Abstract
Four pathogenic bacterial species of the genus ‘Candidatus Liberibacter’, transmitted by psyllid vectors, have been associated with serious diseases affecting economically important crops of Rutaceae, Apiaceae and Solanaceae families. The most severe disease of citrus plants, huanglongbing (HLB), is associated with ‘Ca. Liberibacter asiaticus’ (CaLas), ‘Ca. Liberibacter americanus’ (CaLam) and ‘Ca. Liberibacter africanus’ (CaLaf), while ‘Ca. Liberibacter solanacearum’ (CaLsol) is associated with zebra chip disease in potatoes and vegetative disorders in apiaceous plants. Since these bacteria remain non-culturable and their symptoms are non-specific, their detection and identification are done by molecular methods, mainly based on PCR protocols. In this study, a new quantitative real-time PCR protocol based on TaqMan probe, which can also be performed in a conventional PCR version, has been developed to detect the four known phytopathogenic species of the genus Liberibacter. The new protocol has been validated according to European Plant Protection Organization (EPPO) guidelines and is able to detect CaLas, CaLam, CaLaf and CaLsol in both plants and vectors, not only using purified DNA but also using crude extracts of potato and citrus or psyllids. A comparative analysis with other previously described qPCR protocols revealed that this new one developed in this study is more specific and equally or more sensitive. Thus, other genus-specific qPCR protocols have important drawbacks regarding the lack of specificity, while with the new protocol there was no cross-reactions in 250 samples from 24 different plant and insect species from eight different geographical origins. Therefore, it can be used as a rapid and time-saving screening test, as it allows simultaneous detection of all plant pathogenic species of ‘Ca. Liberibacter’ in a one-step assay.
Similar content being viewed by others
Introduction
The genus Liberibacter comprises Gram-negative bacteria belonging to the Rhizobiaceae family (Class: Alphaproteobacteria, order: Rhizobiales)1,2, whose niche is confined to the phloem of host plants and haemolymph and salivary glands of insect vectors that can transmit them. The main vectors of these bacteria are phloem sap-feeding psyllids species (Hemiptera: Psylloidea), in which they multiply in a persistent, circulative form3,4,5,6,7. Most of the species of this genus are associated with important plant diseases such as huanglongbing (HLB), the most devastating citrus disease worldwide, the zebra chip of Solanaceae and vegetative disorders of Apiaceae. A remarkable aspect about these bacteria is the fact that they have not been cultured on laboratory media so far8,9. Indeed, only one species in the genus, Liberibacter crescens, that is non-pathogenic, has been cultivated10. The species 'Candidatus Liberibacter asiaticus' (CaLas), 'Candidatus Liberibacter africanus' (CaLaf)1 and 'Candidatus Liberibacter americanus' (CaLam)11 are associated to HLB. Almost all commercial citrus species and cultivars, regardless of rootstocks, are susceptible to this disease3,12.
Typical symptoms of HLB are rapid tree decline, including yellow shoots with blotchy mottled leaves, lopsided fruits with color inversion, aborted seeds, leaf and fruit drop and shoot dieback; the disease causes tree death within a few years3,13, producing billions of dollars of economic losses worldwide annually14.
HLB is distributed by Asia, America, Oceania and Africa15 and it is transmitted mainly by Diaphorina citri (Hemiptera: Liviidae) in America and Asia11,16,17 and Trioza erytreae (Hemiptera: Triozidae) in North Africa7,18. Europe is free of the disease, but the vector T. erytreae is already present in Spain and Portugal19,20. Very recently, other country of the Mediterranean basin, Israel, has reported the presence of D. citri in its territory21.
The species 'Candidatus Liberibacter solanacearum' (CaLsol)22 is associated with diseases or vegetative disorders in strategic crops, such as potato, tomato, pepper, celery, parsnip and carrot23,24,25,26. The symptoms are dependent of the host, but the most characteristics are leaf chlorosis and discoloration, wilting, twisted stems, swollen nodes and aerial tubers7,27,28, abnormal production of leaves and root sprouts, size reduction and other vegetative disorders25,29,30,31. In short, the commercial quality of the products is devalued, and the economic consequences are important32. CaLsol is distributed by America, Europe, North Africa, Israel and New Zealand. The vectors of CaLsol are Bactericera cockerelli in America, Trioza apicalis in North Europe33 and Bactericera trigonica in South Europe30,31 and North Africa34.
