Abstract
For many machine learning applications in drug discovery, only limited amounts of training data are available. This typically applies to compound design and activity prediction and often restricts machine learning, especially deep learning. For low-data applications, specialized learning strategies can be considered to limit required training data. Among these is meta-learning that attempts to enable learning in low-data regimes by combining outputs of different models and utilizing meta-data from these predictions. However, in drug discovery settings, meta-learning is still in its infancy. In this study, we have explored meta-learning for the prediction of potent compounds via generative design using transformer models. For different activity classes, meta-learning models were derived to predict highly potent compounds from weakly potent templates in the presence of varying amounts of fine-tuning data and compared to other transformers developed for this task. Meta-learning consistently led to statistically significant improvements in model performance, in particular, when fine-tuning data were limited. Moreover, meta-learning models generated target compounds with higher potency and larger potency differences between templates and targets than other transformers, indicating their potential for low-data compound design.
Similar content being viewed by others
Introduction
Predicting new active compounds is one of the major tasks in computer-aided drug discovery, for which machine learning approaches have been widely applied over the past two decades1,2. In recent years, deep learning has also been increasingly applied for compound activity and property predictions1,2. The prediction of compounds exhibiting a desired biological activity (that is, activity against a target of interest) is mostly attempted using machine learning models for binary classification (that is, a compound is predicted to have or not to have a specific activity)3,4,5. For this purpose, models for class label prediction (active versus inactive compounds) are typically derived based on training sets of known specifically active compounds and randomly selected compounds assumed to be inactive. These qualitative activity predictions mostly involve virtual screening of compound databases to identify new hits. In addition to qualitative predictions of biological activity, predicting compounds that are highly potent against a given target also is of interest. Compound potency prediction can be quantitative or semi-quantitative in nature. Quantitative predictions aim to specify numerical potency values using, for example, quantitative structure–activity relationship (QSAR)6,7 or free energy methods8,9. Different from qualitative predictions and virtual screening, quantitative potency predictions are usually carried out for small compound sets or structural analogues from lead series. Furthermore, semi-quantitative approaches aim to predict new potent compounds, that is, compounds having higher potency than known actives. For example, such predictions might focus on activity cliffs10, which are defined as pairs of structurally similar compounds or structural analogues with large potency differences10. Prediction of activity cliffs fall outside the applicability domain of standard QSAR methods4.
While quantitative potency predictions are widely carried out, they are difficult to evaluate in benchmark settings. It has been observed that benchmark predictions of different machine learning models and randomized predictions are typically only separated by small error margins11, which makes it difficult to non-ambiguously assess relative method performance11. Therefore, we currently prefer semi-quantitative approaches focusing on the prediction of potent compounds (rather than trying to predict compound potency values across wide potency ranges). Semi-quantitative predictions can be attempted by deep generative modeling2. For example, transformer models have been derived based on pairs of active structural analogues with varying potency to predict activity cliffs and design potent compounds12,13. Therefore, the transformer models were conditioned on observed potency differences. This generative design approach successfully reproduced highly potent compounds for different activity classes based on weakly potent input compounds13. Transformer models have also been derived for other compound property predictions14,15,16 and generative compound design applications17,18,19 as well as for the prediction of drug-target interactions20,21,22.
Notably, all compound activity and potency predictions depend on available data for learning. Like many other data in early-phase drug discovery, high-quality compound potency measurements for given targets are generally sparse, which limits generative design. Therefore, we are considering machine learning approaches for low-data regimes to enable predictions of potent compounds for targets, for which only little compound data is available. Among learning strategies for sparsely distributed data, active learning23,24 and transfer learning25,26 have been investigated for machine learning in drug discovery in various studies24,26. Transfer learning attempts to use information obtained from related prediction tasks to streamline model derivation for such tasks, while active learning focuses on the selection of most informative training instances for iterative model building. Meta-learning including few-shot learning represents another low-data approach that is relevant for drug discovery27,28,29,30. In artificial intelligence, meta-learning is a sub-discipline of machine learning27. It aims to combine the output of different machine learning models and/or meta-data from these models such as parameters derived from training instances to generate models for other prediction tasks27. Alternatively, the same algorithm might be applied to generate models for individual prediction tasks whose outputs are then used to iteratively update a meta-learning model. Hence, meta-learning can also be regarded as a form of ensemble learning. The general aim of meta-learning is achieving transferability of models to related prediction tasks, including the application of prior model knowledge to limit the number of training instances required for new tasks. Given the use of meta-data for learning, the approach is well-suited for parameter-rich deep learning architectures28 and -compared to transfer learning- principally applicable to a wider spectrum of predictions tasks. However, in compound design and property prediction, the exploration of meta-learning is still in its early stages. Therefore, we have explored meta-learning in semi-quantitative potency predictions. To this end, we have adapted a transformer architecture designed for the prediction of potent compounds13 as a base model for deriving meta-learning models and assessed the potential of meta-learning for predicting highly potent compounds for different activity classes and varying amounts of training data.
