Abstract
Although glaucoma is a disease modulated by eye pressure, the mechanisms of pressure sensing in the eye are not well understood. Here, we investigated associations between mechanosensitive ion channel gene variants and primary open-angle glaucoma (POAG). Common (minor allele frequency > 5%) single nucleotide polymorphisms located within the genomic regions of 20 mechanosensitive ion channel genes in the K2P, TMEM63, PIEZO and TRP channel families were assessed using genotype data from the NEIGHBORHOOD consortium of 3853 cases and 33,480 controls. Rare (minor allele frequency < 1%) coding variants were assessed using exome array genotyping data for 2606 cases and 2606 controls. Association with POAG was analyzed using logistic regression adjusting for age and sex. Two rare PIEZO1 coding variants with protective effects were identified in the NEIGHBOR dataset: R1527H, (OR 0.17, P = 0.0018) and a variant that alters a canonical splice donor site, g.16-88737727-C-G Hg38 (OR 0.38, P = 0.02). Both variants showed similar effects in the UK Biobank and the R1527H also in the FinnGen database. Several common variants also reached study-specific thresholds for association in the NEIGHBORHOOD dataset. These results identify novel variants in several mechanosensitive channel genes that show associations with POAG, suggesting that these channels may be potential therapeutic targets.
Similar content being viewed by others
Introduction
Primary open-angle glaucoma (POAG) is a progressive, neurodegenerative disease leading to loss of retinal ganglion cells (RGCs) and irreversible blindness1. Intraocular pressure (IOP) is the only modifiable risk factor, and is the target of current medical and surgical approaches for glaucoma treatment1. Despite the known importance of IOP in disease progression, we only have limited understanding of the proteins and molecules that regulate IOP, and the mechanisms by which elevated IOP leads to RGC loss. As the primary sensors that mediate many responses to mechanical signals2, mechanosensitive ion channels may play a role in both IOP regulation and the pathologic responses of RGCs to IOP.
In recent years, significant advances have been made in our understanding of mechanosensitive ion channels, including the discovery of the PIEZO family of mechanically activated channels3,4, and the validation of the TRAAK/TREK two-pore domain K + (K2P) channels and TMEM63 (hyperosmolality-gated calcium-permeable) channel families as mechanosensitive5,6. In addition, many members of the TRP (transient receptor potential) channel family have been implicated in sensing pressure and stretch in mammals7, including TRPV18,9, TRPV210, TRPV411, TRPC112,13, TRPC314, TRPC515, TRPC616,17, TRPA118,19, TRPP120, TRPM321, TRPM422 and TRPM723. Mechanosensitive channel families play diverse physiological roles in human health and disease, ranging from touch, blood-pressure sensing and hearing4,24,25,26,27,28. However, their roles in the eye are not entirely clear. Several in vitro and mouse studies have suggested that mechanosensitive ion channels may play a role in glaucoma, either by regulating aqueous humor outflow and IOP29,30,31,32,33,34,35,36,37, or by modulating RGC survival38,39,40,41. However, clinical evidence in humans implicating the role of mechanosensitive channels in glaucoma is lacking.
In this study, we surveyed mechanosensitive ion channel genes in the K2P, TMEM63, PIEZO and TRP channel families and asked whether common and rare genetic risk variants in these genes are associated with POAG using data from independent human genetic datasets.
Results
Association results for common variants
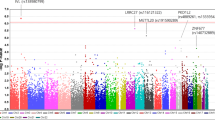
In this study we investigated association results for all single nucleotide polymorphisms (SNPs) with minor allele frequencies (MAF) greater than 5% in individuals with European Ancestry located within the genomic regions harboring the genes of interest including 50 Kb upstream and downstream of the gene to capture regulatory elements. Table 1 lists the SNPs corresponding to each genomic region with the best association result for POAG overall and also for the high-tension glaucoma (HTG) and normal-tension glaucoma (NTG) subgroups. Association results for 4 SNPs reached the significance threshold for POAG overall (rs2841593, KCNK2 (TREK1); rs8016340, TMEM63C; rs3124515, TRPM3; and rs34419652, TRPV2). The TRPV2 SNP is an expression quantitative trait loci (eQTL) in at least one tissue type in the Gene-Tissue Expression (GTEx) database, suggesting that this SNP can impact gene expression. Two SNPs reached the significance threshold for HTG (rs112014893, PIEZO2; rs4738210, TRPA1) and 4 SNPs for NTG (rs12709104, PIEZO1; rs2026109, TRPM3; rs9901098, TRPV2; rs12423752, TRPV4). We next investigated association for these SNPs and glaucoma phenotypes in FinnGen. While none of these SNPs reached nominal (P < 0.05) significance for disease association, two SNPs showed consistent direction of effects including rs34419652 (TRPV2 and POAG) and rs2026109 (TRPM3 and NTG) (Supplemental Table 1).