There are no curative treatments for diseases caused by 'Ca. Liberibacter' species (CaLspp). Early and specific detection is essential to prevent the entry of these pathogens into a disease-free zone and limit the dispersion in infected zones. The European and Mediterranean Plant Protection Organization (EPPO) have established standard diagnostic protocols (PM 7) for the molecular detection of HLB associated bacteria35,36 and CaLsol37. In both cases, real-time PCR methods are recommended for the analysis of vectors and symptomatic and asymptomatic plants35,36,37. These methods are more sensitive, specific, and faster than conventional PCR protocols previously described1,22,38,39,40,41, and the results are more reproducible42,43. In the recently revised PM 7/121 (2) diagnostic protocol for HLB36, two screening tests are recommended: the real-time PCR adapted from Li et al.44, and a conventional duplex PCR adapted from Teixeira et al.11 and Hocquellet et al.45, whereas 'Candidatus Liberibacter' spp. universal real-time PCR by Bertolini et al.46 has been removed as it produces false-positive results36. However, recent works have demonstrated that both real-time PCRs by Bertolini et al.46 and Li et al.44, although with a good sensitivity, produced undesired amplifications in environmental samples due to their highly conserved nature of the 16S rRNA gene used as target47. Other universal real-time PCR protocols, such as Ananthakrishnan et al.48, based on other genes, have been described but not validated according to EPPO requirements49. The removal of a universal real-time PCR from diagnostic protocols, such as that of Bertolini et al.46, implies the loss of a rapid screening test (RST) that detect all 'Candidatus Liberibacter' species, including CaLsol, and does not require DNA purification. Therefore, in the present study, our aim was to develop a new highly specific, sensitive and validated qPCR protocol for accurate detection of 'Ca. Liberibacter' pathogenic species. This protocol could be used as a new genus specific RST tool, suitable for a first screening, as it allows simultaneous detection of all phytopathogenic species of ‘Ca. Liberibacter’ in a single step assay. Moreover, it could be used for plant and insect vector sampling with or without DNA extraction, reducing cost and time.
Results
Primer and probe design
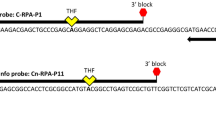
Primers and probe sequences were designed on a highly conserved region corresponding to the housekeeping gene of the RNA polymerase beta subunit, rpoB, with a single copy gen in the genome. The sequence alignment of rpoB gene from the nine whole genomes of CaLspp bacteria used in this study showed an Alignment Identity Percentage (AIP) of 91.2%. Additionally, another alignment of 106 sequences of rpoB gene region of CaLspp available in NCBI database was done. A graphical representation of the alignment performed with 64 sequences, including two non-pathogenic CaLspp species ('Ca. Liberibacter ctenarytaina' (CaLct) and 'Ca. Liberibacter brunswickensis' (CaLbru)), is shown in Supplementary Fig. S1. Two primers were designed which amplify a region of 156 bp located in the 5’ end of the rpoB gene (RefSeq genomic positions 28,333 to 28,178, accession number NC_012985): CaL_rpoB-F (5’-CCT GYA AAC CYT CAT TAG GAC G-3’) and CaL_rpoB-R (5’-TTG TGT TCA ATG GTC TCG GGC GTG- 3’). Primer pairwise identity in the alignment was 96.9% for CaL_rpoB-F and 98.8% for CaL_rpoB-R. Furthermore, a specific TaqMan probe was designed: CaL_rpoB-P (5’-FAM-AGA TCA GGT ATG TCA ATT ATC TCA GG-ZNA4-BHQ1-3’). Probe pairwise identity in the alignment was 99.9%. In order to improve the affinity to the targets and adjust the melting temperature, a ZNA modification (Zip Nucleic Acids, Metabion, Germany) was included in 3’ end.
Validation of the new qPCR protocol
Analytical specificity
Results of new PCR in conventional version for the detection of CaLspp species (CaLas, CaLam, CaLaf and CaLsol) in different hosts (Solanum tuberosum, Daucus carota, Citrus spp., Vinca sp., Bactericera trigonica, Trioza apicalis and Diaphorina citri) from several origins (Brazil, Spain, Costa Rica and Finland) showed amplification of a single fragment with a size of 156 bp, and non-specific amplifications were not observed.
