarditis, pyuria and fever manifestation, respectively. Rs3087459 (CC) and rs9369217 (TC) were related to SLE patients with positive anti-SSB antibody. Rs5369 (AA) was associated with IgG and CRP levels in SLE patients. In conclusion, elevated serum ET-1 in SLE patients may be a potential disease marker, and its gene polymorphisms were related to SLE susceptibility.
Similar content being viewed by others
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by dysregulated production of autoantibodies and inflammatory cytokines1. Clinical manifestations caused by systemic inflammation and tissue damage in SLE are complex and different, for instance, damages in skin, kidney, joint, blood vessel2. Clear pathogenesis of SLE remains obscure, but genetic susceptibility, environmental factors and sex hormones have been demonstrated to play pivotal roles in. To date, genome-wide association studies (GWASs) identified about 180 genetic loci associated with SLE susceptibility3.
Endothelin-1 (ET-1), the main component of endothelin family, consists of 21 amino acids and two intramolecular disulphide bonds. ET-1 is expressed in vascular endothelial cells, smooth muscle cells, cardiomyocytes, fibroblasts and macrophages4. Being an endogenous long-acting vasoconstrictor, ET-1 promotes growth factors production and stimulates proliferation and contraction of vascular endothelial cells, smooth muscle cells and fibroblasts5. Interestingly, SLE is associated with vascular damages resulted from endothelial dysfunction, such as vasculitis and Raynaud’s phenomenon. ET-1 derived from endothelial cells may lead to endothelial dysfunction6. In rheumatoid arthritis (RA) patients, higher serum ET-1 levels were detected when compared to controls, and were positively related to C-reactive protein (CRP) levels7. Levels of ET-1 were increased in both psoriasis vulgaris and systemic sclerosis (SSc) patients8,9. These findings indicated that ET-1 is excessively expressed in inflammatory autoimmune diseases. To date, there is limited discussion regarding ET-1 and SLE, and it is still unclear whether ET-1 gene polymorphisms are associated with increased risk of SLE.
Nephroblastoma overexpressed (NOV, CCN3) is highly expressed in some immune cells, such as regulatory T cells10. CCN3 is capable of inducing angiogenesis by binding ligands αvβ3 and α5β1, and thereby promoting adhesion and migration of endothelial cells11. Elevated levels of CCN3 were reported in RA, SSc patients12. Type I interferon is associated with development of SLE13. When expression of type I interferon and its receptors are inhibited in lupus mice, autoantibodies production and disease development are suppressed13. Type III interferons, including IL-28A, IL-28B, IL-29, are functionally similar to type I interferon. IL-28B involves in antiviral and antitumor responses, and contributes to development of immune-related disorders14. For example, levels of IL-28B were increased in SLE patients and were related to disease activity15. However, the relationship between ET-1 and CCN3, as well as the relationship between ET-1 and IL-28B, in SLE remains obscure.
Therefore, role of ET-1 in SLE is not clear, and whether ET-1 regulates CCN3, IL-28B, and then contributes to SLE pathogenesis is not discussed. In this study, we conducted a case–control study with SLE patients and healthy controls to reveal serum levels of ET-1 in SLE, gene polymorphisms of ET-1 in SLE. Second, we examined serum levels of CCN3, IL-28B in SLE patients, and discussed association of ET-1 and CCN3, IL-28B. If the two inflammatory components (CCN3, IL-28B) were abnormally expressed in SLE patients and there is significant association of ET-1 and CCN3, ET-1 and IL-28B, in the future, we will conduct functional study to clarify how ET-1 contributes to SLE development, and whether ET-1 contributes to SLE development by regulating CCB3, IL-28B.
Results
ET-1 serum levels in SLE from training cohort
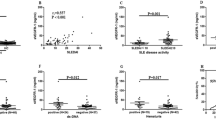
To study association between serum levels of ET-1 and SLE, a training cohort with 53 SLE patients and 80 healthy controls was analyzed. Clinical and laboratory characteristics of all subjects are shown in Table 1. Age of SLE patients and controls was 38.00 (27.00–49.00) years and 37.00 (34.00–40.00) years, and there was no difference in age of the two groups (Z = − 0.145, P = 0.884). Moreover, gender of SLE and healthy controls was matched (χ2 = 2.847, P = 0.092). ET-1 serum levels were significantly higher in SLE patients compared to healthy controls (74.11 (40.00–117.82) vs 17.35 (12.18–29.28) pg/ml, Z = − 8.029, P < 0.001, Fig. 1A). Patients with alopecia (N = 15) had increased serum levels of ET-1 when compared with that in patients without alopecia (N = 38) (P = 0.031, Fig. 1B). Patients with proteinuria (N = 31) had increased serum levels of ET-1 when compared with that in patients without proteinuria (N = 22) (P = 0.007, Fig. 1C). Similarly, SLE patients with positive anti-Sm antibody (N = 16) revealed elevated levels of ET-1 in serum than that in patients with negative anti-Sm antibody (N = 37) (P = 0.003, Fig. 1D). Moreover, SLE patients with active disease (SLEDIA ≥ 10) had higher ET-1 levels compared to patients with less active disease (SLEDIA < 10) (P < 0.001, Fig. 1E). Correlation analysis revealed positive correlation between serum ET-1 levels and SLEDAI score (rs = 0.463, P < 0.001, Fig. 1F), ESR levels (rs = 0.479, P = 0.005, Fig. 1G). Other clinical and laboratory features in SLE patients are not associated with serum ET-1 levels (Supplementary table 1 and 2). To evaluate ability of serum ET-1 to distinguish SLE patients from healthy individuals, we performed ROC curve analysis, which showed area under ROC curve (AUC) 0.912 (95% CI = 0.866–0.959) (P < 0.001, Fig. 1H).