Association results for exome array variants
DNA variants impacting protein structure (coding variants), while often rare in populations, can have large effects on human traits and disease. To evaluate potential roles for protein coding variants in the mechanosensitive genes in POAG, we evaluated rare (MAF < 1%) variants effecting protein structure (missense, stop-loss and splice-sites) using exome array data available for two of the NEIGHBORHOOD cohorts, NEIGHBOR and MEE (total 2606 cases and 2606 controls).
We first evaluated gene-based association using SKAT-O and did not identify significant association with any of the selected mechanosensitive genes that met the criteria of having at least 3 variants available for analysis (Supplemental Table 2). Interestingly one gene, TRPC1, has interesting evidence of association (P = 6.34E−3) with POAG in the UK Biobank (defined as ICD-10 code H40, N = 11,588 cases and 383,253 controls) using SKAT-O and only missense alleles (Supplemental Table 2).
In the single-variant analysis, 55 variants met the inclusion criteria (MAF < 1%, CADD > 15) (Supplemental Table 3). Two of these variants, both in PIEZO1, demonstrated interesting association with POAG in the NEIGHBOR and MEE samples. Both PIEZO1 variants were found more commonly in controls than in cases suggesting protective effects. The first of these is a missense allele, R1527H (OR 0.17, P = 0.0018). This allele is enriched in the Finnish population and the direction of effect was consistent in FinnGen (beta − 0.11, P = 0.05) and also in the UK Biobank (beta − 0.149, P = 0.347) (Table 2). The second PIEZO1 allele alters a canonical splice donor site (genomic location 16-88737727-C-G Hg38). The distribution of this variant also suggests protective effects in POAG (OR 0.38, P = 0.02) and the direction of effect was also consistent in the UK Biobank (beta = − 0.1, P = 0.696) (Table 2). This variant was not present in the FinnGen dataset. The splice variant is predicted to alter splicing (Splice AI = 0.970 (donor_loss)), and incorrect splicing is likely to cause the inclusion of 27 amino acids due to retention of the short intron number 9 (of 50 in the gene) (Fig. 1).
Consequence of PIEZO1 canonical donor splice variant (16-88737727-C-G). The C > G change disrupts the canonical donor splice sequence GT (the minus strand is shown in the image), leading to retention of the short intron (81 base pairs). A stop codon is not encountered in the reading frame, and as a result 27 amino acids are included in the polypeptide sequence. The amino acids potentially included in the protein are listed below the sequence at the top of the image. Image is rendered from the UCSC genome browser (https://genome.ucsc.edu/).
In addition to the PIEZO1 variants, two other coding variants of interest were identified. The first is a low frequency variant in TMEM63B (V307M) that showed nominal association with POAG in the NEIGHBOR and MEE samples (OR = 1.13, P = 0.04). This variant also showed consistent evidence of association with POAG in the UK Biobank and also in FinnGen (Table 2). A missense allele in TRPA1 (A122V) was also nominally associated with POAG in the NEIGHBOR and MEE samples (OR 4.3, P = 0.02). However, this variant had the opposite direction of effect in the UK Biobank (beta − 0.326, P = 0.58) (Table 2). This variant was not present in the FinnGen dataset.
Single-cell and single-nucleus RNA sequence data
Expression of the mechanosensitive genes in ocular tissues was determined using publicly available single-cell and single-nucleus RNA sequence data42,43 (Fig. 2). PIEZO1 and PIEZO2 have relatively strong expression in lymphatic endothelial cells in the ocular anterior segment, tissue related to Schlemm’s canal endothelia42. TMEM63B, TMEM63C, TRPM3 and TRPV2 all have relatively strong expression in retinal ganglion cells.
Expression of selected genes with interesting associations with POAG in ocular tissues. Single-cell43 and single-nucleus42 sequencing data were extracted for anterior segment cell types, neuronal cell types in the retina including retinal ganglion cells, and non-neuronal retinal cell types. Dot plots show expression of selected genes in anterior segment and retinal cell types. Dot size indicates fraction of expressing cells, and color indicates mean expression in expressing cells.
Discussion
Although mechanosensitive channels have been implicated in glaucoma pathogenesis based on experimental studies, the association between mechanosensitive channels genetic variants and glaucoma risk has not been well studied. Here, we surveyed common and rare genetic variants for 20 mechanosensitive channel genes in the K2P, TMEM63, PIEZO and TRP channel families for association with POAG.
