Abstract
Wheat stripe rust caused by Puccinia striiformis f. sp. tritici (Pst) is an airborne disease that endangers wheat during its entire growth period. In this study, the Pst134EA_003354 uncharacterized protein (GenBank: XM_047941824.1) of Pst was used as the target sequence, and the primers PS-RPA-F and PS-RPA-R, as well as the probe PS-LF-probe, were designed for recombinase polymerase amplification (RPA) technology. Flow chromatography was combined with the process to establish an RPA detection method for Pst. This method successfully established visual detection within 10 min under a constant temperature of 39 °C, and the detection results were consistent with those of ordinary PCR analysis. However, it only had high specificity for Pst, and the detection limit was 10 fg/μL. In addition, this rapid method successfully detected Pst from wheat leaves during the field incubation period, indicating substantial benefits for applied use. In summary, the RPA detection method established in this study has the favourable characteristics of high efficiency, simple functionality, and rapid and universal practicability, providing a theoretical basis for the early detection and prevention of Pst.
Similar content being viewed by others
Introduction
Wheat (Triticum aestivum L.) is the second-largest food crop in China and is one of the most important cereals grown in arid and semiarid regions. It provides 20% of the total calorie and protein intake of the human diet. Wheat stripe rust caused by Puccinia striiformis f. sp. tritici is one of the most destructive diseases in wheat, causing severe yield loss worldwide1. The mass occurrence of wheat diseases caused by phytopathogens is a critical factor that can negatively impact wheat yield2,3. In recent years, wheat stripe rust caused by Puccinia striiformis f. sp. tritici (Pst) has caused significant damage in wheat production. Wheat stripe rust is a disease that can occur in plants from the seedling stage to maturity, as Pst urediospores infect wheat leaves and undermine their ability to photosynthesize normally, which in turn affects wheat yield4. The production of teliospores can occur at all wheat growth stages, but especially when plants reach maturity5. Pathogens in plant tissues can be detected using many methods. Plant pathogens causing crop diseases require accurate and rapid detection6. Accurate and rapid detection of pathogens is important for the effective management of plant pathogens7. Currently, the detection of Pst is mainly accomplished by real-time quantitative fluorescent PCR8, LAMP detection9, molecular labelling methods10,11, or ordinary PCR detection methods. However, these technologies impose stringent requirements on personnel and equipment, place high demands on laboratories and are typically not conducive for on-site detection of plant pathogens in environmental or agricultural settings. A rapid and straightforward procedure for detecting Pst has traditionally been challenging to develop. RPA, which was invented in 2006, has been experiencing rapid development12 and has become a novel isothermal DNA amplification and detection technique13.
Recombinase polymerase amplification (RPA) offers a rapid and particular isothermal alternative to PCR14. The whole RPA process is very fast. Generally, detectable amplification products can be obtained in approximately 20 min15. The technology mainly uses recombinase, single-stranded binding protein, and strand displacement polymerase to specifically recognize and amplify the template at any constant temperature within the range of 25–43 °C. The final target product can be visualized on RPA (lateral flow dipstick, LFD) by adding specific probes16,17,18. Since RPA technology has a fast amplification speed, strong specificity, and high sensitivity, the LFD-RPA product after constant temperature amplification is added to the quantitative buffer solution and reacted with the side stream paper chromatography test strip for 3–5 min, and the test results can be observed and judged by the naked eye. Nucleic acid amplification can be completed by maintaining the activity of the enzyme within a suitable temperature range of 37–42 °C, which does not require high-temperature denaturation, annealing, or other steps. The mild reaction conditions and high amplification efficiency of RPA make it very suitable for rapid clinical diagnosis, food detection, epidemic prevention and control, industrial application, and on-site real-time detection19. RPA is gaining popularity because of its unique characteristics, including a low reaction temperature for amplification and a lack of sensitivity to plant inhibitors20. Because RPA does not require any advanced laboratory equipment, it is very suitable for on-site testing21. At present, RPA technology has not been widely used mainly because it is not a widely available technology and is only used for scientific research15. It is important to perform more assessments in the field on isothermal amplification technologies, including LAMP and RPA techniques22. Furthermore, RPA is more tolerant than PCR to inhibitors and background DNA23. Most components in RPA are supplied by the manufacturer in a freeze-dried pellet that allows components to be taken on site without refrigeration24. Hence, compared to previously reported methods, the advantages of the RPA method are obvious25. It has been used to detect citrus yellow pulse virus, broad bean virus, citrus leaf spot virus, wheat blight, and wheat sheath rot caused by various pathogens26,27,28,29,30. However, the detection of Pst with RPA has not yet been reported.
