Abstract
Pancreatic ductal adenocarcinoma (PDAC) is associated with a very poor prognosis. Therefore, there has been a focus on identifying new biomarkers for its early diagnosis and the prediction of patient survival. Genome-wide RNA and microRNA sequencing, bioinformatics and Machine Learning approaches to identify differentially expressed genes (DEGs), followed by validation in an additional cohort of PDAC patients has been undertaken. To identify DEGs, genome RNA sequencing and clinical data from pancreatic cancer patients were extracted from The Cancer Genome Atlas Database (TCGA). We used Kaplan–Meier analysis of survival curves was used to assess prognostic biomarkers. Ensemble learning, Random Forest (RF), Max Voting, Adaboost, Gradient boosting machines (GBM), and Extreme Gradient Boosting (XGB) techniques were used, and Gradient boosting machines (GBM) were selected with 100% accuracy for analysis. Moreover, protein–protein interaction (PPI), molecular pathways, concomitant expression of DEGs, and correlations between DEGs and clinical data were analyzed. We have evaluated candidate genes, miRNAs, and a combination of these obtained from machine learning algorithms and survival analysis. The results of Machine learning identified 23 genes with negative regulation, five genes with positive regulation, seven microRNAs with negative regulation, and 20 microRNAs with positive regulation in PDAC. Key genes BMF, FRMD4A, ADAP2, PPP1R17, and CACNG3 had the highest coefficient in the advanced stages of the disease. In addition, the survival analysis showed decreased expression of hsa.miR.642a, hsa.mir.363, CD22, BTNL9, and CTSW and overexpression of hsa.miR.153.1, hsa.miR.539, hsa.miR.412 reduced survival rate. CTSW was identified as a novel genetic marker and this was validated using RT-PCR. Machine learning algorithms may be used to Identify key dysregulated genes/miRNAs involved in the disease pathogenesis can be used to detect patients in earlier stages. Our data also demonstrated the prognostic and diagnostic value of CTSW in PDAC.
Similar content being viewed by others
Introduction
With 496,000 newly diagnosed cases globally and 466,000 related deaths in 20201, pancreatic cancer is categorized among the malignancies with the poorest prognostic outcome2. According to the cancer statistics of the International Agency for Research on Cancer (IARC) GLOBOCAN, the incidence rate of pancreatic cancer has been rising in recent decades, and it accounts for 4.9% and 4.5% of worldwide cancer incidence and related deaths, respectively1. Pancreatic ductal adenocarcinoma (PDAC), the most common subtype of pancreatic cancer, accounts for over 90% of the cases3. Despite being the 10th most prevalent cancer, PDAC is the seventh most common cause of cancer-related deaths worldwide due to its poor prognosis4. Although the 5-year survival rate of pancreatic cancer differs regionally, it is < 10% due to a lack of clear clinical manifestations until advanced stages5. The primary reasons for the low survival rate of pancreatic patients are that the disease remains asymptomatic until advanced stages due to the anatomical position of the pancreas in the retroperitoneum and the lack of valuable biomarkers for early stages can be considered as other reasons6,7. Clinical biomarkers play a pivotal role in diagnosing and managing various cancers, including pancreatic cancer. CA-19-9 is one such biomarker commonly used for pancreatic cancer. CA-19-9 is a carbohydrate antigen that can be detected in the blood of some pancreatic cancer patients. Elevated levels of CA-19-9 may indicate the presence of pancreatic cancer, but it is important to note that this biomarker is not specific to pancreatic cancer. Other conditions, such as liver disease, bile duct obstruction, and certain gastrointestinal tumors, can also cause increased CA-19-9 levels8. Although the primary aetiology of pancreatic cancer has not been identified, some genes have been previously shown to be associated with the various cancer subtypes, treatment responses, or poor prognosis in pancreatic cancer9,10,11. Many cancers cannot be effectively treated in the advanced stages of disease, therefore developing novel biomarkers for the early stage is a potential approach for diagnosis, prognosis, and treatment of pancreatic cancer12. Currently, the K-RAS gene is known to be one essential gene playing a crucial role in pancreatic cancer, with a prevalence of more than 85%. Furthermore, P53 and P16, as tumor suppressor genes, are inactivated in approximately 95% of pancreatic cancer patients6. To activate or inactivate proto-oncogenes and other related genes like those functioning as tumor suppressors, including HER2, MYB, AKT2, BRCA2, FHIT, CDKN2A, PALB2, STK11, and PRSS1 are involved. Furthermore, the analysis of mutations in BRCA1/2, MMR(mismatch repairing system), and NTRK1–3 fusions was performed for pancreatic cancer patients receiving the treatment of pembrolizumab, entrectinib, and larotrectinib13. Advanced technologies such as bioinformatics and artificial intelligence are developed to provide cancer research opportunities14,15. Machine learning is a part of artificial intelligence that can improve the accuracy of cancer diagnosis, prediction, and prognosis by employing various statistical techniques16,17,18,19,20.