Comparison of ET-1 levels between SLE patients and healthy controls in the training cohort. (A) Serum ET-1 levels in 53 SLE patients and 80 healthy individuals were examined by ELISA. Each symbol stands for an independent sample. (B–D) ET-1 levels in SLE patients distributed according to alopecia, proteinuria and anti-Sm. (E) Difference of serum levels of ET-1 in SLE patients with less active disease and active disease. (F–G) Relationship between ET-1 levels and SLEDAI, ESR levels. (H) Receiver operating characteristic curve analysis of serum ET-1 as a biomarker for SLE.
Elevated serum levels of ET-1 in SLE from validation cohort
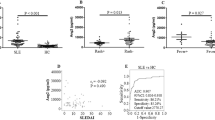
To evaluate potential of serum ET-1 as a biomarker for SLE, a validation cohort including 102 SLE patients and 252 non-SLE diseases patients was discussed further (90 RA, 95 osteoarthritis (OA), 55 sjogren syndrome (SS), 38 anlylosing spondylitis (AS), 17 SSc). Information about age and sex of these diseases patients is shown in Table 1. Results indicated that serum ET-1 levels in SLE patients were significantly higher than that in other rheumatic diseases (all P < 0.001, Fig. 2A). AUC was 0.803 by ROC curve analysis when comparing serum levels of ET-1 between SLE with RA patients (Fig. 2B). Similarly, serum ET-1 in SLE patients compared to that in OA, SS, AS, SSc, showed AUC 0.979, 0.856, 0.892 and 0.903, respectively (Fig. 2C–F). We compared serum ET-1 levels between SLE patients and non-SLE patients, showing that there was a significant difference (P < 0.001, Fig. 2G) and AUC was 0.900 (95% CI = 0.869–0.932) (Fig. 2H).
ET-1 levels in SLE patients from validation cohort. (A) Comparison of serum ET-1 levels in SLE patients (N = 102) and diseases controls (N = 252, including 90 RA, 95 OA, 55 SS, 38 AS, 17 SSc) by ELISA. (B–F) Receiver operating characteristic curve analysis was used to assess potential of serum ET-1 in differentiating SLE from RA, OA, SS, AS, and SSc. (G) Analysis of difference in serum ET-1 between SLE and non-SLE patients. (H) Receiver operating characteristic curve analysis of serum ET-1 between SLE and non-SLE patients.
Connection between ET-1 and CCN3, IL-28B levels in SLE
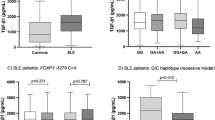
There were increased serum CCN3 levels in SLE patients compared with that in healthy controls (733.67 (553.57–988.08) vs 587.94 (430.06–748.02) pg/ml, Z = − 2.769, P = 0.006, Fig. 3A). Lupus patients with thrombocytopenia had higher levels of CCN3 (P = 0.020, Fig. 3B) (Supplementary table 3). SLEDAI score and other clinical characteristics in SLE patients were not significantly correlated with CCN3 levels (Supplementary table 4). Correlation between serum levels of ET-1 and CCN3 showed significant correlation (rs = 0.338, P = 0.007, Fig. 3C). For IL-28B, its levels were elevated in serum of SLE patients as compared to that in healthy controls (23.30 (16.93–39.12) vs 9.80 (5.34–15.92) pg/ml, Z = -6.974, P < 0.001, Fig. 3D). Lupus patients with hematuria, proteinuria, cylindruria had higher levels of IL-28B (P = 0.014, Fig. 3E; P = 0.012, Fig. 3F; P = 0.032, Fig. 3G) (Supplementary table 5). Higher serum IL-28B levels in SLE patients with active disease (SLEDIA ≥ 10) were obtained when compared to patients with less active disease (SLEDIA < 10) (P = 0.047, Fig. 3H). Correlation analysis reported that SLEDAI score, ESR levels were positively correlated with IL-28B levels, respectively (rs = 0.406, P = 0.003, Fig. 3I; rs = 0.461, P = 0.007, Fig. 3J) (Supplementary table 6). ET-1 levels were positively correlated with IL-28B levels in SLE patients (rs = 0.441, P = 0.001, Fig. 3K).
CCN3 and IL-28B levels in SLE patients and healthy controls. (A) Serum CCN3 levels were tested by ELISA in 53 SLE patients and 80 healthy controls. (B) Serum ET-1 in SLE patients distributed in accordance with thrombocytopenia. (C) Correlation between ET-1 and CCN3 levels in SLE patients. (D) Serum IL-28B levels were detected by ELISA in 53 SLE patients and 80 healthy controls. (E–G) Distribution of serum IL-28B in SLE patients with hematuria, proteinuria and cylindruria. (H) Difference of serum IL-28B in SLE patients with active disease and less active disease. (I–K) Relationship between IL-28B levels and SLEDAI score, ESR levels. (H) Correlation between ET-1 and IL-28B levels in SLE patients.