In this experiment, a wheat stripe rust Pst134EA_003354 uncharacterized protein was used as the target sequence, specific RPA primers and probes were designed and combined with lateral flow layer test strips, and an RPA-LFD rapid detection system for Pst was established. No specialized software has been developed for the design of RPA primers, and only PCR software can be used for design and screening19. This system was used to detect wheat stripe rust in the field and provides a theoretical basis for the early prevention of Pst. The abovementioned nucleic acid detection of Pst has achieved good results, which demonstrates that the detection efficiency of the RPA method is obviously better than that of the PCR method, and the reaction time is faster, which provides a rapid and accurate detection method for the disease prediction and field investigation of wheat seedlings in the later growth stage and provides a good basis for the prevention and control of Pst to reduce the decline in production and to mitigate economic losses.
Results
Primer screening and probe design
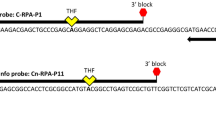
The resulting eight primer sets were used for RPA detection and agarose gel electrophoresis. The results showed that RPA PS4 primers produced detection bands, while other primer pairs produced only quality control bands. The electrophoresis gel detection results also showed that the PS4 primer pair could amplify a 300 bp target band, primer PS1 produced multiple nontarget fragments, and primers PS2–3 and PS5–8 produced obvious primer dimers. Therefore, the primer PS4 set was selected as the primer for this study. Additionally, probes were designed using fragments with the lowest homology between the amplified product and the downstream sequence (Fig. 1A,B. Fig. S1 shows the original electrophoretic gels and blots.). The primer pairs and probe information used are shown in Table 1.
Specific detection of Puccinia striiformis f. sp. tritici
The RPA results showed that only the RPA amplification products of Pst CYR32, CYR33, and CYR34 DNA showed detection bands and quality control bands on the flow chromatography test strip. Of the five other pathogens tested, only wheat DNA and the blank control showed a quality control zone in ddH2O. In addition, PS-RPA-F and PS-RPA-R were primers for ordinary PCR detection, and the agarose gel electrophoresis results showed that Pst was amplified to a specific target of 300 bp with no other pathogen amplification occurring. This indicates that the primer had high specificity for Pst (Fig. 2A,B. Fig. S2 shows the original electrophoretic gels and blots.).
Specific detection of Puccinia striiformis f. sp. tritici by RPA (A) and conventional PCR (B). 1. CYR32, 2. CYR33, 3. CYR34, 4. Puccinia graminis, 5. Puccinia recondita, 6. Blumeria graminis, 7. Puccinia striiformis West. f. sp. hordei, 8. Puccinia recondita. f. sp. agropyri, 9. wheat leaf gDNA, 10. ddH2O, M: marker-500.
RPA sensitivity detection
Following a tenfold dilution of Pst gDNA, the flow chromatography test strips showed positive results for 100 ng/μL, 10 ng/μL, 1 ng/μL, 100 pg/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL and 10 fg/μL concentrations, but 1 fg/μL and 100 ag/μL concentrations only produced the quality control band, meaning the result was negative. This illustrated that RPA was successful in detecting as little as 10 fg/μL of Pst DNA. The same concentrations of Pst gDNA were used for ordinary PCR detection. The results showed that the lower limit of DNA concentration detected by ordinary PCR was also 10 fg/μL. This indicates that the RPA detection system had high sensitivity for Pst (Fig. 3A,B. Fig. S3 shows the original electrophoretic gels and blots.).
Detection of Puccinia striiformis f. sp. tritici isolates
Thirty-five samples from Datong, Huzhu, Ledu, Jianzha, Hualong and Gui-De counties were collected. Thirty samples were found to be infected with Pst using the RPA test, and five samples were not found to be infected with Pst, which was consistent with the results from ordinary PCR analysis. The test results are shown in Fig. 4A,B and Table 2. (Fig. S4 shows the original electrophoretic gels and blots.).