Two rare PIEZO1 coding variants with consistent protective effects for POAG were identified: a missense allele (R1527H) and a variant that alters a canonical splice donor site (genomic location 16-88737727-C-G Hg38). These variants also showed similar effects in the UK Biobank and the R1527H also in the FinnGen database. A PIEZO1 gain-of-function polymorphism (e756del) common in African Americans was previously not found to be associated with glaucoma phenotypes44. It is unclear if the two alleles we identified confer gain or loss of function. Future studies are needed to elucidate the effects of these alleles on channel function.
Common noncoding SNPs located in the PIEZO1 and PIEZO2 genomic regions showed interesting associations with NTG and POAG respectively, although neither of these SNPs had evidence of association in the FinnGen dataset.
Published single-cell and single-nucleus RNA sequencing data42,43 shows that PIEZO1 and PIEZO2 are expressed in both the ocular anterior segment and the retina, with strongest expression in the lymphatic endothelium, recently recognized as contributing to Schlemm’s canal function45. Previous in vitro studies have implicated PIEZO1 and PIEZO2 in aqueous outflow regulation. PIEZO1 activity in trabecular meshwork cells has been proposed to increase conventional aqueous ouflow32,34,35. In contrast, modulating PIEZO2 expression in iridocorneal tissues does not affect the conventional outflow pathway or IOP, but may affect unconventional outflow pathways or aqueous humor production36. PIEZO1 and PIEZO2 are also expressed in astrocytes of the optic nerve head46. PIEZO1 has been suggested to contribute to astrocyte reactivity47, and PIEZO2 expression in the optic nerve head was upregulated after transient IOP elevation46. Future studies are needed to fully elucidate the roles of PIEZO1 and PIEZO2 in different ocular tissues.
Rare missense alleles meeting the threshold for nominal POAG association (P < 0.05) in the NEIGHBOR and MEE samples were also found in TRPA1 (A122V) and in TMEM63B (V307M). However, neither of these were associated with disease risk in the UK Biobank or in the FinnGen dataset. The TMEM63B variant did show consistent direction of effect in both the UK Biobank and in FinnGen. TMEM63B is expressed in retinal ganglion cells (Fig. 2), but the role of these channels in the eye is unknown.
In addition to rare variant association, we also found interesting associations for SNPs located in the genomic regions of TRPV2, TRPM3, TRPA1 and KCNK2 (TREK1). A TRPV2 SNP (rs34419652), an eQTL, was significantly associated with POAG risk in NEIGHBORHOOD and showed a consistent direction of effect in FinnGen. A TRPM3 SNP (rs3124515) was significantly associated with POAG in NEIGHBORHOOD. A KCNK2 (TREK1) SNP (rs2841593) was associated with POAG risk in the NEIGHBORHOOD dataset, although this result did not replicate in FinnGen. KCNK2 has been suggested to sense mechanical stretch in the trabecular meshwork and regulate IOP29,30. The TRP channel genes are expressed in both the anterior segment and the retina (Fig. 2), and have been suggested to play multiple roles in the eye that may contribute to glaucoma risk48. TRPV2 contributes to the myogenic constriction of retinal arterioles49, and possibly regulates blood flow in the retina. TRPM3 is expressed in the ciliary body, lens, and retinal pigment epithelium50,51, and a missense mutation in TRPM3 has been linked to congenital cataracts and glaucoma52. TRPA1 mediates oxidative stress after retinal ischemia53.
Multiple in vitro and animal studies have also suggested roles for TRPV1 and TRPV4 in glaucoma pathogenesis, including roles in IOP regulation31,37,54,55, RGC apoptosis and survival38,40,41, and regulation of inflammatory cytokines39. In this study, we did not find significant associations between TRPV1 and TRPV4 SNPs with POAG, although a TRVP4 SNP (rs12423752) reached threshold significance for NTG in NEIGHBORHOOD.
We selected to investigate a subset of TRP channels that have been implicated in mechanosensation in this study. It remains controversial whether these channels are directly or indirectly activated by mechanical forces56. Nonetheless, loss-of-function mutations in humans suggests their involvement in mechanosensory processes57.
We have presented an overview of the expression of mechanosensitive genes using previously published human single-cell and single-nucleus RNA sequence data42,43 (Fig. 2). Additional histological validation of gene expression would complement this data in elucidating the roles of mechanosensitive channel genes in the eye.
For this gene survey, we used the NEIGHBORHOOD dataset to assess common variation, and two of the NEIGHBORHOOD cohorts (NEIGHBOR and MEE) with exome array data to assess rare coding variation. A strength of our study is that the cases and controls have had clinical examinations. While the sample size is relatively large, it is also potentially limiting for very rare variants and for common variants with small effects. For replication, we used two additional independent datasets: FinnGen, a large population dataset with both common and rare variant association results for a range of glaucoma phenotypes, and the UK Biobank, also a population dataset with glaucoma association results for rare coding variants derived from whole exome sequence data. A limitation of both the FinnGen and the UK Biobank datasets is that glaucoma classification is based on diagnosis codes, and for the UK Biobank also self-report. This may create some variability in glaucoma diagnosis. Another limitation of our analysis is that we used exome array data for assessing rare coding variants. Because the exon array is comprised of selected rare coding variants, it is not equivalent to the full set of variants that can be identified by exome sequencing. Further evaluation of the genes of interest identified in this study using whole exome or whole genome sequencing would be of interest. It would also be valuable to include ethnically diverse populations in future studies.