Association of ET-1 gene single nucleotide polymorphisms (SNPs) with SLE
To discuss whether polymorphisms of ET-1 gene correlate with SLE risk, a total of 314 lupus patients and 500 age, sex-matched healthy volunteers were selected. Hardy–Weinberg equilibrium (HWE) test showed no deviation in SLE patients and controls for all 10 polymorphisms (P > 0.05, Table 2). Information about alleles and genotypes of the SNPs was shown in Table 3. Frequency of rs5370 genotype TG was higher in SLE patients compared to healthy controls (TG vs. GG: OR = 1.443, 95% CI 1.072–1.943, P = 0.016). In the dominant model (TT + TG vs GG), frequency of genotypes TT + TG of rs5370 was increased in SLE patients (OR = 1.334, 95% CI 1.005–1.770, P = 0.046). For rs1476046, frequency of genotype GG in SLE patients was markedly lower than that in controls [GG vs GA + AA (recessive model): OR = 0.693, 95% CI 0.521–0.920, P = 0.011]. Compared with healthy controls, frequency of rs2070699 allele T was declined in SLE patients (T vs G: OR = 0.811, 95% CI 0.664–0.990, P = 0.039). Similarly, decreased frequencies of TT, TG, TT + TG genotypes of rs2070699 were found in SLE patients [TT vs GG: OR = 0.669, 95% CI 0.452–0.986, P = 0.042; TG vs GG: OR = 0.674, 95% CI 0.475–0.957, P = 0.027; TT + TG vs GG (dominant model): OR = 0.672, 95% CI 0.485–0.931, P = 0.017]. Frequency of GG genotype for rs2071942 was increased in SLE patients when compared with that in healthy subjects (GG vs GA: OR = 1.705, 95% CI 1.033–2.812, P = 0.037). With respect to rs2071943, there was decreased frequency of GG in SLE patients than that in control group [GG vs. GA + AA (recessive model): OR = 0.707, 95% CI 0.532–0.938, P = 0.016]. In the dominant model, higher frequencies of CA + CC genotypes of rs3087459 were found in patients with SLE (CA + CC vs AA: OR = 1.352, 95% CI 1.013–1.805, P = 0.040). In addition, SLE patients with rs6458155 had an enhanced frequency of TC genotype (TC vs CC: OR = 1.392, 95% CI 1.021–1.899, P = 0.0037). For rs5369, rs4145451, rs9369217 polymorphisms, we discussed the allele and genotypes distribution between SLE cases and healthy controls, showing no significant differences.
Relationship of ET-1 gene polymorphisms with clinical, laboratory features in SLE patients
Association of qualitative and quantitative clinical, laboratory features in SLE patients with ET-1 gene polymorphisms was shown in Supplementary table 7, 8, 9 and 10, respectively. Compared to patients without these clinical features, frequency of rs5070 genotypes TT + TG was elevated in patients with pericarditis and positive ANA (P = 0.034, P = 0.045), and was decreased in patients with fever (P = 0.005). Moreover, there was a higher frequency of allele T of rs5370 in patients with pericarditis, and a lower frequency of allele T of rs5370 in patients with pyuria compared with those in patients without the features (P = 0.013, P = 0.047). For rs1476046, decreased G allele frequency in patients with pericarditis and increased G allele frequency in patients with pyuria was observed when compared to patients without these features. A lower frequency of genotype GA of rs1476046 in SLE patients with fever and pyuria was noted (P = 0.013, P = 0.028). Interestingly, there were significant differences for genotypes and allele frequencies of rs2070699 polymorphism between SLE patients with alopecia and SLE patients without alopecia (P = 0.013, P = 0.035). Distribution of GG, GA, AA genotypes of rs2071942 and rs2071943 was different between SLE patients with and without fever (P = 0.025, P = 0.010). Patients with pericarditis had a declined frequency of G allele of rs2071942 and rs2071943 compared to patients without pericarditis (P = 0.019, P = 0.025). Frequency of G allele of rs2071942 and rs2071943 in patients with pyuria was higher (P = 0.035, P = 0.028). With respect to rs3087459, decreased frequencies of C allele and CC genotype in patients with positive ANA and increased frequency of CA genotype in patients with positive anti-SSB were observed when compared to patients without these features (P = 0.045, P = 0.029, P = 0.027). In addition, distribution of TT, TC and CC genotypes of rs9369217 was different in SLE patients with and without positive anti-SSB (P = 0.029) (Supplementary table 7 and 8). Other SNPs were not related to clinical and laboratory manifestations in SLE patients (Supplementary table 9).
When discussing quantitative indicators of clinical, laboratory features, levels of IgG between SLE patients with GG + GA genotype and AA genotype for rs5369 was different (P = 0.025). SLE patients carrying rs5369 AA genotype had higher levels of CRP as compared to the patients carrying GG + GA genotype (P = 0.012) (Supplementary table 10).