MiRNAs are non-coding RNAs with a length of 19–24 nucleotides that regulate gene expression of more than 30% of human genes following transcription. They pair to their target's untranslated 3′(3′-UTR) region of mRNAs, resulting in inhibition or degradation of the mRNAs21. Up or down-regulation or misplacement of miRNAs may play crucial roles in cancer development, tumor cell proliferation, migration, invasion, and chemical resistance22,23,24,25,26. These modifications and abnormalities in the miRNA transcription levels have previously been reported in several human malignancies27,28,29. It is hypothesized that genes and miRNAs might be evaluated as biomarkers to initiate better diagnostic or predictive approaches for pancreatic cancer. Previous studies have targeted KRAS and other genes in pancreatic cancer. Some miRNAs, including hsa-miR-217, hsa-miR-96, miR-216a, and miR-148a/b, are reported downregulated, and some, such as the miR-221, miR-210, miR-155, and miR-21 upregulated in pancreatic cancer30,31,32,33,34.
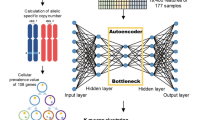
The Cancer Genome Atlas (TCGA) is a project that maps out the genome variation of human cancerous cells by RNA sequencing and using a non-malignant cell as a reference. These maps have identified many core genetic pathways activated in various cancers35,36. Therefore, in the current study, we performed gene expression proofing of pancreatic cancer using the TCGA database and Machine learning to identify differential expression genes (DEGs) and differentially expressed miRNAs (DEmiRNA). Survival was assessed using Kaplan–Meier analysis to predict prognostic biomarkers and the risk model. Additionally, the protein–protein interaction (PPI), the molecular pathways, the co-expression of DEGs, and the correlation between candidate genes and pancreatic cancer with clinical data were evaluated. Furthermore, the diagnostic markers were detected based on machine learning technology (Fig. 1A).
Material and method
Data collection
The TCGA database (http://tcga-data.nci.nih.gov/tcga/) was utilized to extract pancreatic cancer gene and miRNA data from 183 and 193 samples including healthy and tumor samples. RNA gene expression, microRNA, and clinical data were downloaded.
Data preprocessing and the identification of DEGs (differential expression genes)
In the pre-processing step, gene expression data were filtered to eliminate the gene and miRNA with zero expression and duplicates. Then, data was normalized with Limma and DESEQ2 packages in R 4.0.3 software. Filtering and normalization are the most important step in data analysis performed before machine learning. Genes and miRNAs were adjusted between pancreatic cancer samples and healthy tissue samples based on the particular criteria, including P < 0.05 and − 1.5 <|Log2FC (fold change) |< 1.5, to evaluate the upregulated and downregulated genes of the data integrity and subsequent analysis. The heatmap was created by “cluster, dendextend, circlize, RcolorBrewer, ComplexHeatmap, d3heatmap, gplots, pheatmap, and gplots” packages in R software version 4.3.1.
Identifying predictive markers
Machine learning methods can be used to analyze the data collected from various biological data, such as genomics, transcriptomics, and metabolic data27. Our study used machine learning algorithms, including Random Forest (RF), Max Voting, Adaboost, Gradient Boosting machines(GBM), and Extreme Gradient Boosting (XGB), for the analysis of DEGs and identifying novel biomarkers.
Machine learning by stage
Ensemble learning
This method performs better than using a simple algorithm alone because it employs many algorithms to provide poor predictive outcomes in accordance with the features taken from various estimations of data and integrates the results using various voting methods37.
Random Forest (RF): A technique that involves a set of decision trees that naturally incorporate feature selection and interactions into the learning process and report their average as an acceptable label. This algorithm is also the most popular machine learning method.
Max voting
One of the well-known methods in decision-making is max voting. This process is done independently, and as the best class vote is estimated, the outcome with the highest vote is carried out38.
Adaboost
One of the most efficient recognition algorithms in machine learning is Adaptive Boosting, aka. Adaboost. This algorithm makes a pile of weak learners by keeping a set of weights over training data and modifying them after each weak cycle adaptively to make more precise and strong learners out of a collection of weak learners. This recognition algorithm is used for ensemble learning as it has outstanding classification performance that is beneficial in estimating fruit biochemical parameters, image recognition, and complex change prediction modeling39.