Methods
Compounds, activity data, and analogue series
Bioactive compounds with high-confidence activity data were collected from ChEMBL (release 29)31. Only compounds with direct interactions (assay relationship type: "D") with human targets at the highest assay confidence level (assay confidence score 9) were considered. In addition, potency measurements were restricted to numerically specified equilibrium constants (Ki values), which were recorded as (negative decadic logarithmic) pKi values. When multiple measurements were available for the same compound, the geometric mean was calculated as the final potency annotation, provided all values fell within the same order of magnitude. If not, the compound was disregarded. Qualifying compounds were organized into target-based activity classes.
In activity classes, analogue series (AS) with one to five substitution sites were identified using the compound-core relationship (CCR) algorithm32. The core structure of an AS was required to consist of at least twice the number of non-hydrogen atoms as the combined substituents. For each AS, all possible pairs of analogues were generated, termed All_CCR pairs. For each activity class, ALL_CCR pairs from all AS were pooled. All_CCR pairs were then divided into CCR pairs with a potency difference of less than 100-fold and activity cliff (AC)-CCR pairs with a potency difference of at least 100-fold.
On the basis of the specified data curation criteria and AS distributions, 10 activity classes were assembled that consisted of at least ~ 500 qualifying compounds and ~ 50 AS, as summarized in Table 1. These activity classes included ligands of various G protein coupled receptors and inhibitors of different enzymes. Figure 1 shows exemplary AC_CCR pairs for each class.
Analogue pairs representing activity cliffs. For each activity class, exemplary AC_CCR pairs are shown and their potency differences are reported. Numbers on arrows identify activity classes according to Table 1. Core structures and substituents are colored blue and red, respectively.
Meta-learning approach
The basic premise of meta-learning, as investigated herein, is parameterizing a model on a series of training tasks by combining and updating parameter settings across individual tasks. This process aims to improve the ability of the model to adapt to new prediction tasks through the use of meta-data.
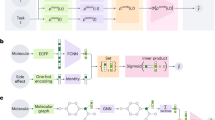
For designing the meta-learning module of Meta-CLM, we adopted the model-agnostic meta-learning (MAML) framework28 for an activity class-specific prediction task distribution p(T). Given its model-agnostic nature, the only assumption underlying the MAML approach is that a given model is parameterized using a parameter vector θ. Accordingly, a meta-learning model is considered as a function fθ with parameter vector θ. The model aims to learn parameter settings θmeta that are derived for individual training tasks and updated across different tasks such that they can be effectively adjusted to new prediction tasks. Therefore, for each of a series of prediction tasks, training data are randomly divided into a support set and a query set Accordingly, when the meta-learning module is applied to a new prediction task Ti such as an activity class the current parameter vector θmeta is updated for task Ti with activity class-specific parameters θi obtained by gradient descent optimization minimizing training errors.
During meta-training, as summarized in Fig. 2, the model fθ is first updated to a task-specific model fθ′ using its support set. Then, the corresponding query set is used to determine the prediction loss of model fθ′ for this task. The procedure is repeated for all prediction tasks (activity classes). Finally, model parameters are further adjusted for testing by minimizing the sum of the prediction loss over all activity classes. Model derivation based on the support sets and evaluation based on query sets are implemented as inner and outer loops, respectively. For meta-testing, the trained meta-learning module is fine-tuned on a specific activity class, for which parameters are adjusted, as also illustrated in Fig. 2. For each class, an individual fine-tuned model is generated.