ET-1 gene haplotypes and SLE risk
Considering genetic linkage disequilibrium, we analyzed correlation between ET-1 gene haplotypes and SLE risk. There were two blocks constructed, one consisting of rs6458155, rs4145451 and the other one including rs2071942, rs2071943, rs5370 (Fig. 4). Results revealed that frequency of haplotype CA was lower in SLE patients compared to healthy controls (P = 0.012). Reduced frequency of haplotype AGG was found in patients with SLE (P = 0.041). No significant differences were observed in other haplotypes (Table 4).
Linkage disequilibrium (LD) analysis for ten SNPs in ET-1 gene. The color and numerical value (D’) of each box represent the Te intensity of LD. Red and pink indicate significant linkage, light blue and white indicate no linkage. The value of D’ varies from 0 to 1, by which value of 1 represents the maximum link. Block 1 consists of rs6458155 and rs4145451. Block 2 consists of rs2071942, rs2071943 and rs5370.
Discussion
SLE is a rheumatic disease with heterogeneous clinical symptoms that may result from excessive endothelial cells activation16. When activated by inflammatory stimuli, endothelial cells exhibit increased levels of surface adhesion molecules, which promote the migration and accumulation of leukocytes to the endothelial cells. Then, vascular obstruction and tissue hypoxia were induced, leading to apoptosis and tissue fibrosis17. Therefore, measuring biomarkers related to endothelial cells activation is useful for diagnosis of SLE. Endothelium-derived ET-1 plays a pathogenic role in connective tissue diseases, pulmonary hypertension, and cancer by affecting angiogenesis, inflammation, and fibrosis18. In OA mice, ET-1 induces increased levels of IL-18 through ET-1/ETAR axis and PI3K-dependent manner, promoting osteoblasts proliferation and exacerbating the disease19. As compared to control group, there were increased ET-1 levels in RA patients, especially in the patients with kidney and cardiovascular damages20. ET-1 levels were also elevated in other autoimmune diseases such as SSc, psoriasis and type 1 diabetes21,22. According to our study, serum ET-1 levels were significantly higher in SLE patients in training cohort when compared with healthy controls, which is consistent with previous findings6,23,24. Moreover, we found that serum ET-1 levels were associated with SLE patients complicated with cylindruria, alopecia and positive anti-Sm antibody. Similarly, Yoshio et al. showed that serum ET-1 in SLE patients correlated with IgM antibody levels. A study from Poland SLE patients reported that serum ET-1 levels were higher in active SLE patients than in inactive SLE patients and healthy controls23. SLE patients with visceral manifestation showed higher ET-1 levels as compared to that in patients without the feature23. In our study (the training cohort), we also found a positive correlation between serum ET-1 levels and SLEDAI score, and SLE patients with active disease activity had higher ET-1 levels. Urinary ET-1 may be a useful measurement of renal inflammatory activity and may serve as a marker in lupus nephritis disease activity25. In the present study, we also explored potential of serum ET-1 as a biomarker for SLE. In training cohort, AUC of ET-1 was 0.912 (95% CI 0.866–0.959), indicating that serum ET-1 could distinguish SLE patients from healthy subjects. In the validation cohort, ET-1 levels were higher in SLE patients than in other rheumatic diseases, including RA, OA, SS, AS, SSc. Compared with non-SLE diseases patients, AUC of serum ET-1 in SLE was higher than 0.800. Therefore, serum ET-1 is a promising marker for SLE. How ET-1 contributes to SLE or what mechanisms of ET-1 may involve in development of the disease? ET-1 could induce levels of connective tissue growth factor (CTGF/CCN2) by activating the MEK/ERK kinase pathway after binding to ETA and ETB receptor26. Indeed, these signalling pathways are pro-inflammatory pathways in SLE pathogenesis. After treatment with ET-1 receptor antagonist (bosentan), anti-dsDNA levels were reduced, glomerulosclerosis and renal T cells infiltration were improved in lupus mice27. These findings suggest that ET-1 may promote SLE progression. IL-17 and Th17 cells are important cytokine and cells in SLE pathogenesis. It is known that ET-1/ETAR signaling pathway regulates production of IL-17 in Th17 cells by an autocrine or paracrine manner28. To date, no study has discussed effects of ET-1 on Th17 cells in SLE. Thus, in future studies, we will consider to explore whether highly expressed ET-1 in SLE will contribute to lupus development by regulating Th17 cells. In patients with lupus nephritis, IgM anti-endothelial cell antibody and immune complexes can bind to endothelial cells in glomerular capillary rings and stimulate production of ET-1, leading to vascular injury24.