Conclusion
We surveyed 4 large mechanosensitive ion channel families—K2P, TMEM63, PIEZO and TRP—to discover associations between genetic variants in these genes and POAG3,5,6,24. We find novel variants in several mechanosensitive ion channel genes that show interesting associations with POAG. Future functional studies are warranted to determine the effects of these variants on channel function, as well as on glaucoma phenotypes.
Methods
NEIGHBORHOOD study participants
NEIGHBORHOOD case and control recruitment and definitions have been reported previously58,59,60. Briefly, the NEIGHBORHOOD dataset includes eight independent datasets with a total of 3,853 cases and 33,480 controls. A harmonized POAG definition was adopted across these data sets based on the following criteria: (1) open anterior segment angles, (2) reproducible glaucomatous visual field loss on reliable tests or (3) an eye with cup-disc ratio of at least 0.7 with one visual field showing glaucomatous loss, and (4) no identifiable secondary cause for optic nerve disease. Elevated intraocular pressure (IOP) was not a criterion for POAG definition, but if present, there had to be no secondary causes on anterior segment examination.
Sixty-seven percent of cases had a history of elevated IOP (≥ 22 mm Hg) measured in a clinical setting and were classified as HTG (high-tension glaucoma). Cases with IOP < 22 mm Hg (without treatment) measured in the clinic at the time of study enrollment were classified as NTG (normal-tension glaucoma). Cases undergoing IOP-lowering therapy at the time of enrollment were included in the HTG group if they had a documented history of IOP > 22 mm Hg prior to treatment, and cases undergoing IOP-lowering therapy at the time of enrollment were included in the NTG group if they did not have recorded pressures > 22 mm Hg before treatment. Pretreatment IOP measurements were not available for all cases.
NEIGHBORHOOD GWAS data
Association data for SNPs located within genomic regions that contain the selected mechanosensitive channel genes was extracted from the NEIGHBORHOOD GWAS data58. For the GWAS case and control samples were genotyped on Illumina 660W (Illumina, San Diego, CA, USA), Affymetrix 500 K, Affymetrix Mapping 5.0, or Affymetrix 6.0 arrays (Affymetrix, Santa Clara, CA, USA). The genotype imputation was based on 1000 Genomes panel (March 2012). For each dataset, age, sex, and study-specific principal components were adjusted in logistic regression models using ProbABEL61. In the meta-analysis, inverse variance weighted method was performed in METAL (2011-03-25 release) with genomic control correction62. Association analyses were done for POAG overall as well as the HTG and NTG subgroups58.
To examine the NEIGHBORHOOD association results for genomic regions that include the mechanosensitive genes of interest, we extracted data for all SNPs located within the genomic region that included the gene as well as 50 Kb on both the 5’ and 3’ ends of the gene of interest. We did not include TRPC5 because it is located on the X chromosome and the X chromosome was not included in the NEIGHBORHOOD GWAS analysis. The SNP with the smallest P value for association was selected as representative of the overall result for each genomic region. We used a p value correction for the number of genomic regions examined to determine statistical significance (P = 0.05/20 = 0.0025).
The Gene-Tissue Expression (GTEx) database (https://www.gtexportal.org/home/) was used to identify SNPs that are expression quantitative trait loci (eQTLs) and hence may influence expression levels of the genes of interest.
Exome array genotyping and association analysis
2606 cases and 2606 controls from the NEIGHBORHOOD NEIGHBOR and MEE cohorts were submitted to the Center for Inherited Disease Research (CIDR) for genotyping using the Illumina HumanExome BeadChip (Illumina, Inc., San Diego, CA). Illumina Genome Studio (Illumina, Inc.) and PLINK63 was used for all QC steps, except where noted. Basic QC for samples included screens for call rate (≥ 98.5%) and high (≥ 95%) concordance with previous Illumina 660 K Beadchip genotypes59 where available (about 80% of samples). Recorded sex in the clinical records was verified with genotyped sex by two criteria: mean fluorescence intensity on the X and Y chromosomes, plus genotype heterozygosity on the X chromosome and call rate on the Y, allowing male and female samples to have heterozygous X-linked and successful Y-linked genotypes, respectively. Samples were tested for pairwise relationships and unexpected duplication using KING64, testing overall genomic sharing by robust kinship estimation, and proportion shared 0, 1 and 2 alleles identical by descent under the assumption of population homogeneity.