Amplification products from positive samples were sequenced. The identity with CaLspp was confirmed using BLAST. CaLaf, CaLam, CaLas and CaLsol infected samples have an AIP ranged 97.06% to 100% with the sequences GenBank accession numbers: CP004021.150, CP006604.151, CP010804.252, CP002371.153 and EU834131.140 respectively. New obtained partial rpoB gene sequences were submitted to GenBank database under the following GenBank accession numbers: OM831078 to OM831100.
The new real-time qPCR protocol designed in this study was able to detect CaLas, CaLaf, CaLam and CaLsol on a wide range of samples from different origins and hosts. Specifically, the new protocol was able to detect CaLas in 35 citrus samples from Brazil and Costa Rica, and in 10 samples of the vector D. citri. Also was able to detect CaLam in 11 citrus samples from Brazil and from the plant collection of National Research Institute for Agriculture, Food and the Environment (INRAE), and in eight samples of D. citri. CaLaf was detected in one plant sample (Vinca sp.) from the plant collection of INRAE. CaLsol was detected in 39 Daucus carota (including seeds), S. tuberosum and Cuscuta campestris samples from Spain and Finland; and in 14 samples of the insect vectors T. apicalis and B. trigonica.
The positive samples were confirmed by previously validated qPCR protocols25,44,54. All of them were positive by new protocol, with Cq \(\left( {{\overline{\text{X}}}_{{{\text{Cq}}}} \pm {\text{SE}}} \right)\) which ranged from 23.0 ± 2.9 to 33.2 ± 4.2 (data not shown). No amplification was detected in the 105 non-target plant and insect samples analyzed, including the strain BT-1 of L. crescens and those samples that gave undesired amplifications with other PCR protocols, as described in Morán et al.47.
Limit of detection (LOD) and standard curve
Absolute quantification by the qPCR protocol designed in this study was performed analyzing known amounts of dsDNA target using serial dilutions of gBlocks (Integrated DNA Technologies, Inc., USA). The results of the linear regression analysis were used to calculate the equation of the standard curve, as the average of the three replicates examined, obtaining a slope of -3.4358 and a correlation coefficient (R2) of 0.9991 (Fig. 1), with an efficiency of 95.5%. The LOD, corresponding to the lowest amplified concentration, was 6 copies µl−1.
Analytical sensitivity
Total DNA purified from selected CaLas- or CaLsol-infected samples of C. sinensis (CC140, CC116 and CC114) and S. tuberosum (PA), respectively, were quantified by the new real-time qPCR protocol. Results showed that the samples contained 299.8 (CC140), 178.5 (CC116), 149.0 (CC114) and 275.7 (PA) copies·µl−1 of target bacteria. These samples were used to prepare spiked crude extracts of C. lemon, C. reticulata and S. tuberosum.
The results of analytical sensitivity of the new qPCR designed are shown in Table 1. CaLas was detected in all direct samples of citrus extract, except for the most concentrated one (1/2). CaLsol was detected in all potato extract samples. The average of Cq values \(\left( {{\overline{\text{X}}}_{{{\text{Cq}}}} \pm {\text{SE}}} \right)\) obtained at the limit of detection in C. lemon, C. reticulata and S. tuberosum spiked crude extracts were 37.2 ± 0.2, 34.8 ± 0.0 and 35.1 ± 0.1, corresponding to 4.5, 3.7 and 6.9 copies·µl−1, respectively (Table 1). CaLsol was detected with Cq values ranging between 30.4 ± 0.2 and 35 ± 0.1 (Table 1).
Repeatability and reproducibility
Both parameters, repeatability and reproducibility, were evaluated in the context of a Test Performing Study (TPS 2020 HLB, data unpublished) organized by the European Union Reference Laboratory (EURL) for Bacteriology in the field of Plant Health. Evaluation was performed by six different laboratories, which analyzed 20 DNA samples from CaLas, CaLam, CaLsol and X. citri pv. citri infected and healthy plants with different protocols. The participating laboratories had sufficient technical expertise for the diagnostic tests. Results showed repeatability and reproducibility of 100% for the new real-time qPCR protocol designed, obtaining agreement on all samples from all TPS participants.