CCN3 involves in some autoimmune diseases pathogenesis12. Serum CCN3 levels were elevated in RA patients and positively correlated with levels of IL-629. CCN3 levels were increased in SSc, multiple sclerosis patients30,31. For OA, CCN3 could inhibit PI3K/AKT/mTOR pathway by reducing HMGB1 levels and decrease extracellular matrix catabolism32. In our study, we found that serum CCN3 levels were significantly higher in SLE patients compared to normal subjects and were associated with thrombocytopenia. In addition, we observed a positive correlation between ET-1 levels and CCN3 levels in SLE patients. CCN3 inhibits levels of vascular adhesion molecules and reduces monocytes adhesion. CCN3 negatively regulates activation of NF-κB pathway, affecting inflammatory cytokines production of endothelial cells and cardiovascular homeostasis33. Type III interferons (IFNs) and type I IFNs may promote THP-1 cells differentiation, thereby contributing to the activation of follicular B cells and participating in development of autoimmune diseases34. IL-28B regulates innate and adaptive immune responses. SSc patients with pulmonary fibrosis had higher serum levels of IL-28B and IL-28B gene polymorphism (rs12979860) is associated with risk of pulmonary fibrosis in Caucasian SSc patients35. With respect to SLE, IL-28B gene SNPs (rs8099917, rs12979860) are risk factors for lupus nephritis in Taiwanese. Moreover, serum levels of IL-28B were elevated in SLE patients compared to healthy controls, which were related to complement levels and SLE disease activity36. IL-28B levels are associated with SLE disease activity15. According to our study, IL-28B levels in SLE patients were positively associated with disease activity and ESR levels. SLE patients with hematuria, proteinuria, cylindruria showed higher levels of IL-28B, which are typical clinical manifestations of SLE. We analyzed correlation between ET-1 and IL-28B levels and observed that serum levels of ET-1 were positively correlated with IL-28B levels. Therefore, in the future, we will conduct functional study (for example, knock out ET-1 gene in animal models) to discuss whether ET-1 regulates CCN3 or IL-28B, and then contributes to SLE development.
It is now accepted that SNPs are new genetic markers that may be used for discovery of high risk patients. ET-1 gene polymorphisms are widely discussed in vascular-related diseases and cancer, such as hypertension, coronary atherosclerosis, and papillary thyroid cancer37,38. However, ET-1 gene polymorphisms are less studied in autoimmune diseases. In this study, we explored relationship between ET-1 gene polymorphisms and SLE risk by a case–control study from Chinese Han population. We found that genotypes of rs5370 (TG,TT + TG), rs1476046 (GG), rs2070699 (TT,TG), rs2071942 (GA), rs2071943 (GG), rs3087459 (CA + AA), rs6458155 (TC) and allele of rs2070699 (T) were associated with SLE susceptibility. For rs5370, TG, TT + TG genotype frequencies were higher in SLE patients, suggesting that rs5370 polymorphism may increase the risk of SLE. Frequency of rs2071942 GG genotype was increased in SLE patients compared to healthy subjects, which was positively associated with SLE risk. Mantaka et al. discussed association of ET-1 gene rs2071942 and rs5370 polymorphisms with primary biliary cirrhosis (PBC), showing that distribution of genotypes and alleles of both loci were not significantly different between controls and PBC patients, whereas rs2071942 allele A and rs5370 allele T were associated with stage of disease progression39. These inconsistency may be due to differences in sample size, ethnicity, disease type. Similarly, frequencies of genotypes and alleles of rs5370 and rs18000541 were not significantly different between RA patients and healthy controls, but TT genotype of rs5370, rs18000541 was related to RA patients complicated with hypertension, indicating that rs5370 and rs18000541 loci were associated with cardiovascular risk in RA40. In our study, rs5370, rs1476046, rs2070699, rs2071942, rs2071943 rs5369, rs3087459, and rs9369217 polymorphisms correlated with some clinical, laboratory features in SLE patients. For example, SLE patients carrying rs2070699 T allele and TT genotype were more likely to develop symptoms of alopecia. Regarding rs2071942 and rs2071943, allele G was associated with pericarditis and pyuria, and genotype GA was related to fever. Moreover, significant correlation between rs5370 T allele, rs1476046 G allele and pericarditis, pyuria in SLE cases was observed. Rs3087459 (CC) and rs9369217 (TC) were associated with SLE patients with positive anti-SSB antibody. Rs5369 AA genotype correlated with IgG and CRP levels, suggesting that mutations at the rs5369 locus may affect levels of these disease markers in SLE patients. Indeed, CRP is an acute phase protein produced by hepatocytes in response to inflammation. CPR has important roles in active lupus nephritis. In SLE patients or mice models, IgG immune complexes are deposited in the spleen, causing damage to immune barrier of the spleen, and then numbers of autoantibodies are produced41. A study discussed association of graves’ disease (GD) risk and ET-1 gene polymorphisms (rs5370 and rs1800541), revealing that the polymorphisms were not associated with disease susceptibility, but were related to autoantibodies production in GD patients42. To the best of our knowledge, this study is the first to discuss relationship between ET-1 gene polymorphisms and SLE, which may offer new insights and basis for further discussion of ET-1 genetic mutation and SLE in the future.
This study has several limitations. First, a larger number of SLE patients across multiple centers are required to confirm serum ET-1 as a potential disease marker for SLE. Second, the underlying mechanism by which ET-1 regulates CCN3, IL-28B, and subsequently influences pathogenesis of SLE requires further clarification.
In conclusion, ET-1 is associated with pathogenesis of SLE, may be a potential disease biomarker, and ET-1 gene polymorphisms were related to SLE susceptibility in the Chinese Han population.