RPA practicality test
Twenty-one wheat leaf samples were collected from seven areas in Qinghai Province. The leaves were tested, and the results from the Dashijia, Ahetan, Shangduoba, and Sheren villages were positive. However, Pst was not detected in samples from Heerjia, Xiaduoba, and Baiwujia villages. The results from the ordinary PCR tests were comparable. Pst was detected on the first day after inoculation. However, the RPA and ordinary PCR analysis showed weak bands, indicating that the amount of Pst was relatively low on the first day after infection. Puccinia striiformis f. sp. tritici was detected from Days 2–8, and observation stopped on the ninth day when sporadic urediospores appeared on the surface of the leaves. This shows that the established RPA system can detect Pst at the seedling stage (Fig. 5A–D, and Table 3. Fig. S5 shows the original electrophoretic gels and blots.).
RPA (A) and conventional PCR (B) detection of Puccinia striiformis f. sp. tritici isolates after different quantities of days of infection. 1–8: 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, 7 days, 8 days; M: marker-500; Detection of Puccinia striiformis f. sp. tritici isolates by RPA (C) and conventional PCR (D). The Puccinia striiformis f. sp. tritici isolates were collected from 1–3: Dashijia, 4–6: Heerjia, 7–9: Ahetan, 10–12: Xiaduoba, 13–15: Sheren, 16–18: Shangduoba, and 19–21: Baiwujia; M: marker-500.
Discussion
In this study, we designed and screened specific RPA primers and probes based on the Pst134EA_003354 uncharacterized protein and established a rapid detection method for Pst using RPA technology in combination with lateral flow chromatography test strips to detect Pst visually during the incubation period.
Primer and probe design are key factors in the success of RPA detection. The length of RPA primers is generally between 30 and 35 bp; however, TwistDx has recently announced that PCR primers (18 bases and higher) can successfully be used31. The target band of the amplified product in this study was between 100 and 500 bp. The primers PS-RPA-F and PS-RPA-R and the probe PS-LF-Probe used in this study specifically detected the physiological races CYR32, CYR33, and CYR34 and can be used to detect the pathogens of other wheat leaf diseases. This illustrates that the specificity of the test was strong. It has been reported that the detection limit of the LAMP detection system for Pst is 1 pg/μL9, and the dual real-time quantitative fluorescent system established by Pan et al. can detect Pst at a minimum of 0.4 pg/μL32. When using RPA technology to detect other diseases, the detection sensitivities have been reported as follows: Wei et al. reported a sensitivity of 75 fg/μL for the detection of the causal agent of tomato leaf bacterial spot pathogen33, the RPA detection sensitivity of Pythium aphanidermatum established by Zhao et al. was 3.75 fg/μL34, and Shen constructed an RPA for the detection of tobacco and found the sensitivity of bacterial wilt to be 1 pg/μL35. Relatively speaking, the PS-RPA-F, PS-RPA-R, and PS-LF-Probe established in this study had a lower detection limit of 10 fg/μL for Pst genomic DNA, and the sensitivity was relatively high.
LAMP detection of Pst requires the design of four primers and a loop primer for amplification36, while RPA technology requires only forwards and reverse primers to detect the target band. Additionally, the complexity of RPA primer design is relatively low compared to LAMP detection technology, and the RPA technique can specifically detect Pst within 20 min at a constant temperature of 39 °C. The method is simple, fast, and can be conducted at room temperature, and the detection buffer can be divided into aliquots to avoid false-positive results. The practicality of RPA technology is an important factor warranting the promotion of this method.
In this study, thirty mixed isolates of Pst and five uninfected samples were collected from the Datong, Huzhu, Ledu, Jianzha, Hualong and Gui-DeGui-De counties of Qinghai Province, and 21 self-inoculated leaves of spring wheat seedlings were collected from areas with a high incidence of Pst that were infected for 1–8 days. A small amount of Pst was found in the mixed isolates on the first day after infection. In addition, 12 samples were detected in areas with a high incidence of Pst. This shows that the application of this system can rapidly and efficiently detect Pst in leaves during the incubation period. This study provides a theoretical basis for the early prevention and control of wheat stripe rust.
The technique has been applied to detect wheat take-all32 and wheat sheath blight33. RPA detection is of great significance for the early diagnosis, prediction, and comprehensive control of various wheat diseases. In the future, we will use this technique to distinguish the physiological races of wheat stripe rust. The traditional method is to use differential hosts. This process is long and complex. However, if RPA detection technology is used, the physiological race can be identified quickly, saving time, material resources, and workforce. This will be the direction of our subsequent efforts.