None of the six laboratories obtained amplifications from healthy plant samples, while for ‘Ca. Liberibacter’ spp. infected plants they obtained mean Cq values and standard error ranging from 21.84 ± 7 to 32.81 ± 1.20. The coefficient of variation obtained was less than 0.06 in all samples (Table 2).
Comparison of the new qPCR real-time with other PCR protocols and evaluation in random sampling survey
The new real-time qPCR designed for the detection of all ‘Ca. Liberibacter’ pathogenic species was compared with two real-time PCR protocols previously described by Bertolini et al.46 and Ananthakrishnan et al.48, both based in TaqMan probes. For this comparison, direct samples, DNA purifications from C. sinensis cv. Valencia infected with CaLas (sample CC123) and S. tuberosum infected with CaLsol (sample PB) diluted in healthy plant material were analyzed (Fig. 2).
Relative sensitivity (average of Cq values) of the new real-time qPCR protocol designed in this study, performed with both direct samples (solid texture) and DNA purifications (dotted texture), in comparison with another two real-time PCR protocols described by Ananthakrishnan et al.48 and Bertolini et al.46. (A) CaLas-infected Citrus sinensis cv. Valencia sample (CC123) diluted in healthy plant material. (B) CaLsol-infected Solanum tuberosum sample (PB) diluted in healthy plant material. Bars represent standard deviations.
Comparative results of relative sensitivity showed that, in both C. sinensis and S. tuberosum samples, the mean Cq values obtained in all dilutions tested were higher in the direct samples than in the DNA purifications. In the case of direct C. sinensis samples (Fig. 2A left), the new real-time qPCR revealed mean Cq values ranging from 27.5 ± 0.3 to 37.7 ± 0.5, similar to the values obtained by Bertolini et al.46 (26.2 ± 0.4 and 35.1 ± 0.5) and earlier than those obtained using the protocol described by Ananthakrishnan et al.48 (28.8 ± 0.8 and 39.5 ± 1.1). For direct S. tuberosum samples (Fig. 2B left), the values obtained with the new real-time qPCR protocol ranged from 31.9 ± 0.6 to 34 ± 0.4, again similar to those obtained using Bertolini et al.46 (from 32 ± 0.4 to 34.9 ± 0.1), and earlier than those obtained using Ananthakrishnan et al.48 (from 35.5 ± 0.3 to 38.7 ± 0.5). For purified DNAs, the new real-time PCR protocol showed mean Cq values from 21.5 ± 0.5 to 31.8 ± 0.3 in C. sinensis (Fig. 2A right), again like those obtained by Bertolini et al.46 (from 21.1 ± 0.7 to 31.3 ± 0.6) and better than those obtained by Ananthakrishnan et al.48 (from 24.5 ± 0.7 to 37.1 ± 0.4) except for potato sample PB, where only Bertolini was able to detect the target at dilution 1:10,000.
And in the case of purified DNA in S. tuberosum (Fig. 2B right), the mean Cqvalues of the new qPCR protocol ranged from 31.9 ± 0.7 to 34.0 ± 0.4, those of Bertolini et al.46 from 32.0 ± 0.5 to 34.9 ± 0.2 and those of Ananthakrishnam et al.48 from 35.5 ± 1.0 to 38.7 ± 0.7.
Relative sensitivity was also comparatively evaluated in direct samples from ten CaLsol-infected B. trigonica specimens (Fig. 3). With the new real-time qPCR, mean Cqvalues ranging from 19.8 to 29.2 were obtained, with the protocol of Li et al.54 from 19.7 to 30.6, and with that of Teresani et al.25 later values from 26.1 to 35.1. In four of the ten insect samples, the Cqvalues reached by the new real-time protocol were earlier than those with the other two protocols described above, while in the other six samples it was Li et al.54 that reached earlier Cqvalues (Fig. 3). However, it should be noted that, due to technical limitations, this comparison could only be made with 10 specimens, so the conclusions that can be drawn from these results are preliminary.
Relative sensitivity (average of Cqvalues) of the new real-time qPCR protocol designed in this study, performed with direct samples of ten specimens of Bactericera trigonica infected with ‘Candidatus Liberibacter solanacearum’, in comparison with real-time PCRsprotocols described by Teresani et al.25 and Li et al.54.