Gradient boosting machines(GBM): As decision trees develop, a group forms gradient boosting machines using the information previously generated by growing trees. This way, each decision tree stems from an original training set focused on the parts where earlier model iterations deliver poor prediction40.
Extreme Gradient Boosting (XGB): Extreme Gradient Boosting (XGB) is considered one of the applications of gradient boosted decision trees. To have optimized memory usage and get the most out of hardware computing power, we can use XGBoost. It also reduces the processing time with enhanced performance compared to other machine learning algorithms and deep learning models41.
Performance of machine learning methods
In both true positive and true negative machine learning, accuracy is a measure of an algorithm's effectiveness and performance. F1score is a measure mostly used in unbalanced data to evaluate the algorithm's performance in a false positive and false negative. Auc_curve is a measure to evaluate the correct performance of the algorithm in classifying each class. The confusion matrix is a table that identifies four types of classifications (TN, TP, FN, FP) and shows the algorithm's overall performance. R2 is mainly used in regression algorithms to evaluate the performance of machine learning methods.
Investigation of the correlations of Clinical/Demographic with cancer
To explore the relationships between variables, R 4.1.3 was used to create a cancer correlation matrix to investigate the association between clinical data, including age, tumor size, lymph node involvement, distant metastasis, and stage. A correlation matrix visualizes connections by showing the coefficient of correlation between variables. The correlation coefficient is evaluated on a scale of − 1 to 1. While a negative correlation shows the variables moving in opposite directions, a positive correlation indicates that the variables are moving in the same direction. The cut-off for statistical significance was set ata p < 0.05.
Functional enrichment analysis of the genes and miRNAs
Functional analysis of Gene Ontology (GO) and Reactom, Do, GSEA pathways signaling pathways was performed. In these two analyzes, the categories that include biological process (BP), cellular components (CC), and molecular function (MF) are used. Results with enrichment score > 1, FDR < 0.25, and adjust p < 0.05 were determined as statistically significant results.
PPI network construction
The STRING v11.5 database (http://string-db.org/) was obtained to evaluate the interactions between the target genes of the selected miRNAs. The highest confidence score was set at 70.7 and was considered significant. Proteins were selected based on their interaction with other proteins. Cytoscape software was utilized to view the protein–protein interaction networks (PPIs). Selected miRNAs with several connections to other target genes propose their essential part in PPI.
Identifying prognostic markers
Kaplan‐Meier survival curves and Cox proportional hazard ratio (HR) were plotted for top-selected genes and miRNAs using SPSS version 20 and 95% CI . All the data were analyzed under screening criteria, including the cut-off threshold of HR > 1 and P < 0.05. The candidate genes and miRNAs presented as “prognostic genes”.
Identifying diagnostic markers
Diagnosing PDAC before the tumor spreads provides the best chance for treatment and survival. Here, we assessed the candidate genes, miRNAs, and every combination discovered through survival analysis and machine learning algorithms. In order to evaluate the diagnostic potency and create diagnostic models, a generalized linear model, and combined receiver operating characteristic (ROC) curve analysis were used. Additional diagnostic parameters such as sensitivity, specificity, cut-off value, positive predictive value, negative predictive value, and area under the ROC curve were assessed to evaluate the discrimination of individual or combined biomarkers. The entire procedure was applied using R 4.1.3’s combioROC package.
Quantitative real-time PCR
RNA was isolated from twenty-one Formalin-Fixed Paraffin-Embedded (FFPE) tissue samples using a Parstous kit (Parstous, Tehran, Iran). The extraction quality was evaluated on 1.5% agarose gel, and the quantity was assessed by a Nanodrop 2000 spectrophotometer (BioTek, USA EPOCH). cDNA was synthesized according to the manufacturer's instructions (Parstous, Tehran, Iran). Quantitative real-time PCR was performed using specific primers (Macrogene Co., Seoul, South Korea) and SYBR green master mix (Parstous Co. Tehran, Iran) using an ABI-PRISM StepOneTM instrument (Foster City, CA)18. To identify tissue-specific housekeeping genes for gene expression analysis and to avoid single control normalization error, accurate normalization of qRT-PCR data based on the geometric means of multiple internal control genes was performed. The housekeeping gene which was used as an internal control was GAPDH.
Statistical analysis
The RNA-Seq data analysis, including quality control, preprocessing, and identifying differential expression genes, was performed by R software version 4.3.1. The data were compared by paired t-test and were expressed as mean ± standard deviation (SD). A p-value < 0.05 was considered statistically significant.