European ancestry was verified from the first two principal components derived from genotypes at 9000 ancestry-informative markers by means of the SNPweights program65, including representative HapMap CEU, YRI, CHB and JPT samples as reference populations. A principal components analysis was conducted using over 52,040 independent (pairwise r2 < 0.1), common (MAF ≥ 0.005) SNPs and the smartpca program in EIGENSOFT to detect finer population structure. Of the first 20 principal components, the first, sixth and eighth were significantly associated (P < 0.05 by logistic regression) with POAG status.
Initial QC screens for markers included call rate (≥ 98%) and consistency with Hardy–Weinberg proportions (P > 10−6 by Fisher exact test). Genotype clustering was confirmed for rare (MAF < 0.02) variants using genotype calls from zCall66 run with a stringent Z-score threshold of Z = 21 for calling heterozygous genotypes, from a GenomeStudio report containing genotype calls and X and Y intensity values as input. Every rare SNP with two or more additional heterozygous calls by zCall than by GenCall was reviewed in Genome Studio, and, if necessary, cluster locations were adjusted manually.
Association between single variants and POAG case/control status was tested by logistic regression, including age at exam, sex, and three principal components observed to be significantly associated with POAG as covariates. For the single-variant association test we selected SNPs with minor allele frequencies (MAF) less than 1% and CADD (Combined Annotation Disruption Depletion) scores of > 15. A CADD score of 15 indicates that the variant is in the top 5% of deleterious alleles within the human genome67. Gene-based association tests were done using the optimized kernel association test implemented in SKAT-O68 for genes with at least 3 variants identified in the exome array data. Covariate adjustments were the same as for the single-variant analysis. We considered the rare variant association exploratory and identified variants of interest as those with P-values for association equal to or less than 0.05.
GWAS and exome array association results were compared to association analyses completed using exome data from the UK Biobank69 (https://app.genebass.org/) and association analyses using genome-wide genotypes from FinnGen (https://r8.finngen.fi/).
Splice AI (https://spliceailookup.broadinstitute.org/) was used to predict splicing alteration as a consequence of PIEZO1 canonical donor splice variant (16-88737727-C-G).
Data availability
The summary statistics for the NEIGHBORHOOD genotype data are available at http://eaglep.case.edu/glaucomagenetics_web/ and also through reasonable request. All variants analyzed in this report are included in the manuscript either in tables or as supplemental data.
References
Weinreb, R. N., Aung, T. & Medeiros, F. A. The pathophysiology and treatment of glaucoma: A review. JAMA 311, 1901–1911. https://doi.org/10.1001/jama.2014.3192 (2014).
Kefauver, J. M., Ward, A. B. & Patapoutian, A. Discoveries in structure and physiology of mechanically activated ion channels. Nature 587, 567–576. https://doi.org/10.1038/s41586-020-2933-1 (2020).
Coste, B. et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60. https://doi.org/10.1126/science.1193270 (2010).
Murthy, S. E., Dubin, A. E. & Patapoutian, A. Piezos thrive under pressure: Mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell. Biol. 18, 771–783. https://doi.org/10.1038/nrm.2017.92 (2017).
Brohawn, S. G., Su, Z. & MacKinnon, R. Mechanosensitivity is mediated directly by the lipid membrane in TRAAK and TREK1 K+ channels. Proc. Natl. Acad. Sci. U. S. A. 111, 3614–3619. https://doi.org/10.1073/pnas.1320768111 (2014).
Murthy, S. E. et al. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 7, e41844. https://doi.org/10.7554/eLife.41844 (2018).
Eijkelkamp, N., Quick, K. & Wood, J. N. Transient receptor potential channels and mechanosensation. Annu. Rev. Neurosci. 36, 519–546. https://doi.org/10.1146/annurev-neuro-062012-170412 (2013).
Ciura, S. & Bourque, C. W. Transient receptor potential vanilloid 1 is required for intrinsic osmoreception in organum vasculosum lamina terminalis neurons and for normal thirst responses to systemic hyperosmolality. J. Neurosci. 26, 9069–9075. https://doi.org/10.1523/jneurosci.0877-06.2006 (2006).
Sharif Naeini, R., Witty, M. F., Séguéla, P. & Bourque, C. W. An N-terminal variant of Trpv1 channel is required for osmosensory transduction. Nat. Neurosci. 9, 93–98. https://doi.org/10.1038/nn1614 (2006).
Muraki, K. et al. TRPV2 is a component of osmotically sensitive cation channels in murine aortic myocytes. Circ. Res. 93, 829–838. https://doi.org/10.1161/01.Res.0000097263.10220.0c (2003).