Materials and methods
Test materials and reagents
The test pathogens in this study were Puccinia striiformis f. sp. tritici races CYR32, CYR33, CYR34; isolates of Pst; Puccinia graminis Pers tritici Eriks and E Henn; Puccinia recondita, Blumeria graminis f. sp. tritici, Puccinia striiformis West. f. sp., Puccinia recondita. f. sp. agropyri. The pathogens were provided by the Key Laboratory of Comprehensive Management of Agricultural Pests of Qinghai Province and the College of Plant Protection, Northwest A&F University.
The test materials in this study were Mingxian 169 wheat seedling leaves collected from the Dashijia, Heerjia, and Chada villages in Gui-De County; from the Shangduoba, Xiaduoba, Xingfu, and Longshang villages in Hualong County; from Ahetan village in Xunhua County; from Baiwujia village in Minhe County; from the Kangjia, Hanjia, Baojia, Maojiazhai and Benkang villages in Datong County; from Gelong village in Huzhu County; from Xiakou village in Ledu County; and from Jianzha County in Yangjiacun (Fig. 6). There was a distance of at least 50 m between samples in a single wheat field to avoid duplication of any original isolate during the season.
The reagents used in this study were as follows: RAA-nfo nucleic acid amplification reagent (type test strip) was acquired from Qinghai Baisai Trading Co., Ltd.; HybriDetect was purchased from Milenia Biotec Versailler Straße, Gießen Milenia; Premix Ex TaqTM II was purchased from TaKaRa; Shanghai Sangon Biotech Co., Ltd. synthesized all the primers that were used. Ordinary PCR is in vitro enzymatic synthesis of specific DNA fragments, which is based on the level of DNA concentration. In this experiment, the detection band began to weaken when the DNA concentration was lower than 1 pg/μL. The reaction mode of the RPA technology is different from that of PCR in that it does not require thermal cycle steps such as denaturation and annealing. This technology mainly relies on recombinase, polymerase, and single-stranded binding protein (SSB).
Extraction of pathogenic fungus DNA
Wheat stripe rust samples were stored at 4 °C in a desiccator. There were 10 blades of Mingxian 169 and 30 blades of each sample at each sampling point. The genomic DNA of Pst and other control pathogens was extracted using the (Cetyltrimethylammonium Bromide)CTAB method37. The concentration of the obtained DNA was measured using a NanoDrop™ One ultramicro ultraviolet spectrophotometer, and the DNA integrity was checked using a 2% agarose gel. It was stored at − 80 °C until use.
RPA reaction system establishment
Preparation began by adding 40.9 μL of buffer A, 2 μL of forwards and reverse primers (10 μmol/L), and 0.6 μL of probe (10 μmol/L) to the detection tube containing the test dry enzyme preparation. Then, 2.0 μL of the test sample DNA and 2.5 μL of buffer B were added. This was mixed thoroughly by inverting the tube 5–6 times; then, the solution was centrifuged at a low speed for 10 s. The PE tube was then placed in a water bath at a constant temperature of 39 °C and incubated for 10 min. After the reaction, 50 μL DNA extract (volume ratio = 24:25:1) was added for extraction, the tube was centrifuged at 12,000 rpm for 5 min, and the supernatant was collected. Finally, 20–40 μL of the supernatant was diluted 20–50 times with sterile water or PBS, resulting in a dilution volume that was not less than 100 μL. RPA-LFD technology is based on the principle of RPA amplification. The primers with the biotin marker and the probe with the carboxyl fluorescein (FAM) marker are used to amplify the target nucleic acid so that the final amplification product carries both FAM and biotin marker LFD. The front end is coated with gold nanoparticles with FAM antibody, and the detection line is covered with a biotin antibody. When the reaction solution enters the test strip, the amplification product with FAM and biotin forms a biotin antibody-nucleic acid-gold nanoparticle complex on the detection line through antigen–antibody binding and colour development. There is also a quality control line on the LFD, which is coated with a fixed antibody, that can be directly combined with gold nanoparticles with FAM antibody and coloured on the quality control line to ensure the effectiveness of the test strip.