Material and methods
Subjects
This study included 314 SLE patients, 252 non-SLE patients, and 500 healthy controls. All patients were from Department of Rheumatology and Immunology of Affiliated Hospital of Southwest Medical University, and healthy controls came from Examination Center of Center for Disease Control and Prevention in Jiangyang District, Luzhou. SLE was diagnosed by 2019 American College of Rheumatology (ACR) revised criteria43. Healthy controls were the participants who did not have any diseases. Based on SLE disease activity index 2000 (SLEDAI-2 K) score44, SLE patients were divided into less active disease (SLEDIA < 10) and active disease (SLEDIA ≥ 10). RA, OA, SS, AS, SSc were diagnosed by 2010 ACR criteria for RA45, 1986 ACR criteria for OA46, 2016 ACR criteria for SS47, modifed New York criteria for AS48, 2013 ACR/European League Against Rheumatism criteria for SSc49, respectively. Participants were all Chinese Han population. This study was endorsed by Ethics Committee of Affiliated Hospital of Southwest Medical University and according to Declaration of Helsinki formulated by World Medical Association. Informed consent from each participant was obtained.
The study has two parts. In the first part, we evaluated serum levels of ET-1 in SLE patients, and discussed association of ET-1 with SLE pathogenesis, including 53 SLE patients and 80 healthy controls (training cohort). Similarly, we detected serum CCN3 and IL-28B levels in 53 SLE patients and 80 healthy individuals, and discussed correlation between ET-1 and CCN3, ET-1 and IL-28B. Then, another cohort of 102 SLE patients, 90 patients with RA, 95 with OA, 55 with SS, 38 with AS, and 17 with SSc, were included to discuss potential of serum ET-1 as a biomarker for SLE (validation cohort). In the second part, we genotyped 10 SNPs of ET-1 gene (rs5369, rs5370, rs1476046, rs2070699, rs2071942, rs2071943, rs3087459, rs4145451, rs6458155, rs9369217) to discuss genetic susceptibility of SLE, including 314 SLE patients and 500 healthy controls.
DNA preparation, screening and detection of single-nucleotide polymorphisms
Peripheral blood was centrifuged to obtain serum, which was stored at -80℃ until use. Genomic DNA was extracted by TIANamp Blood DNA kits (TIANGEN) on the basis of instructions. Ensemble database and 1000 genomes project (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/) were used to find out information of ET-1 gene polymorphisms in Chinese Han population. Criteria for SNP selection were as follows: (1) MAF > 0.05 in Chinese Han population. (2) SNPs were prior to be selected as they were located at functional positions such as 5’UTR, 3’UTR, frameshift mutation and non-synonymous mutation. ET-1 gene polymorphisms (rs5369, rs5370, rs1476046, rs2070699, rs2071942, rs2071943, rs3087459, rs4145451, rs6458155, rs9369217) were genotyped using Kompetitive Allele-Specific PCR (KASP) method. Information of KASP primers for 10 SNPs was listed in Supplementary table 11. Briefly, KASP genotyping mix was prepared, consisting of 5ul DNA, 5ul 2 × KASP Master mix, and 0.14ul Assay mix. Then, the mix was added to PCR plates for PCR cycling reactions: (a) 94 °C (15 min); (b) 94 °C (20 s), 61 °C (60 s), for a total of 10 cycles; (c) 94 °C (20 s), 55 °C (60 s), for a total of 26 cycles. Finally, fluorescence data were read using an enzyme labeler with FRET function.
Measurement of ET-1, CCN3, IL-28B by enzyme-linked immune sorbent assay (ELISA)
Levels of ET-1, CCN3 and IL-28B in SLE patients and control groups were determined by ELISA. All ELISA kits were purchased from CUSABIO (Wuhan, China, serial numbers CSB-E07007h (ET-1), CSB-EL015956HU (CCN3), CSB-E13296h (IL-28B)). According to the instructions, serum samples were added to the wells and incubated at 37 °C for 2 h. After removing the liquid, biotin-conjugate was added and incubated at 37 °C for 1 h. Subsequently, the plates were washed 3 times with washing buffer and HRP-avidin was added and incubated at 37 °C for 1 h. After 5 times’ washes, TMB substrate was added to each well and the termination solution was added after incubation at 37 °C for 15 min in the dark. The data were measured at 450 nm and converted into concentration by linear standard curve.
Statistics
SPSS 23.0 and GraphPad Prism 5.01 were used for analysis of all data. For quantitative data, if the data followed normal distribution, means ± standard deviation (SD) and independent samples t-test were used to describe and analyze. Otherwise, median (interquartile range) and Wilcoxon ranks sum test were used. For categorical data, we described them by frequency and percentage, and adopted chi-square test for comparison. Potential of serum ET-1 as the biomarker for lupus was evaluated by area of receiver operating characteristic (ROC) curve. Hardy–Weinberg equilibrium assessed deviation of individual polymorphism in SLE patients and healthy controls. Distribution of genotypes and alleles between SLE patients and healthy controls was compared using chi-square test, then odds ratio (OR) and 95% confidence interval (CI) were analyzed by logistic regression model. HaploView 4.1 software was used to calculate linkage disequilibrium and haplotype analysis. The level of significance was P < 0.05 (two-sided).