Ethics approval and consent to participate
The data was downloaded from TCGA portal (https://tcga-data.nci.nih.gov/). TCGA generates over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data. The data will remain publicly available for anyone in the research community to all procedures consisting of Ethical issues followed by the TCGA committee. This article does not contain any studies with animals performed by any of the authors. This study was approved by the Ethical Committee of Mashhad University of Medical Sciences (IR.MUMS.MEDICAL.REC.1401.430).
Results
Data description and Identification of differentially expressed genes (DEGs) and differentially expressed miRNAs (DEmiRNA)
The clinical features of the patients are shown in Supplement Table 1. TCGA database containing 193 patients was used as a source to download our required data. Then the data were filtered and finally normalized with the DEseq package. Genes compliant with criteria 1 | LogFC |> and p-value < 0.05 were selected. Using five different machine learning methods, including SVM, DTS, RF, LR, and KNN, some key genes were nominated and analyzed by five various criteria: Accuracy, f1score, r2score, auc_curve, and Confusion matrix. During each step, the best classification algorithm was introduced.
Identifying predictive markers for genes and miRNAs
As shown in Fig. 1B,C, three genes (ABCA12, B3GNT3, and BMF) and eight miRNA (hsa.miR.577, hsa.miR.503, hsa.miR.3613, hsa.miR.19a, hsa.miR.19b.2, hsa.miR.365a, hsa.miR.365b, and hsa.miR.4668) were found to be dysregulated in four different stages of pancreatic cancer.
Investigation of the correlations of Clinical/Demographic with cancer
No association was found between DEG and clinical data for the patients from whom the RNA samples were obtained; only age was significantly associated with prior malignancy. The correlation is considered low when less than 0.3, moderate between 0.3 and 0.6, and strong when more than 0.6 (Fig. 1D,E). The heat map depicted for visualizing DEGs and DEmiRNA across the samples based on the specific criteria (Fig. 2A–F).
Functional enrichment analysis of the RNAs and miRNAs
A list of genes was generated, then the gene enrichment to determine the functionally related genes involved in different pathways was calculated, and the expression of other genes was adjusted by R software. Finally, the key genes were enriched to study the Reactom, Do, Go, GSEA pathways. In stage 1, the highest number of genes in the biological process (BP) portion is involved in regulating leukocyte activation and cell activation. As in cellular component (CC), most genes play a role in the receptor complex and side of the membrane pathways. Moreover, during stage 1 in the Molecular Function (MF), the highest number of genes are involved in the pathways of NAD + nucleosidase activity and hydrolyzing N-glycosyl compounds hydrolase activity (Fig. 3). In stage 2, most genes in the BP were involved in the positive regulation of cell death. In CC, the two pathways of hydrolyzing N-glycosyl mitochondrial membrane and mitochondrial envelope are affected by most of the genes, and in MF, the highest number of genes are involved in oxidoreductase activity (Fig. 4). During stage 3, in the BP portion, the highest number of genes are in the immune response-regulating signaling pathway; in the CC stage, the highest number of genes in the side of the membrane pathway, and in the MF section, the genes are equally involved in 3 molecular pathways including, protein and molecular sequestering activity, and NAD(P) H oxidase H2O2-forming activity (Fig. 5). Stage 4 in the BP involved three inflammatory response pathways, cell activation and activation regulation of leukocytes. Also, in the CC, the highest number of genes are involved in the two pathways of hydrolyzing N-glycosyl compounds, hydrolase activity, and NAD + nucleosidase activity (Fig. 6).
PPI network construction
Figure 7 illustrates the interaction of DEGs checked and plotted using the STRING (interaction score: 0.4). In accordance with PPI network, the CD22 gene has the highest binding capacity, followed by CTSW and BTNL9 (Figs. 7C,D, 8B).
Identifying prognostic markers for RNAs and miRNAs
Kaplan Meier analysis was applied to identify key prognostic signature genes in pancreatic cancer. The outcome revealed survival is associated with three genes, including BTNL9 (HR = 1.02), CD22 (HR = 1.7), and CTSW (HR = 2.03) and five miRNAs, including hsa.miR.539 (HR = 1.3), hsa.miR.412 (HR = 1.04), hsa.miR.153.1 (HR = 1.5), hsa.miR.642a (HR = 1.00), and hsa.miR.363 (HR = 1.5) in PDAC patients. All analyses were performed by R software (Figs. 7, 8A).