The ordinary PCR system
A specific primer of Pst, PS4F/PS4R, was used for ordinary PCR detection. The reaction system was as follows: MasterMix 12.5 μL, primers (10 μmol/L) 1 μL each, template 1 μL, and ddH2O 9.5 μL. The amplification program was as follows: 95 °C predenaturation for 3 min; 35 cycles of 95 °C denaturation for 1 min, 67.2 °C annealing for 30 s, and a 72 °C extension for 1 min; extension at 72 °C for 5 min; and storage at 4 °C. A sample of 4 μL was taken for electrophoresis.
Design of the RPA primers and probes
In this study, the Pst134EA_003354 uncharacterized protein was used as the target sequence (GenBank: XM_047941824.1) in conjunction with the characteristics of RPA primers. The primers and probe were designed using the Twist Amp nfo assay design manual guidelines (http://www.twistdx.co.uk). Eight sets of specific RPA primer pairs for Pst were designed based on the Pst134EA_003354 uncharacterized protein sequence (GenBank: XM_047941824.1) of Pst using Primer Premier 3.0 software (Table 4). The standard parameters of RPA primer design were taken into consideration, and primer specificity was considered using primer-BLAST software (http://www.ncbi.nlm.nih.gov). CYR32 was selected as the source of Pst. DNA containing specific fragments of Pst was amplified by PCR, and the PCR stock solution of Pst was sequenced by amplification. The PCR stock solution was sequenced and analysed utilizing the NCBI database. After initial screening by conventional PCR and RPA, one optimally performing primer set (PS-RPA-F/PS-RPA-R) that consistently amplified an ~ 300 bp specific region was selected. The reverse primer (PS-RPA-R) was conjugated with the antigenic biotin molecule at the 5′ end. DNAMAN V6 software was used to compare the sequences to confirm the accuracy of the products and design the probes. The corresponding TwistAmp nfo probe (PS-LF-Probe) was designed by modifying the 31st nucleotide with a base analogue tetrahydrofuran (THF) residue and the 5ˈ terminus labelled with FAM, whereas the 3′ terminus was designed to contain a C3-spacer polymerase extension blocker. Shanghai Sangon Biological Engineering Co., Ltd. performed the fragment design probe, sequencing, and primer design.
RPA-specific detection of Puccinia striiformis f. sp. tritici
In this study, the physiological races CYR32, CYR33, and CYR34 of Pst were selected. These are currently the main common physiological Pst species in wheat production in China, and the above physiological races have been identified via differential Chinese hosts of Pst that consisted of 19 wheat varieties, with the isolates identified being Puccinia striiformis f. sp. tritici; Puccinia graminis Pers tritici Eriks and E Henn; Puccinia recondita; Blumeria graminis f. sp. Tritici; Puccinia striiformis West. f. sp.; and Puccinia recondita. f. sp. agropyri. Wheat gDNA served as the control bacteria, and water was used as the negative control. PS-RPA-F and PS-RPA-R primers and the probe PS-LF-Probe were used to test the specificity of RPA, which was verified by conventional PCR.
RPA-sensitivity testing of Puccinia striiformis f. sp. tritici
A 1 μL sample of CYR33 gDNA was taken, and a tenfold serial dilution was conducted to obtain the appropriate concentration as follows: 100 ng/μL, 10 ng/μL, 1 ng/μL, 100 pg/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL, 10 fg/μL, 1 fg/μL, and ultimately, 100 ag/μL. The RPA and an ordinary PCR test were performed, and the results were compared.
Detection of Puccinia striiformis f. sp. tritici isolates
Wheat volunteer (self-sown) seedlings and late-autumn seedlings were collected in the Datong, Huzhu, Ledu, Jianzha, Hualong and Gui-De Counties of Qinghai Province. Pst had occurred in the standard samples. Obvious Pst urediospores appeared on some of the leaves, but the physiological races of the standard samples were not yet determined. Therefore, RPA and PCR analyses were performed to assess the presence of mixed fungi.
The practicality of the RPA testing system
Mingxian 169 wheat leaves were inoculated with CYR34, and samples were taken after 24 h of humidification culture at 5–10 °C between inoculations. After 1–8 days, leaves were collected to extract DNA. Historically, Gui-De, Xunhua, Hualong, and Minhe counties have frequently suffered from wheat stripe rust outbreaks38. The diseased plants were marked in late November 2020, and the marked seedling leaves were collected at the end of March 2021, when no Pst uredospores had appeared on the surface of the leaves. Twenty-one leaf samples were collected for DNA extraction and stored at − 80 °C. RPA detection and conventional PCR detection methods were used to detect whether the samples were infected with Pst.