Data availability
Datasets are available from the corresponding author on reasonable request.
References
He, N., Leng, X. & Zeng, X. Systemic lupus erythematosus following human papillomavirus vaccination: A case-based review. Int. J. Rheum. Dis. 25, 1208–1212 (2022).
Yang, C., Li, R., Xu, W. D. & Huang, A. F. Increased levels of sirtuin-1 in systemic lupus erythematosus. Int. J. Rheum. Dis. 25, 869–876 (2022).
Ha, E., Bae, S. C. & Kim, K. Recent advances in understanding the genetic basis of systemic lupus erythematosus. Semin. Immunopathol. 44, 29–46 (2022).
Torres Crigna, A. et al. Endothelin-1 axes in the framework of predictive, preventive and personalised (3P) medicine. EPMA J. 12, 265–305 (2021).
Kostov, K. The causal relationship between endothelin-1 and hypertension: Focusing on endothelial dysfunction, arterial stiffness, vascular remodeling, and blood pressure regulation. Life Basel 1, 986 (2021).
Hajialilo, M. et al. Relationship between serum vascular cell adhesion molecule-1 and endothelin-1 levels with organ involvement and disease activity in systemic lupus erythematosus patients. Lupus 27, 1918–1925 (2018).
Alvarez-Cienfuegos, A. et al. Endothelin-1 serum levels in women with rheumatoid arthritis. Acta Reumatol. Port. 44, 250–257 (2019).
Borska, L. et al. Roles of miR-31 and endothelin-1 in psoriasis vulgaris: Pathophysiological functions and potential biomarkers. Physiol. Res. 66, 987–992 (2017).
Stochmal, A., Czuwara, J., Zaremba, M. & Rudnicka, L. Altered serum level of metabolic and endothelial factors in patients with systemic sclerosis. Arch. Dermatol. Res. 312, 453–458 (2020).
Dombrowski, Y. et al. Regulatory T cells promote myelin regeneration in the central nervous system. Nat. Neurosci. 20, 674–680 (2017).
Lin, C. G. et al. CCN3 (NOV) is a novel angiogenic regulator of the CCN protein family. J. Biol. Chem. 278, 24200–24208 (2003).
Peng, L. et al. The emerging roles of CCN3 protein in immune-related diseases. Med. Inflamm. 18(2021), 5576059 (2021).
Li, J. et al. Deficiency of type I interferon contributes to Sle2-associated component lupus phenotypes. Arthritis Rheum 52, 3063–3072 (2005).
Lin, S. C., Kuo, C. C., Tsao, J. T. & Lin, L. J. Profiling the expression of interleukin (IL)-28 and IL-28 receptor α in systemic lupus erythematosus patients. Eur. J. Clin. Invest. 42, 61–69 (2012).
Amezcua-Guerra, L. M. et al. Type III interferons in systemic lupus erythematosus: Association between interferon λ3, disease activity, and anti-Ro/SSA antibodies. J. Clin. Rheumatol. 23, 368–375 (2017).
Ci, W. et al. Characteristics and risk factors of severe coronary artery disease in systemic lupus erythematosus: A multicenter, Chinese Rheumatism Date Center database study. Int. J. Rheum. Dis. 25, 1186–1195 (2022).
Mostmans, Y. et al. The role of endothelial cells in the vasculopathy of systemic sclerosis: A systematic review. Autoimmun. Rev. 16, 774–786 (2017).
Davenport, A. P. & Maguire, J. J. Endothelin. Handb. Exp. Pharmacol. 176, 295–329 (2006).
Zhong, X., Wang, H. & Huang, S. Endothelin-1 induces interleukin-18 expression in human osteoblasts. Arch. Oral. Biol. 59, 289–296 (2014).
Haq, A., El-Ramahi, K., Al-Dalaan, A. & Al-Sedairy, S. T. Serum and synovial fluid concentrations of endothelin-1 in patients with rheumatoid arthritis. J. Med. 30, 51–60 (1999).
Raschi, E. et al. Scleroderma-specific autoantibodies embedded in immune complexes mediate endothelial damage: An early event in the pathogenesis of systemic sclerosis. Arthritis Res. Ther. 22, 265 (2020).
Saleh, M. A. et al. Free radical scavenging decreases endothelin-1 excretion and glomerular albumin permeability during type 1 diabetes. Physiol. Rep. 4, e13055 (2016).
Kuryliszyn-Moskal, A., Klimiuk, P. A., Ciolkiewicz, M. & Sierakowski, S. Clinical significance of selected endothelial activation markers in patients with systemic lupus erythematosus. J. Rheumatol. 35, 1307–1313 (2008).
Yoshio, T. et al. Endothelin-1 release from cultured endothelial cells induced by sera from patients with systemic lupus erythematosus. Ann. Rheum. Dis. 54, 361–365 (1995).
Dhaun, N. et al. Urinary endothelin-1 in chronic kidney disease and as a marker of disease activity in lupus nephritis. Am. J. Physiol. Renal. Physiol. 296, F1477–F1483 (2009).
Leask, A. The role of endothelin-1 signaling in the fibrosis observed in systemic sclerosis. Pharmacol. Res. 63, 502–503 (2011).