Meta-learning. The illustration summarizes training, fine-tuning, and testing of the meta-learning module of Meta-CLM using exemplary AC-CCR pairs. For each activity class, the support set is used for the initial parameterization of the model (θ). The support loss \(\mathscr{L}\)support is calculated for updating model parameters (θ′). Then, the query set is used to calculate the prediction loss \({\mathscr{L}}\)′query for this task. The process is repeated for all training classes, followed by summation of \(\mathscr{L}\)′query over all tasks to further adjust the parameter settings. The trained module then enters the fine-tuning and testing phase. Solid and dashed lines indicate inner and outer loops, respectively, for meta-training and -testing including fine-tuning.
The meta-learning process aims to capture prior training information through initial parameter vector adjustments, followed by updates through monitoring of the joint loss across all training tasks29. Capturing prior training knowledge should enable the model to more effectively adapt to new prediction tasks based on advanced parameter settings available for initialization and shorter optimization paths with reduced training data requirments33,34.
This algorithmic approach differs from conventional multi-task learning where a single model is trained on multiple tasks, aiming to share representations and knowledge between these tasks to collectively improve the basis for learning. Hence, the primary goal of multi-task learning is to improve predictive performance for all tasks by leveraging commonalities between them. Accordingly, model weights are updated based on a combination of the losses from all tasks in a single optimization step. Shared representations for multiple tasks support the model’s ability to simultaneously learn features common to these tasks.
Transformer models
Base model
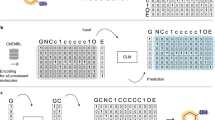
For meta-learning, the transformer architecture derived previously for the prediction of highly potent compounds based on weakly potent templates was adopted13. Figure 3 illustrates the architecture of the base CLM. The transformer consisted of multiple encoder-decoder modules with attention mechanism35 and was designed for translating string-based representations of chemical structure. Accordingly, the transformer can be perceived as a chemical language model (CLM). The base model (referred to as CLM in the following) was devised to predict compounds with higher potency for given input compounds13. An encoder module consisted of encoding sub-layers including a multi-head self-attention sub-layer and a fully connected feed-forward network sub-layer. The encoder compressed an input sequence into a context vector in its final hidden state, providing the input for the decoder module composed of a feed-forward sub-layer and two multi-head attention sub-layers. The decoder transformed the context vector into a sequence of tokens. Both the encoder and decoder modules utilized the attention mechanism during training to effectively learn from the underlying feature space.
During training, the CLM was challenged to learn mappings of template/source compounds (SCs) to target compounds (TCs) conditioned on potency differences (ΔPot) resulting from replacements of substituent(s):
Hence, training focused on structural analogues with specific potency differences. Then, given a new (SC, ΔPot) test instance, the model generated a set of structurally related TCs with putatively higher potency than SCs.
For transformer modeling, compounds and potency differences must be tokenized. Accordingly, compounds were represented as molecular-input line-entry system (SMILES) strings36 generated using RDKit37. Tokenization was facilitated by representing atoms with single-character tokens (e.g., "C"or "N"), two-character tokens (e.g., "Cl" or "Br"), or tokens enclosed in brackets (e.g. "[nH]" or "[O-]"). Potency differences were subjected to binning tokenization12,13,38,39 by dividing the global range of potency differences (-6.62 to 6.52 pKi units) into 1314 bins with a constant width of 0.01. Each bin was encoded by a single token and each potency difference was assigned to the corresponding token12,13. In addition, two special "start" and "end" tokens were defined as the start and end points of a sequence, respectively.
The model was pre-trained using a large set of 881,990 All_CCR pairs originating from 496 public activity classes13. For pre-training, All_CCR triples (CpdA, CpdB, PotB-PotA) were generated in in which CpdA and CpdB represented the SC and TC, respectively, and (PotB-PotA) their potency difference.
CLM was implemented using Pytorch40. Default hyperparameter settings were used for the transformer architecture together with a batch size of 64, learning rate of 0.001, and encoding dimension of 256. During training, the transformer model minimized the cross-entropy loss between the ground-truth and output sequence. The Adam optimizer was used41. The model was trained for a maximum of 1000 epochs. At each epoch, a checkpoint was saved, and the final model was selected based on the minimal loss.
The base model achieved a reproducibility of 0.857 for the entire test set (corresponding to 10% of pre-training set). Hence, the base CLM model regenerated ~ 86% of the target compounds from CCR-triples not used for training.