Wu, L., Gao, X., Brown, R. C., Heller, S. & O’Neil, R. G. Dual role of the TRPV4 channel as a sensor of flow and osmolality in renal epithelial cells. Am. J. Physiol. Renal Physiol. 293, F1699-1713. https://doi.org/10.1152/ajprenal.00462.2006 (2007).
Chen, J. & Barritt, G. J. Evidence that TRPC1 (transient receptor potential canonical 1) forms a Ca(2+)-permeable channel linked to the regulation of cell volume in liver cells obtained using small interfering RNA targeted against TRPC1. Biochem. J. 373, 327–336. https://doi.org/10.1042/bj20021904 (2003).
Maroto, R. et al. TRPC1 forms the stretch-activated cation channel in vertebrate cells. Nat. Cell. Biol. 7, 179–185. https://doi.org/10.1038/ncb1218 (2005).
Quick, K. et al. TRPC3 and TRPC6 are essential for normal mechanotransduction in subsets of sensory neurons and cochlear hair cells. Open Biol. 2, 120068. https://doi.org/10.1098/rsob.120068 (2012).
Gomis, A., Soriano, S., Belmonte, C. & Viana, F. Hypoosmotic- and pressure-induced membrane stretch activate TRPC5 channels. J. Physiol. 586, 5633–5649. https://doi.org/10.1113/jphysiol.2008.161257 (2008).
Inoue, R. et al. Synergistic activation of vascular TRPC6 channel by receptor and mechanical stimulation via phospholipase C/diacylglycerol and phospholipase A2/omega-hydroxylase/20-HETE pathways. Circ. Res. 104, 1399–1409. https://doi.org/10.1161/circresaha.108.193227 (2009).
Spassova, M. A., Hewavitharana, T., Xu, W., Soboloff, J. & Gill, D. L. A common mechanism underlies stretch activation and receptor activation of TRPC6 channels. Proc. Natl. Acad. Sci. U. S. A. 103, 16586–16591. https://doi.org/10.1073/pnas.0606894103 (2006).
Zhang, X. F., Chen, J., Faltynek, C. R., Moreland, R. B. & Neelands, T. R. Transient receptor potential A1 mediates an osmotically activated ion channel. Eur. J. Neurosci. 27, 605–611. https://doi.org/10.1111/j.1460-9568.2008.06030.x (2008).
Kwan, K. Y., Glazer, J. M., Corey, D. P., Rice, F. L. & Stucky, C. L. TRPA1 modulates mechanotransduction in cutaneous sensory neurons. J. Neurosci. 29, 4808–4819. https://doi.org/10.1523/jneurosci.5380-08.2009 (2009).
Nauli, S. M. et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137. https://doi.org/10.1038/ng1076 (2003).
Grimm, C., Kraft, R., Sauerbruch, S., Schultz, G. & Harteneck, C. Molecular and functional characterization of the melastatin-related cation channel TRPM3. J. Biol. Chem. 278, 21493–21501. https://doi.org/10.1074/jbc.M300945200 (2003).
Morita, H. et al. Membrane stretch-induced activation of a TRPM4-like nonselective cation channel in cerebral artery myocytes. J. Pharmacol. Sci. 103, 417–426. https://doi.org/10.1254/jphs.fp0061332 (2007).
Numata, T., Shimizu, T. & Okada, Y. Direct mechano-stress sensitivity of TRPM7 channel. Cell. Physiol. Biochem. 19, 1–8. https://doi.org/10.1159/000099187 (2007).
Yue, L. & Xu, H. TRP channels in health and disease at a glance. J. Cell. Sci. 134, jcs258372. https://doi.org/10.1242/jcs.258372 (2021).
Yan, H. et al. Heterozygous variants in the mechanosensitive ion channel TMEM63A result in transient hypomyelination during infancy. Am. J. Hum. Genet. 105, 996–1004. https://doi.org/10.1016/j.ajhg.2019.09.011 (2019).
Du, H. et al. The cation channel TMEM63B Is an osmosensor required for hearing. Cell Rep. 31, 107596. https://doi.org/10.1016/j.celrep.2020.107596 (2020).
Lee, L. M., Müntefering, T., Budde, T., Meuth, S. G. & Ruck, T. Pathophysiological role of K(2P) channels in human diseases. Cell. Physiol. Biochem. 55, 65–86. https://doi.org/10.33594/000000338 (2021).
Ranade, S. S., Syeda, R. & Patapoutian, A. Mechanically activated ion channels. Neuron 87, 1162–1179. https://doi.org/10.1016/j.neuron.2015.08.032 (2015).
Carreon, T. A., Castellanos, A., Gasull, X. & Bhattacharya, S. K. Interaction of cochlin and mechanosensitive channel TREK-1 in trabecular meshwork cells influences the regulation of intraocular pressure. Sci. Rep. 7, 452. https://doi.org/10.1038/s41598-017-00430-2 (2017).