Identifying diagnostic markers for RNAs and miRNAs
For stages 1 and 2, GLM model analysis for HCK and SIGLEC7 combination in diagnostic biomarkers with coefficients of 1.2920 and − 0.5562 (AUC of 0.74, 95% CI with sensitivity of 0.85 and specificity of 0.66). For stage 3, the combination of B3GNT3, ABCA12, and ADAP2 with 0.8409 (AUC of 0.86, 95% CI with sensitivity of 0.8 and specificity of 1. In stage 4, our finding showed that the Coefficients of combination AIF1 and RASGRP3 were 4.233 and − 7.841 (AUC of 0.86, 95%CI with 0.8 sensitivity and one specificity). Furthermore, three miRNAs (Has.mir.194.2, hsa.mir.194.1, and hsa.mir.192) had the highest AUC value, sensitivity, and specificity and coefficients of 4.932, 5.531, and 3.584, respectively (Supplement Table 1).
Validation of CTSW in an additional cohort of PDAC
The clinical data are shown in Supplement Table 2; our population consisted of 52.4% males and 47.6% females. The mean age was 61.66 years and 52.4% underwent advanced stage. We further evaluated the value expression of CTSW in PDAC cases using RT-PCR. This data showed the significant downregulation of this gene in tumor tissue (P < 0.05) (Fig. 8C).
Discussion
To the best of our knowledge, this is the first study showing the potential of downregulation of hsa.miR.642a, hsa.mir.363, CD22, BTNL9, and CTSW and overexpression of hsa.miR.153.1, hsa.miR.539, hsa.miR.412 with shorter survival of patients with PDAC (Supplement Fig. 1) The result indicated the diagnostic value of the combination of AIF1 and RASGRP3 in an advanced stage with the Coefficients of combination AIF1 and RASGRP3 were 4.233 and -7.841 (AUC of 0.86, 95%CI with 0.8 sensitivity and one specificity). The result of the survival analysis showed that the CTSW gene is a novel prognostic marker. CTSW (Cathepsin W), also known as LYPN is a novel human cysteine proteinase member of the C1 peptidase family expressed in CD8 + T and NK cells and regulated by interleukin-2. This gene has a specific function in the cytotoxicity-mediated mechanism by NK cells and CD8 + T cells. Various T cell populations can act differently in regulating a tumor's degree, stage, and ability to invade endometrial cancer. CTSW is an immunomodulatory gene that functions similarly to the CTSF gene42. In research done by Song and colleagues, the expression of CTSF in non-small cell lung cancer was evaluated, and downregulated levels of CTST were observed in NSCLC samples despite normal tissues and good prognosis of NSCLC being correlated with high expression of CTSF. Besides using GeneMANIA, the gene–gene interaction network was established for CTSF and showed that CTSF had a similar function as CTSW genes43. A study on endometrial cancer reported that the CTSW gene had a positive correlation with tumor infiltration levels of B cells, CD8 + T cells, CD4 + T cells, macrophages, and dendritic cells42.
BMF (Bcl-2 modifying factor) is a proapoptotic protein that belongs to the BCL-2 protein family. This gene has been identified in the BH3-only proteins subgroup and initiates the innate apoptotic pathway44. Consequently, BMF is linked with various cellular activities, including chemical sensitivity. For example, the YAP/TEAD/SLUG axis suppressed apoptosis by suppressing BMF transcription45. Badr et al. reported that upregulation of livin and downregulation of BMF and p53 expression are significantly correlated with more tumor aggressiveness (advanced TNM stage), making metastasis progress more rapidly and decreasing overall survival in colon cancer patients. Thus, we can apply these genes as crucial prognostic markers related to poor results46. Another research showed that STARD13 3′UTR could play as a ceRNA for BMF to enhance apoptosis and be used as a potential therapeutic target in breast cancer cells47. FERM is a superfamily of proteins, and one of its members is FERM domain-containing 4A (FRMD4A); these proteins are ubiquitous parts of the cytocortex and are involved in cell transport cell structure and signaling functions. Moreover, tumor progression and metastasis are the cellular events in which the proteins of the FERM family are involved. These proteins function as regulators or scaffolding units and are involved in many membrane-associated factors' functions48. In another study on tongue squamous cell carcinoma and squamous cell carcinoma, the expression of FMRD4A was increased, contrasting our findings.48,49. ArfGAP with dual PH domains 2 (ADAP2) belongs to the ArfGAP family of genes, which is the GTPase activating protein. This gene is expressed for ARF6, which acts as a scaffold in the innate and membrane immunosuppressive phosphate signaling pathways50. It is reported that the ADAP2 gene expression was decreased in primary lower-grade glioma51. Contrary to this, the expression of this gene was increased in radiation-resistant esophageal cancer cells52. Protein phosphatase 1 regulatory subunit 17(PPP1R17), also known as C7orf16, is a negative regulator that inhibits phosphatase activities of protein phosphatase 1 (PP1) and protein phosphatase 2A (PP2A) complexes which their substrates are the S6 ribosomal protein53. Contrary to our results, research in lung cancer adenocarcinoma has demonstrated that PPP1R17 can be used as biomarkers as it was specifically detected in stage III, which can help us detect cancer stage in tumor progression through cleft junction incompatibility, Wnt signaling, and GPCR signaling pathways54. Another study reported that PPP1R17 is a HAR-regulated gene that slows the progression of the neural precursor cell cycle while increasing cell cycle length, which is mainly observed in the neural growth of primates, especially humans.55. The CACNG3 gene encodes a transient AMPA regulatory protein (TARP) known as an auxiliary subunit of the calcium channel γ3. This gene is involved in the neurons formation, and has also been reported as a potential source of epilepsy56. In line with our results, several other studies have shown that the CACNG 3 gene in Gliomas has been predicted as an oncogene and significantly dysregulated in glioblastoma tissue compared to healthy controls57. Other studies have also reported dysregulation of CACNG3 gene in breast cancer58. In our study, hsa.miR.153.1, also known as MIRN153-1, was found to be a new microRNA that had not been used in any other diseases or cancers and had increased expression in pancreatic cancer.