Model for meta-learning
The CLM variant for meta-learning was also implemented using Pytorch following the protocol described above. The meta-learning model, designated Meta-CLM, consisted of two modules including the base model for generating mappings of SCs to TCs conditioned on potency differences and the meta-learning module (the design of which is detailed below). For derivation of the metal-learning module, a subset of 176 of the 496 activity classes was selected for which at least 300 All-CCR pairs per class were available, amounting to a total of 491,688 qualifying All_CCR triples. For meta-learning, each activity class was considered a separate training task (see below). Therefore, All_CCR triples from each class were randomly split into support set (80%) and query set (20%). The Adam optimizer was used for gradient descent optimization during meta-learning.
Model fine-tuning
For fine-tuning and comparative evaluation of CLM and Meta-CLM, the 10 activity classes in Table 1 were used. Fine-tuning was separately carried out using AC-CCR pairs from each class. The AC-CCR pairs from each class were randomly divided into fine-tuning (80%) and test instances (20%). In each case, it was confirmed that the fine-tuning and test pairs had no core structure overlap (otherwise, a new partition was generated). For fine-tuning, AC_CCR pairs were exclusively used. AC_CCR triples were ordered such that TC was the highly potent compound. To assess the ability of CLM and Meta-CLM to learn in low-data regimes, model variants were derived based on 10%, 25%, 50% and 100% of the training data. To adapt to differently sized training sets, the pre-trained model was fine-tuned with a smaller learning rate of 0.0001. With a maximum of 200 training epochs, the final fine-tuned model was selected based on minimal cross-entropy loss.
Model evaluation
For each activity class, CCR pairs sharing core structures with the fine-tuning set were excluded, then the final test set was generated by adding the remaining CCR pairs to test AC-CCR pairs. Test set CCR and AC-CCR pairs yielded class-dependent numbers of unique CCR and AC-CCR test compounds. To evaluate the performance of each fine-tuned CLM and corresponding Meta-CLM, test compounds were divided into two categories: SCs with a maximum potency of 1 μM (corresponding to a pKi value of 6) and TCs with a potency greater than 1 μMol (pKi > 6). These test TCs were termed known target compounds (KTCs), which represented highly potent test compounds. Table 2 reports the test composition for each activity class. Depending on the activity class, 139 to 3838 KTCs were available.
For each test set SC, 50 hypothetical TCs were sampled and compared to available KTCs. The ability of a model to reproduce KTCs was considered as the key criterion for model validation.
Results
Reproducibility of known target compounds
We first analyzed the ability of Meta-CLM to reproduce KTCs in comparison to CLM. The results are reported in Table 3. For all activity classes, Meta-CLM and CLM correctly reproduced multiple KTCs over all fine-tuning conditions, thus providing non-ambiguous proof for the models’ ability to predict potent compounds. From correctly predicted SC-KTC pairs, unique KTCs were extracted (a given KTC can occur in multiple pairs). The number of correctly predicted SC-KTC pairs and unique KTCs varied depending on the activity class. Importantly, Meta-CLM consistently predicted more SC-KTC pairs and unique KTCs than CLM across all activity classes, without an exception. For Meta-CLM, the number of SC-KTC pairs varied from 71 to 5102 pairs when utilizing 100% of the training samples and the number of unique KTCs varied from 27 to 287, corresponding to a reproducibility ratio of ~ 7% to ~ 45% of available KTCs per class. For comparison, CLM, the base model, generated from 53 to 4385 SC-KTC pairs, with 23 to 241 unique KTCs and a corresponding reproducibility ratio of ~ 5% to ~ 36% per class. Moreover, for decreasing numbers of fine-tuning samples, Meta-CLM consistently reproduced more KTCs than CLM. For complete fine-tuning sets, Meta-CLM and CLM reached mean reproducibility rates of ~ 21% and ~ 14%, respectively. For only 10% of the fine-tuning samples, Meta-CLM reached a mean reproducibility rate of ~ 15% compared to only ~ 7% for CLM. Thus, Meta-CLM learned more effectively from sparse data than CLM, consistent with the aims of meta-learning.
Figure 4 illustrates the differences in KTC reproducibility rates between Meta-CLM and CLM. Independent-samples t-tests were carried out to assess the statistical significance of the observed differences. For complete fine-tuning sets, increases in reproducibility detected for Meta-CLM were statistically significant for three of 10 activity classes. However, for fine-tuning sets of deceasing size, 25 of 30 increases across all activity classes were statistically significant, thus providing further evidence for the ability of Meta-CLM to more effectively learn from sparse data. For most classes, there was a sharp decline in CLM reproducibility rates when 25% or 10% of the fine-tuning samples were used.