Yarishkin, O. et al. TREK-1 channels regulate pressure sensitivity and calcium signaling in trabecular meshwork cells. J. Gen. Physiol. 150, 1660–1675. https://doi.org/10.1085/jgp.201812179 (2018).
Ryskamp, D. A. et al. TRPV4 regulates calcium homeostasis, cytoskeletal remodeling, conventional outflow and intraocular pressure in the mammalian eye. Sci. Rep. 6, 30583. https://doi.org/10.1038/srep30583 (2016).
Yarishkin, O. et al. Piezo1 channels mediate trabecular meshwork mechanotransduction and promote aqueous fluid outflow. J. Physiol. 599, 571–592. https://doi.org/10.1113/jp281011 (2021).
Uchida, T. et al. Mechanical stretch induces Ca(2+) influx and extracellular release of PGE(2) through Piezo1 activation in trabecular meshwork cells. Sci. Rep. 11, 4044. https://doi.org/10.1038/s41598-021-83713-z (2021).
Morozumi, W. et al. Piezo 1 is involved in intraocular pressure regulation. J. Pharmacol. Sci. 147, 211–221. https://doi.org/10.1016/j.jphs.2021.06.005 (2021).
Zhu, W. et al. The role of Piezo1 in conventional aqueous humor outflow dynamics. iScience 24, 102042. https://doi.org/10.1016/j.isci.2021.102042 (2021).
Fang, J. et al. Piezo2 downregulation via the Cre-lox system affects aqueous humor dynamics in mice. Mol. Vis. 27, 354–364 (2021).
Patel, P. D. et al. Impaired TRPV4-eNOS signaling in trabecular meshwork elevates intraocular pressure in glaucoma. Proc. Natl. Acad. Sci. U. S. A. https://doi.org/10.1073/pnas.2022461118 (2021).
Weitlauf, C. et al. Short-term increases in transient receptor potential vanilloid-1 mediate stress-induced enhancement of neuronal excitation. J. Neurosci. 34, 15369–15381. https://doi.org/10.1523/jneurosci.3424-14.2014 (2014).
Sappington, R. M. & Calkins, D. J. Contribution of TRPV1 to microglia-derived IL-6 and NFkappaB translocation with elevated hydrostatic pressure. Invest. Ophthalmol. Vis. Sci. 49, 3004–3017. https://doi.org/10.1167/iovs.07-1355 (2008).
Ryskamp, D. A. et al. The polymodal ion channel transient receptor potential vanilloid 4 modulates calcium flux, spiking rate, and apoptosis of mouse retinal ganglion cells. J. Neurosci. 31, 7089–7101. https://doi.org/10.1523/JNEUROSCI.0359-11.2011 (2011).
Taylor, L., Arnér, K. & Ghosh, F. Specific inhibition of TRPV4 enhances retinal ganglion cell survival in adult porcine retinal explants. Exp. Eye Res. 154, 10–21. https://doi.org/10.1016/j.exer.2016.11.002 (2017).
van Zyl, T. et al. Cell atlas of the human ocular anterior segment: Tissue-specific and shared cell types. Proc. Natl. Acad. Sci. U. S. A. 119, e2200914119. https://doi.org/10.1073/pnas.2200914119 (2022).
Yan, W. et al. Cell Atlas of the human fovea and peripheral retina. Sci. Rep. 10, 9802. https://doi.org/10.1038/s41598-020-66092-9 (2020).
Baxter, S. L. et al. Investigation of associations between Piezo1 mechanoreceptor gain-of-function variants and glaucoma-related phenotypes in humans and mice. Sci. Rep. 10, 19013. https://doi.org/10.1038/s41598-020-76026-0 (2020).
Thomson, B. R. et al. Angiopoietin-1 is required for Schlemm’s canal development in mice and humans. J. Clin. Invest. 127, 4421–4436. https://doi.org/10.1172/jci95545 (2017).
Choi, H. J., Sun, D. & Jakobs, T. C. Astrocytes in the optic nerve head express putative mechanosensitive channels. Mol. Vis. 21, 749–766 (2015).
Liu, J., Yang, Y. & Liu, Y. Piezo1 plays a role in optic nerve head astrocyte reactivity. Exp. Eye Res. 204, 108445. https://doi.org/10.1016/j.exer.2021.108445 (2021).
Ma, L. et al. The roles of transient receptor potential ion channels in pathologies of glaucoma. Front. Physiol. 13, 806786. https://doi.org/10.3389/fphys.2022.806786 (2022).
McGahon, M. K. et al. TRPV2 channels contribute to stretch-activated cation currents and myogenic constriction in retinal arterioles. Invest. Ophthalmol. Vis. Sci. 57, 5637–5647. https://doi.org/10.1167/iovs.16-20279 (2016).