In conclusion, we have identified some specific genes that are differentially expressed at different stages of pancreatic cancer. CTSW gene was reported as a novel prognostic biomarker and validated by Real-time PCR in pancreatic tumor tissue. Eventually, we highly recommend using machine learning to detect biomarkers in other cancers as well.
Data availability
The data was downloaded from TCGA portal (https://tcga-data.nci.nih.gov/). TCGA generated over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data. The data will remain publicly available for anyone in the research community.
References
Ferlay, J. et al. Cancer statistics for the year 2020: An overview. Int. J. Cancer 149(4), 778–789 (2021).
Jagadeesan, B., Haran, P. H., Praveen, D., Chowdary, P. R. & Aanandhi, M. V. A comprehensive review on pancreatic cancer. Res. J. Pharm. Technol. 14, 552–554 (2021).
Jin, C. & Bai, L. Pancreatic cancer—Current situation and challenges. Gastroenterol. Hepatol. Lett. 2(1), 1–3 (2020).
Menini, S., Iacobini, C., Vitale, M., Pesce, C. & Pugliese, G. Diabetes and pancreatic cancer—A dangerous liaison relying on carbonyl stress. Cancers 13(2), 313 (2021).
Hu, J. X. et al. Pancreatic cancer: A review of epidemiology, trend, and risk factors. World J. Gastroenterol. 27(27), 4298–4321 (2021).
Kamisawa, T., Wood, L.D., Itoi, T., & Takaori, K.J.T.L. Pancreatic Cancer. Lancet. 388(10039), 73–85 (2016).
Kanno, A. et al. Multicenter study of early pancreatic cancer in Japan. Pancreatology 18(1), 61–67 (2018).
Ballehaninna, U. K. & Chamberlain, R. S. Biomarkers for pancreatic cancer: Promising new markers and options beyond CA 19-9. Tumor Biol. 34, 3279–3292 (2013).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321(5897), 1801–1806 (2008).
Yang, J., Shi, W., Zhu, S. & Yang, C. Construction of a 6-gene prognostic signature to assess prognosis of patients with pancreatic cancer. Medicine 99(37), e22092 (2020).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518(7540), 495–501 (2015).
De Dosso, S. et al. Treatment landscape of metastatic pancreatic cancer. Cancer Treat. Rev. 96, 102180 (2021).
Nevala-Plagemann, C., Hidalgo, M. & Garrido-Laguna, I. From state-of-the-art treatments to novel therapies for advanced-stage pancreatic cancer. Nature Rev. Clin. Oncol. 17(2), 108–123 (2020).
Kolodziejczyk, A. A., Kim, J. K., Svensson, V., Marioni, J. C. & Teichmann, S. A. The technology and biology of single-cell RNA sequencing. Mol. Cell 58(4), 610–620 (2015).
Chinnappan, J. et al. Integrative bioinformatics approaches to therapeutic gene target selection in various cancers for nitroglycerin. Sci. Rep. 11(1), 22036 (2021).
Hornbrook, M. C. et al. Early colorectal cancer detected by machine learning model using gender, age, and complete blood count data. Dig. Dis. Sci. 62(10), 2719–2727 (2017).
Kinar, Y. et al. Performance analysis of a machine learning flagging system used to identify a group of individuals at a high risk for colorectal cancer. PLoS ONE 12(2), e0171759 (2017).
Dimitriou, N., Arandjelović, O., Harrison, D. J. & Caie, P. D. A principled machine learning framework improves accuracy of stage II colorectal cancer prognosis. NPJ Digit. Med. 1(1), 1–9 (2018).