Reproducibility of known target compounds. For each activity class, the proportion of correctly reproduced KTCs is reported for Meta-CLM and CLM over varying percentages of fine-tuning samples. Mean and standard deviations (error bars) are provided. To assess the statistical significance of observed differences between reproducibility rates, independent-samples t tests were conducted: 0.05 < p ≤ 1.00 (ns), 0.01 < p ≤ 0.05 (*), 0.001 < p ≤ 0.01 (**), 0.0001 < p ≤ 0.001 (***), p ≤ 0.0001 (****). Stars denote increasing levels of statistical significance and “ns” stands for “not significant”.
We also note that both models produced large numbers of novel candidate compounds for SCs. For complete fine-tuning sets, Meta-CLM and CLM generated on average 2375 and 2818 new candidate compounds per activity class (ranging from 119 to 9952 and 234 to 10,779 candidates, respectively). While these new compounds cannot be considered for model validation, they provide large pools of candidates for practical applications in the search for potent compounds.
Compound potency
In addition to reproducing KTCs, the actual potency level of correctly predicted KTCs and potency differences between SCs and corresponding KTCs represented other highly relevant criteria for model assessment. According to our semi-quantitative design approach, ideally, the models should predict highly potent compounds from given SCs. Therefore, we next analyzed the potency of correctly predicted KTCs and potency differences between Meta-CLM and CLM.
Known target compounds
Figure 5 shows the distributions of logarithmic potency values of KTCs reproduced by Meta-CLM and CLM. Importantly, KTCs generated by Meta-CLM were overall consistently more potent than those generated by CLM across all activity classes and fine-tuning conditions. Thirty-eight of the total of 40 observed differences between the respective potency value distributions were statistically significant. Especially for 25% and 10% of the fine-tuning samples, Meta-CLM generated multiple KTCs with low-nanomolar or even sub-nanomolar potency for each activity class, whereas CLM only generated a few KTCs with potency higher than 10 nM (pKi > 8) for three classes.
Potency value distribution of reproduced known target compounds. For all activity classes, boxplots report the distributions of logarithmic potency values of KTCs correctly reproduced by Met-CLM and CLM over varying numbers of fine-tuning samples. To assess the statistical significance of differences between potency value distributions, independent-samples t tests were conducted: 0.05 < p ≤ 1.00 (ns), 0.01 < p ≤ 0.05 (*), 0.001 < p ≤ 0.01 (**), 0.0001 < p ≤ 0.001 (***), p ≤ 0.0001 (****).
Potency differences between source and target compounds
Furthermore, we analyzed potency differences captured by SC-KTC pairs. Following our design strategy, increasingly large potency differences between corresponding SCs and correctly reproduced KTCs were favored. Figure 6 shows the distribution of potency differences between corresponding SCs and KTCs for Meta-CLM and CLM predictions. In the case of Meta-CLM (CLM), four (six) activity classes displayed median potency differences between SCs and corresponding KTCs between one and two orders of magnitude (10- to100-fold) and the remaining six (four) classes displayed median potency differences exceeding two orders of magnitude (> 100-fold) for complete fine-tuning sets. Hence, significant potency differences were generally observed. For half of the activity classes, median potency differences were comparable for all fine-tuning conditions when separately viewed for Meta-CLM and CLM, respectively. However, when Meta-CLM and CLM were compared, potency differences of SC-KTC pairs were consistently larger for Meta-CLM. Again, 38 of 40 observed differences were statistically significant. Overall, many more KTCs with at least 1000-fold higher potency than the corresponding SCs were generated by Meta-CLM compared to CLM. Thus, Meta-CLM predicted KTCs with overall higher potency than CLM and much larger potency differences between SCs and KTCs.
Distribution of potency differences between source and known target compounds. For all activity classes, boxplots report the distributions of logarithmic potency differences for SC-KTC pairs predicted by Meta-CLM and CLM over varying numbers of fine-tuning samples. To assess the statistical significance of differences between the distributions, independent-samples t tests were conducted: 0.05 < p ≤ 1.00 (ns), 0.01 < p ≤ 0.05 (*), 0.001 < p ≤ 0.01 (**), 0.0001 < p ≤ 0.001 (***), p ≤ 0.0001 (****).