Deo, M., Yu, J. Y., Chung, K. H., Tippens, M. & Turner, D. L. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 235, 2538–2548. https://doi.org/10.1002/dvdy.20847 (2006).
Gilliam, J. C. & Wensel, T. G. TRP channel gene expression in the mouse retina. Vis. Res. 51, 2440–2452. https://doi.org/10.1016/j.visres.2011.10.009 (2011).
Bennett, T. M., Mackay, D. S., Siegfried, C. J. & Shiels, A. Mutation of the melastatin-related cation channel, TRPM3, underlies inherited cataract and glaucoma. PLoS ONE 9, e104000. https://doi.org/10.1371/journal.pone.0104000 (2014).
de Souza Monteiro Araújo, D. et al. TRPA1 mediates damage of the retina induced by ischemia and reperfusion in mice. Cell Death Dis. 11, 633. https://doi.org/10.1038/s41419-020-02863-6 (2020).
Jo, A. O. et al. Differential volume regulation and calcium signaling in two ciliary body cell types is subserved by TRPV4 channels. Proc. Natl. Acad. Sci. U. S. A. 113, 3885–3890. https://doi.org/10.1073/pnas.1515895113 (2016).
Uchida, T. et al. TRPV4 is activated by mechanical stimulation to induce prostaglandins release in trabecular meshwork, lowering intraocular pressure. PLoS ONE 16, e0258911. https://doi.org/10.1371/journal.pone.0258911 (2021).
Nikolaev, Y. A. et al. Mammalian TRP ion channels are insensitive to membrane stretch. J. Cell. Sci. https://doi.org/10.1242/jcs.238360 (2019).
Kaneko, Y. & Szallasi, A. Transient receptor potential (TRP) channels: A clinical perspective. Br. J. Pharmacol. 171, 2474–2507. https://doi.org/10.1111/bph.12414 (2014).
Bailey, J. N. et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat. Genet. 48, 189–194. https://doi.org/10.1038/ng.3482 (2016).
Wiggs, J. L. et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 8, e1002654. https://doi.org/10.1371/journal.pgen.1002654 (2012).
Wiggs, J. L. et al. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: Rationale, study design, and clinical variables. J. Glaucoma 22, 517–525. https://doi.org/10.1097/IJG.0b013e31824d4fd8 (2013).
Aulchenko, Y. S., Struchalin, M. V. & van Duijn, C. M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinform. 11, 134. https://doi.org/10.1186/1471-2105-11-134 (2010).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191. https://doi.org/10.1093/bioinformatics/btq340 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. https://doi.org/10.1086/519795 (2007).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873. https://doi.org/10.1093/bioinformatics/btq559 (2010).
Chen, C. Y. et al. Improved ancestry inference using weights from external reference panels. Bioinformatics 29, 1399–1406. https://doi.org/10.1093/bioinformatics/btt144 (2013).
Goldstein, J. I. et al. zCall: A rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics (Oxford, England) 28, 2543–2545. https://doi.org/10.1093/bioinformatics/bts479 (2012).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315. https://doi.org/10.1038/ng.2892 (2014).
Lee, S. et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 91, 224–237. https://doi.org/10.1016/j.ajhg.2012.06.007 (2012).
Karczewski, K. J. et al. Systematic single-variant and gene-based association testing of thousands of phenotypes in 394,841 UK Biobank exomes. Cell Genomics 2, 100168. https://doi.org/10.1016/j.xgen.2022.100168 (2022).
Acknowledgements
We thank Wenjun Yan for help with extracting single cell and single nuclear RNA sequencing data. Funding was provided by NIH KL2TR003143 (WWL), NIH NEI K08 EY034600 (WWL), American Glaucoma Society (WWL), E. Matilda Ziegler Foundation for the Blind (WWL), Research to Prevent Blindness Career Development Award (WWL), Glaucoma Research Foundation (WWL), NIH NEI P30 Vision Research Core EY026877 (Stanford Ophthalmology), an unrestricted grant from Research to Prevent Blindness (Stanford Ophthalmology), NIH NEI R01 EY022305 (JLW), NIH NEI P30 EY014104 (JLW).
Author information
Authors and Affiliations
Consortia
Contributions
Conception: W.W.L. Design: W.W.L. and J.L.W. Analysis: T.G.K., J.N.C.B., Z.X., P.H., and J.L.W. Writing: W.W.L. and J.L.W. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, W.W., Kinzy, T.G., Cooke Bailey, J.N. et al. Mechanosensitive ion channel gene survey suggests potential roles in primary open angle glaucoma. Sci Rep 13, 15871 (2023). https://doi.org/10.1038/s41598-023-43072-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-43072-3
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.