Nazari, E. et al. Identification of potential biomarkers in stomach adenocarcinoma using machine learning approaches. Curr. Bioinform. 18(4), 320–333 (2023).
Khalili-Tanha, G. et al. Identification of ZMYND19 as a novel biomarker of colorectal cancer: RNA-sequencing and machine learning analysis. J. Cell Commun. Signal. 1–17. https://doi.org/10.1007/s12079-023-00779-2 (2023).
Salmaninejad, A., Pourali, G., Shahini, A., Darabi, H. & Azhdari, S. MicroRNA and exosome in retinal-related diseases: Their roles in the pathogenesis and diagnosis. Comb. Chem. High Throughput Screen. 25(2), 211–228 (2022).
Yonemori, K., Kurahara, H., Maemura, K. & Natsugoe, S. MicroRNA in pancreatic cancer. J. Hum. Genet. 62(1), 33–40 (2017).
Waspada, I., Wibowo, A. & Meraz, N. S. Supervised machine learning model for microrna expression data in cancer. Jurnal Ilmu Komputer dan Informasi 10(2), 108–115 (2017).
Savareh, B. A. et al. A machine learning approach identified a diagnostic model for pancreatic cancer through using circulating microRNA signatures. Pancreatology 20(6), 1195–1204 (2020).
Shi, X.-H. et al. A five-microRNA signature for survival prognosis in pancreatic adenocarcinoma based on TCGA data. Sci. Rep. 8(1), 1–10 (2018).
Samami, E. et al. The potential diagnostic and prognostic value of circulating MicroRNAs in the assessment of patients with prostate cancer: rational and progress. Front. Oncol. 11, 716831 (2022).
Xia, T., Chen, X.-Y. & Zhang, Y.-N. MicroRNAs as biomarkers and perspectives in the therapy of pancreatic cancer. Mol. Cell. Biochem. 476(12), 4191–4203 (2021).
Acunzo, M., Romano, G., Wernicke, D. & Croce, C. M. MicroRNA and cancer—A brief overview. Adv. Biol. Regulat. 57, 1–9 (2015).
Pourali, G. et al. Circulating tumor cells and cell-free nucleic acids as biomarkers in colorectal cancer. Curr. Pharm. Des. 29(10), 748–765 (2023).
Xue, Y. et al. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet. 206(6), 217–221 (2013).
Sohrabi, E., Rezaie, E., Heiat, M. & Sefidi-Heris, Y. An integrated data analysis of mRNA, miRNA and signaling pathways in pancreatic cancer. Biochem. Genet. 59(5), 1326–1358 (2021).
Khojasteh-Leylakoohi, F. et al. Association of a genetic variant in the adenosine triphosphate transmembrane glycoprotein and risk of pancreatic cancer. Ann. Pancreatic Cancer. 6, 6 (2023).
Akhlaghipour, I., Fanoodi, A., Zangouei, A.S., Taghehchian, N., Khalili-Tanha, G. & Moghbeli, M. MicroRNAs as the critical regulators of forkhead box protein family in pancreatic, thyroid, and liver cancers. Biochem. Genetics 61(5), 1645–1674 (2023).
Sardarzadeh, N. et al. Association of a genetic variant in the cyclin-dependent kinase inhibitor 2B with risk of pancreatic cancer. Rep. Biochem. Mol. Biol. 11(2), 336 (2022).
Tomczak, K., Czerwińska, P. & Wiznerowicz, M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 19(1a), A68-77 (2015).
Azari, H. et al. Machine learning algorithms reveal potential miRNAs biomarkers in gastric cancer. Sci. Rep. 13(1), 6147 (2023).
Dong, X., Yu, Z., Cao, W., Shi, Y. & Ma, Q. A survey on ensemble learning. Front. Comp. Sci. 14(2), 241–258 (2020).
Usman, M., Shafique, Z., Ayub, S. & Malik, K. Urdu text classification using majority voting. Int. J. Adv. Comput. Sci. Appl. 7(8). https://doi.org/10.14569/IJACSA.2016.070836 (2016).
Wang, J., Xue, W., Shi, X., Xu, Y. & Dong, C. Adaboost-based machine learning improved the modeling robust and estimation accuracy of pear leaf nitrogen concentration by in-field VIS-NIR spectroscopy. Sensors 21(18), 6260 (2021).
Baran, Á., Lerch, S., El Ayari, M. & Baran, S. Machine learning for total cloud cover prediction. Neural Comput. Appl. 33(7), 2605–2620 (2021).