Ethical approval
Our study complies with relevant institutional, national, and international guidelines and legislation. The use of plants in this study, whether cultivated or wild, as well as the collection of plant material, were in line with the policies of the relevant institutions and with national and international norms and laws.
Data availability
All data generated or analysed during this study are included in this published article.
References
Chai, Y., Pardey, P. G., Hurley, T. M., Senay, S. D. & Beddow, J. M. A probabilistic bio-economic assessment of the global consequences of wheat leaf rust. Phytopathology 110, 1886–1896 (2020).
Shiferaw, B. et al. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 5, 291–317 (2013).
Boamah, S. M. et al. The role of Trichoderma species in plant’s response to salt stress. Asian J. Res. Crop Sci. 6, 28–43 (2021).
Hovmoller, M. S. Disease severity and pathotype dynamics of Puccinia striiformis f. sp tritici in Denmark. Plant Pathol. 50, 181–189 (2010).
Chen, W. et al. Field production, germinability, and survival of Puccinia striiformis f. sp. tritici teliospores in China. Plant Dis. 105, 2122–2128 (2021).
Longchar, B., Phukan, T., Yadav, S. & Senthil-Kumar, M. An efficient low-cost xylem sap isolation method for bacterial wilt assays in tomato. Appl. Plant Sci. 8, e11335 (2020).
Chakdar, H. et al. noxB-based marker for Alternaria spp.: A new diagnostic marker for specific and early detection in crop plants. 3 Biotech. 9, 249 (2019).
Pan, J. J. et al. Quantification of latent infections of wheat stripe rust by real-time PCR. Acta Phytopathol. Sin. 40, 504–510 (2010).
Aggarwal, R., Sharm, S., Manjunatha, C., Gupta, S. & Singh, V. K. Development and validation of loop mediated isothermal amplification based detection assay for Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Australas. Plant Pathol. 46, 577–583 (2017).
Shan, W., Chen, S. & Li, Z. Rapid analysis of epidemic races of Puccinia striiformis f. sp. tritici in China. Sci. Agric. Sin. 28, 1–7 (1995).
Xi, L. et al. Evaluation of resistance to stripe rust and molecular detection of resistance gene(s) in 243 common wheat landraces from the Yunnan Province. Sci. Agric. Sin. 54, 684–695 (2021).
Ma, L. et al. Point-of-care diagnostic assay for rapid detection of porcine deltacoronavirus using the recombinase polymerase amplification method. Transbound. Emerg. Dis. 66, 546–551 (2019).
Cao, Y. et al. Rapid and visual detection of milk vetch dwarf virus using recombinase polymerase amplification combined with lateral flow strips. Virol. J. 17, 102 (2020).
Chao, X., Liang, L., Jin, W. J. & Wan, Y. S. Recombinase polymerase amplification (rpa) of camv-35s promoter and nos terminator for rapid detection of genetically modified crops. Int. J. Mol. Sci. 15, 18197–18205 (2014).
Priti, J. S., Baranwal, V. K., Dietzgen, R. G. & Ghosh, A. A rapid field-based assay using recombinase polymerase amplification for identification of Thrips palmi, a vector of tospoviruses. J. Pest Sci. 94, 219–229 (2021).
Gamze, B. et al. Genome-informed recombinase polymerase amplification assay coupled with a lateral flow device for in-field detection of Dickeya species. Plant Dis. 104, 2217–2224 (2020).
Lu, X. et al. A Rapid, equipment-free method for detecting Phytophthora infestans in the field using a lateral flow strip-based recombinase polymerase amplification assay. Plant Dis. 104, 2774–2778 (2020).
Kalantar, M. M. H., Masoud, S., Ali, K. & Pooria, G. A review of the current isothermal amplification techniques: Applications, advantages and disadvantages. J. Glob. Infect. Dis. 3, 293–293 (2011).
Tan, M. et al. Recent advances in recombinase polymerase amplification: Principle, advantages, disadvantages and applications. Front. Cell. Infect. Microbiol. 12, 1019071 (2022).