Rivera-Gonzalez, O., Case, C. T., Wilson, N. A., Speed, J. S. & Taylor, E. B. Endothelin receptor antagonism improves glucose tolerance and adipose tissue inflammation in an experimental model of systemic lupus erythematosus. Am. J. Physiol. Endocrinol. Metab. 324, E73–E84 (2023).
Tanaka, K., Yoshioka, K., Tatsumi, K., Kimura, S. & Kasuya, Y. Endothelin regulates function of IL-17-producing T cell subset. Life Sci. 118, 244–247 (2014).
Wei, Y. et al. Higher Serum CCN3 is associated with disease activity and inflammatory markers in rheumatoid arthritis. J. Immunol. Res. 9(2020), 3891425 (2020).
Henrot, P., Pain, C., Taïeb, A., Truchetet, M. E. & Cario, M. Dysregulation of CCN3 (NOV) expression in the epidermis of systemic sclerosis patients with pigmentary changes. Pigment Cell Melanoma Res. 33, 895–898 (2020).
Naughton, M. et al. CCN3 is dynamically regulated by treatment and disease state in multiple sclerosis. J. Neuroinflamm. 17, 349 (2020).
Huang, X. et al. NOV/CCN3 induces cartilage protection by inhibiting PI3K/AKT/mTOR pathway. J. Cell Mol. Med. 23, 7525–7534 (2019).
Lin, Z. et al. A novel role of CCN3 in regulating endothelial inflammation. J. Cell Commun. Signal 4, 141–153 (2010).
Tanemura, S. et al. Role of interferons (IFNs) in the differentiation of T peripheral helper (Tph) cells. Int. Immunol. 34, 533–544 (2022).
Metwally, M. et al. IFNL3 genotype is associated with pulmonary fibrosis in patients with systemic sclerosis. Sci. Rep. 9, 14834 (2019).
Chen, J. Y. et al. Interferon-λ3/4 genetic variants and interferon-λ3 serum levels are biomarkers of lupus nephritis and disease activity in Taiwanese. Arthritis Res. Ther. 20, 193 (2018).
Ebrahimi, N. et al. The association of endothelin-1 gene polymorphism and its plasma levels with hypertension and coronary atherosclerosis. Arch. Med. Sci. 17, 613–620 (2019).
Aydin, A. F., Vural, P., Doğru-Abbasoğlu, S. & Çil, E. The endothelin 1 and endothelin receptor A gene polymorphisms increase the risk of developing papillary thyroid cancer. Mol. Biol. Rep. 46, 199–205 (2019).
Mantaka, A. et al. Polymorphisms of genes related to endothelial cells are associated with primary biliary cirrhosis patients of Cretan origin. Hum. Immunol. 73, 829–835 (2012).
Panoulas, V. F. et al. Polymorphisms of the endothelin-1 gene associate with hypertension in patients with rheumatoid arthritis. Endothelium 15(4), 203–212. https://doi.org/10.1080/10623320802228708 (2008).
Qi, S., Chen, Q., Xu, D., Xie, N. & Dai, Y. Clinical application of protein biomarkers in lupus erythematosus and lupus nephritis. Lupus 27, 1582–1590 (2018).
Aydın, A. F. et al. Polymorphisms of endothelin 1 (G5665T and T-1370G) and endothelin receptor type A (C+70G and G-231A) in Graves’ disease. Int. Immunopharmacol. 18, 198–202 (2014).
Aringer, M. et al. 2019 European League against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann. Rheum. Dis. 8, 1151–1159 (2019).
Gladman, D. D., Ibañez, D. & Urowitz, M. B. Systemic lupus erythematosus disease activity index 2000. J. Rheumatol. 29, 288–291 (2002).
Aletaha, D. et al. 2010 rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann. Rheum. Dis. 69, 1580–1588 (2010).
Altman, R. et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthr. Rheum 29, 1039–49 (1986).
Shiboski, C. H. et al. 2016 American College of Rheumatology/European League Against Rheumatism classification criteria for primary Sjögren’s syndrome: A consensus and data-driven methodology involving three international patient cohorts. Ann. Rheum Dis. 76, 9–16 (2017).
van der Linden, S., Valkenburg, H. A. & Cats, A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthr. Rheum 27, 361–8 (1984).
van den Hoogen, F. et al. classification criteria for systemic sclerosis: An American college of rheumatology/European league against rheumatism collaborative initiative. Ann. Rheum. Dis. 72, 1747–1755 (2013).
Funding
This work was supported by grants from the Sichuan Provincial Natural Science Foundation (2022NSFSC0697, 2022NSFSC0694).
Author information
Authors and Affiliations
Contributions
Study conception and design: R.L., W.D.X., A.F.H. Acquisition of data, analysis and interpretation of data: Y.Y.T., D.C.W., C.Y. Drafting the article: R.L., W.D.X., A.F.H. Final approval of the version of the article to be published: all authors, and that all authors agree to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, R., Yang, C., Tang, YY. et al. Association between endothelin-1 and systemic lupus erythematosus: insights from a case–control study. Sci Rep 13, 15970 (2023). https://doi.org/10.1038/s41598-023-43350-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-43350-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.