Conclusion
In this work, we have explored meta-learning for the prediction of potent compounds using conditional transformer models. Compound potency predictions are of high interest in drug discovery but high-quality activity data available for machine learning are typically sparse. For these predictions, meta-learning was of particular interest to us because the approach is well-suited for models that are rich in meta-data, yet currently only little explored for drug discovery applications. Therefore, we have adapted a previously investigated transformer architecture to construct a meta-learning model by adding a special meta-learning module to a pre-trained transformer. Then, meta-learning model variants were derived for different activity classes and their performance in the design of potent compounds was compared to reference transformers. For model validation, the ability to reproduce potent KTCs served as the major criterion. All models successfully reproduced KTCs. However, compared to reference models, meta-learning significantly increased the number of correctly predicted KTCs across all activity classes, especially for decreasing numbers of fine-tuning samples. This was an encouraging finding, consistent with expectations for successful meta-learning. Moreover, meta-learning models also produced target compounds with overall higher potency than other transformers and larger potency differences between templates and targets. These improvements were not anticipated but are highly attractive for practical applications. The generative models designed for predicting potent compounds produced large numbers of candidate compounds with novel structures. New candidate compounds predicted by the meta-learning models should represent an attractive resource for prospective applications in searching for potent compounds for targets of interest. Taken together, the results reported herein, provide proof-of-concept for the potential of meta-learning in generative design of potent compounds. Moreover, in light of our findings, we anticipate that meta-learning will also be a promising approach for other compound design applications in low-data regimes.
Data availability
Calculations were carried out using publicly available programs and compound data. Python scripts generated for the study and the activity classes used are available via the following link: https://uni-bonn.sciebo.de/s/kfAQZ0mbCGHtr0m.
References
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug. Discov. 18, 463–477 (2019).
Walters, W. P. & Barzilay, R. Applications of deep learning in molecule generation and molecular property prediction. Acc. Chem. Res. 54, 263–270 (2020).
Lo, Y. C., Rensi, S. E., Torng, W. & Altman, R. B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 23, 1538–1546 (2018).
Patel, L., Shukla, T., Huang, X., Ussery, D. W. & Wang, S. Machine learning methods in drug discovery. Molecules 25, 5277 (2020).
Rodríguez-Pérez, R., Miljković, F. & Bajorath, J. Machine learning in chemoinformatics and medicinal chemistry. Annu. Rev. Biomed. Data Sci. 5, 43–65 (2022).
Lewis, R. A. & Wood, D. Modern 2D QSAR for drug discovery. WIREs Comput. Mol. Sci. 4, 505–522 (2014).
Muratov, E. N. et al. QSAR without borders. Chem. Soc. Rev. 49, 3525–3564 (2020).
Mobley, D. L. & Gilson, M. K. Predicting binding free energies: Frontiers and benchmarks. Annu. Rev. Biophys. 46, 531–558 (2017).
Williams-Noonan, B. J., Yuriev, E. & Chalmers, D. K. Free energy methods in drug design: Prospects of “Alchemical perturbation” in medicinal chemistry. J. Med. Chem. 61, 638–649 (2018).
Stumpfe, D., Hu, H. & Bajorath, J. Evolving concept of activity cliffs. ACS Omega 4, 14360–14368 (2019).
Janela, T. & Bajorath, J. Simple nearest-neighbour analysis meets the accuracy of compound potency predictions using complex machine learning models. Nat. Mach. Intell. 4, 1246–1255 (2022).
Chen, H., Vogt, M. & Bajorath, J. DeepAC–conditional transformer-based chemical language model for the prediction of activity cliffs formed by bioactive compounds. Dig. Discov. 1, 898–909 (2022).
Chen, H. & Bajorath, J. Designing highly potent compounds using a chemical language model. Sci. Rep. 13, 7412 (2023).
Chen, D. et al. Algebraic graph-assisted bidirectional transformers for molecular property prediction. Nat. Commun. 12, 3521 (2021).
Song, Y., Chen, J., Wang, W., Chen, G. & Ma, Z. Double-head transformer neural network for molecular property prediction. J. Cheminform. 15, 27 (2023).
Jiang, Y. et al. Pharmacophoric-constrained heterogeneous graph transformer model for molecular property prediction. Commun. Chem. 6, 60 (2023).