Dhieb, N., Ghazzai, H., Besbes, H., Massoud, Y., (eds). Extreme gradient boosting machine learning algorithm for safe auto insurance operations. In 2019 IEEE international conference on vehicular electronics and safety (ICVES); 2019: IEEE.
Chen, P. et al. Identification of prognostic immune-related genes in the tumor microenvironment of endometrial cancer. Aging 12(4), 3371 (2020).
Song, L. et al. Expression signature, prognosis value and immune characteristics of cathepsin F in non-small cell lung cancer identified by bioinformatics assessment. BMC Pulm. Med. 21(1), 1–17 (2021).
Liew, S. H., Nguyen, Q.-N., Strasser, A., Findlay, J. K. & Hutt, K. J. The ovarian reserve is depleted during puberty in a hormonally driven process dependent on the pro-apoptotic protein BMF. Cell Death Dis. 8(8), e2971 (2017).
Xu, F. et al. RBMS2 chemosensitizes breast cancer cells to doxorubicin by regulating BMF expression. Int. J. Biol. Sci. 18(4), 1724 (2022).
Badr, E. A. et al. A correlation between BCL-2 modifying factor, p53 and livin gene expressions in cancer colon patients. Biochem. Biophys. Rep. 22, 100747 (2020).
Guo, X. et al. Displacement of Bax by BMF mediates STARD13 3′ UTR-induced breast cancer cells apoptosis in an miRNA-depedent manner. Mol. Pharm. 15(1), 63–71 (2018).
Zheng, X. et al. FRMD4A: A potential therapeutic target for the treatment of tongue squamous cell carcinoma. Int. J. Mol. Med. 38(5), 1443–1449 (2016).
Goldie, S. J. et al. FRMD4A upregulation in human squamous cell carcinoma promotes tumor growth and metastasis and is associated with poor prognosis. Cancer Res. 72(13), 3424–3436 (2012).
Pyfrom, S. C., Luo, H. & Payton, J. E. PLAIDOH: A novel method for functional prediction of long non-coding RNAs identifies cancer-specific LncRNA activities. BMC Genomics 20(1), 1–24 (2019).
Zhang, M., Wang, X., Chen, X., Guo, F. & Hong, J. Prognostic value of a stemness index-associated signature in primary lower-grade glioma. Front. Genet. 11, 441 (2020).
Luo, J. et al. mRNA and methylation profiling of radioresistant esophageal cancer cells: The involvement of Sall2 in acquired aggressive phenotypes. J. Cancer 8(4), 646 (2017).
Mosti, F. & Silver, D. L. Uncovering the HARbingers of human brain evolution. Neuron 109(20), 3231–3233 (2021).
Liang, J., Lv, J. & Liu, Z. Identification of stage-specific biomarkers in lung adenocarcinoma based on RNA-seq data. Tumor Biol. 36(8), 6391–6399 (2015).
Girskis, K. M. et al. Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron 109(20), 3239-3251.e7 (2021).
Thompson, C. H., Saxena, A., Heelan, N., Salatino, J. & Purcell, E. K. Spatiotemporal patterns of gene expression around implanted silicon electrode arrays. J. Neural Eng. 18(4), 045005 (2021).
Liu, P. et al. Calcium-related gene signatures may predict prognosis and level of immunosuppression in gliomas. Front. Oncol. 12, 708272 (2022).
Singh, H. N. & Rajeswari, M. R. Identification of genes containing expanded purine repeats in the human genome and their apparent protective role against cancer. J. Biomol. Struct. Dyn. 34(4), 689–704 (2016).
Funding
This research was supported by Mashhad University of Medical Sciences, grant No. 4010928 and National Institute for Medical Research and Development (NIMAD 962782).
Author information
Authors and Affiliations
Contributions
F.K.L., R.M., N.K.T., and AA drafted the manuscript. E.N., H.N., and S.S. participated in data analysis. G.K.T. performed R.T.-P.C.R. G.P., Z.Y., and M.M. designed, and validation study. M.N., S.M.H., M.G.M., G.F., E.G., M.K., A.K.L., J.B., and A.A. designed, supervised, and revised the manuscript. All authors approved the final manuscript. All authors approved the manuscript and gave their consent for submission and publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khojasteh-Leylakoohi, F., Mohit, R., Khalili-Tanha, N. et al. Down regulation of Cathepsin W is associated with poor prognosis in pancreatic cancer. Sci Rep 13, 16678 (2023). https://doi.org/10.1038/s41598-023-42928-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-42928-y
This article is cited by
-
An optimized AdaBoost algorithm with atherosclerosis diagnostic applications: adaptive weight-adjustable boosting
The Journal of Supercomputing (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.