Arif, M., Busot, G. Y., Mann, R., Rodoni, B. & Stack, J. P. Field-deployable recombinase polymerase amplification assay for specific, sensitive and rapid detection of the US select agent and toxigenic bacterium, Rathayibacter toxicus. Biology (Basel) 10, 620 (2021).
Fan, X. et al. Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 11, 1696 (2020).
Sow, D. et al. Molecular diagnosis of urogenital schistosomiasis in pre-school children, school-aged children and women of reproductive age at community level in central Senegal. Parasit. Vectors 16, 43 (2023).
Li, J., Macdonald, J. & Von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 144, 31–67 (2019).
Zou, Y., Mason, M. G. & Botella, J. R. Evaluation and improvement of isothermal amplification methods for point-of-need plant disease diagnostics. PLoS One 15, e0235216 (2020).
Ma, L. et al. Establishment of a Real-Time recombinase polymerase amplification assay for the detection of Avian Reovirus. Front. Vet. Sci. 7, 551350 (2020).
Ma, Z. M. et al. Establishment of RT-RPA for citrus yellow vein clearing virus (CYVCV) detection. Sci. Agric. Sin. 54, 3241–3249 (2021).
Qin, J. C. Pathogen Identification of Broad Bean Virus and Rapid Detection Technology of Common Bean Mosaic Virus (Yangzhou University, 2021).
Duan, Y. et al. Detection of citrus leaf blotch virus by reverse transcription recombinase polymerase amplification (RT-RPA). Sci. Agric. Sin. 54, 1904–1912 (2021).
Ju, Y. L. et al. Development and evaluation of recombinase polymerase amplification for the detection of Gaeumannomyces graminis var. tritici. Plant Prot. 46, 150–155+180 (2020).
Ju, Y. L. et al. Development and application of Rc-RPA-LFD for the rapid detection of Rhizoctonia cerealis. Acta Phytopathol. Sin. 50, 618–621 (2020).
TwistDx, Inc. PCR primers work using standard RPA reagents. Google Scholar (2017).
Pan, Y., Gu, Y. L., Luo, Y. & Ma, Z. H. Establishment and application of duplex real-time PCR quantitative determination method on latent infection of wheat stripe rust. Acta Phytopathol. Sin. 46, 485–491 (2016).
Wei, M., Qian, T., Zhao, W., Li, G. & Kong, J. Rapid detection of Pseudomonas syringae pv. tomato by RPA method. Plant Prot. 42, 150–153 (2016).
Zhao, Y. Y. Establishment of LAMP detection method for Pythium arrhenomanes and RPA assay for Pythium aphanidermatum (Nanjing Agricultural University, 2018).
Shen, P. F. Rapid Detection of Ralstonia solanacearum Using Recombinase Polymerase Amplification and Biocontrol Effect of XCS1 Against R. solanacearum (Anhui Agricultural University, 2020).
Li, J. J., Xiong, C., Liu, Y., Liang, J. S. & Zhou, X. W. Loop-mediated isothermal amplification (LAMP): Emergence as an alternative technology for herbal medicine identification. Front. Plant Sci. 7, 1956 (2016).
Wang, Y. et al. Molecular characterization and RNAi-based functional analysis of obstructor family genes in Locusta migratoria. Acta Phytopathol. Sin. 48, 73–82 (2015).
Yao, Q. Study on the Epidemic Rule of Wheat Stripe Rust in Qinghai Province (Northwest Agricultural and Forestry University, 2018).
Acknowledgements
The authors are grateful for the critical review of the manuscript by Meinan Wang.
Funding
This project was partially funded by the National Key R&D Program of China (2021YFD1401000), the National Natural Science Foundation of China (31960516), and the Key R&D and Transformation Program of Qinghai Province (2022-NK-125).
Author information
Authors and Affiliations
Contributions
Y.L. and J.H. executed the experiments. Q.G. and J.Y. analyzed the bioinformatic data. Y.L. and Q.Y. wrote the manuscript and prepared Figs. 1, 2, 3, 4, 5, 6 and Tables 1, 2, 3, 4. All authors reviewed the manuscript and agreed to publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, Y., Hao, J., Guo, Q. et al. Establishment of a recombinase polymerase amplification detection method for Puccinia striiformis f. sp. tritici. Sci Rep 13, 16133 (2023). https://doi.org/10.1038/s41598-023-42663-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-42663-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.