Bagal, V., Aggarwal, R., Vinod, P. K. & Priyakumar, U. D. MolGPT: Molecular generation using a transformer-decoder model. J. Chem. Inf. Model. 62, 2064–2076 (2022).
Mazuz, E., Shtar, G., Shapira, B. & Rokach, L. Molecule generation using transformers and policy gradient reinforcement learning. Sci. Rep. 13, 8799 (2023).
Wang, Y., Zhao, H., Sciabola, S. & Wang, W. cMolGPT: a conditional generative pre-trained transformer for target-specific de novo molecular generation. Molecules 28, 4430 (2023).
Chen, L. et al. TransformerCPI: Improving compound–protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics 36, 4406–4414 (2020).
Huang, K., Xiao, C., Glass, L. M. & Sun, J. MolTrans: molecular interaction transformer for drug–target interaction prediction. Bioinformatics 37, 830–836 (2021).
Chen, L. et al. Sequence-based drug design as a concept in computational drug design. Nat. Commun. 14, 4217 (2023).
Warmuth, M. K. et al. Active learning with support vector machines in the drug discovery process. J. Chem. Inf. Comput. Sci. 43, 667–673 (2003).
Reker, D. & Schneider, G. Active-learning strategies in computer-assisted drug discovery. Drug Discov. Today 20, 458–465 (2015).
Weiss, K., Khoshgoftaar, T. M. & Wang, D. A survey of Transfer Learning. J. Big Data 3, 9 (2016).
Cai, C. et al. Transfer learning for drug discovery. J. Med. Chem. 63, 8683–8694 (2020).
Vilalta, R. & Drissi, Y. A Perspective View and Survey of Meta-Learning. Artif. Intell. Rev. 18, 77–95 (2002).
Finn, C., Abbeel, P. & Levine, S. Model-agnostic meta-learning for fast adaptation of deep networks. In Proceedings of 34th International Conference on Machine Learning (Eds. Precup, D. & Teh, Y. W.) 1126–1135 (JMLR.org, 2017).
Wang, Y., Yao, Q., Kwok, J. T. & Ni, L. M. Generalizing from a few examples: A survey on few-shot learning. ACM Comput. Surv. 53, 1–34 (2020).
Vella, D. & Ebejer, J. P. Few-shot learning for low-data drug discovery. J. Chem. Inf. Model. 63, 27–42 (2023).
Bento, A. P. et al. The CHEMBL bioactivity database: An update. Nucleic Acids Res. 42, D1083–D1090 (2014).
Naveja, J. J., Vogt, M., Stumpfe, D., Medina-Franco, J. L. & Bajorath, J. Systematic extraction of analogue series from large compound collections using a new computational compound–core relationship method. ACS Omega 4, 1027–1032 (2019).
Raghu, A., Raghu, M., Bengio, S. & Vinyals, O. Rapid learning or feature reuse? towards understanding the effectiveness of MAML. In 8th International Conference on Learning Representations (OpenReview.net, 2020).
Lv, Q., Chen, G., Yang, Z., Zhong, W. & Chen, C. Y. C. Meta learning with graph attention networks for low-data drug discovery. IEEE Trans. Neural Netw. Learn. Syst. 6, 1–13 (2023).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 5, 6000–6010 (2017).
Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
RDKit: Cheminformatics and Machine Learning Software. http://www.rdkit.org (Accessed on 1 July 2021).
He, J. et al. Molecular optimization by capturing chemist’s intuition using Deep Neural Networks. J. Cheminform. 13, 26 (2021).
He, J. et al. Transformer-based molecular optimization beyond matched Molecular Pairs. J. Cheminform. 14, 18 (2022).
Paszke, A. et al. PyTorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 32, 8026–8037 (2019).
Kingma, D.P. & Ba, J. Adam: a method for stochastic optimization. In 3th International Conference on Learning Representations (OpenReview.net, 2015).
Acknowledgements
The authors thank Martin Vogt for many helpful suggestions. H.C. is supported by the China Scholarship Council (CSC).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
All authors contributed to designing and conducting the study, analyzing the results, and preparing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, H., Bajorath, J. Meta-learning for transformer-based prediction of potent compounds. Sci Rep 13, 16145 (2023). https://doi.org/10.1038/s41598-023-43046-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-43046-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.