Discussion
A genus-specific tool to detect pathogenic ‘Ca. Liberibacter’ bacterial species is essential for the management of important plant diseases, such as HLB, the most devastating disease affecting Citrus spp., associated with CaLas, CaLam and CaLaf, and Zebra Chip, an economically important disease of potato associated with CaLsol. Some genus-specific real-time PCR protocols were previously published46,48,55; however, several papers have highlighted the high risk of obtaining non-specific amplification in non-target samples19,43,47. Accurate early detection is imperative to avoid decisions based on false positive and false negative results, which can have very detrimental consequences for the agriculture and related sectors.
In the present study, we have designed, developed and extensively assessed a new accurate one-step qPCR protocol that could be adopted to facilitate the diagnostic work of the important diseases associated with ‘Ca. Liberibacter’ bacterial species.
Moreover, it can detect the target bacteria directly from the plant material without the need for DNA extraction, which reduces the risk of contamination, saves time, decreases the cost per reaction and allows processing a large number of samples. Moreover, the protocol also works in conventional PCR version.
The target region of the new protocol designed is located in the RNA polymerase β subunit gene (rpoB), a monocopy gene with a ranging size from 4140 to 4197 bp for the four pathogenic species of CaLspp analyzed in this study. This housekeeping gene is highly conserved, and it is involved in the transcription process and the regulatory pathways that control gene expression in all living organisms56,57. In addition, rpoB gene has been demonstrated to be useful in providing resolution within groups of closely related bacteria and may refine phylogenetic identification of bacteria58. Most of the CaLspp qPCR protocol are targeted on the 16S rRNA gene46,55. This gene had been the most used gene for primers development in detection of CaLspp1,22,25,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,59,60,61,62,63, because it is a highly conserved gene for cell functions64 and there are three copies presented in the genome of CaLspp bacteria, which increase the PCR sensitivity14,65. However, although the sensitivity of the16S rRNA-based protocols is very high, due to the targeted gene copy number, several papers have reported non-specific amplifications19,43,47.
Regarding the sensitivity of the new qPCR protocol, the standard showed a high efficiency (95.5%) with a high correlation coefficient (R2 = 0.9991). Additionally, it showed the better LOD, 6 synthetic DNA copies µl−1, among the current available protocols, which set their limit in 10–2048,54,55,63,65. The efficiency, much higher than, for example, Ananthakrishnan et al.48 (E CaLas = 82.3%, E CaLam = 78.2% and E CaLaf = 77.8%), is only surpassed by the protocol designed by Orce et al.55 (E = 98.0%), but the advantage of new qPCR method is that it has been tested on many more matrices, and can also be adapted to conventional PCR format.
The specificity of the protocol developed in this study was widely tested for inclusivity, with samples naturally infected with CaLsol or the three bacterial species associated with HLB, and for exclusivity, with non-targeted samples of host plants. This is the first time that a set of universal primers and probe for CaLspp phytopathogenic species detection is tested with almost all its known plant and insect hosts. Orce et al.55 tested their new designed qPCR set of primers in symptomatic citrus tissues and some citrus-related pathogens, such as Xanthomonas citri subsp. citri among others. Ananthakrishnan et al.48 tested their primers and probe with CaLsol infected plants, psyllids, HLB-associated species and eight endophytic bacteria: Paenibacillus glucanolyticus, Microbacterium sp., Pantoea agglomerans, Pseudomonas sp., Enterobacter cloacae, Rhizobium sp., Agrobacterium tumefaciens, Sinorhizobium sp., and the two citrus pathogens Xanthomonas axonopodis pv. citri and X. axonopodis pv. citri Aw; and they did not find unspecific amplifications.
Recently, a paper by Morán et al.47 focused on the unspecific amplifications obtained in citrus plant and insect samples, revealing that almost 10% of the samples amplified by Bertolini et al.46 were, in fact, non-infected; besides, authors also found cross reactions in samples by using the protocol developed by Li et al.44. Some of these samples, which show unspecific amplifications, described by Morán et al.47 were analyzed in the present study, and the result by the new qPCR protocol was negative, providing evidence of the high specificity of the new designed primers and probe. After sequencing, Morán et al.47 found that unspecific amplifications obtained in the samples, which have also been used in the present study, were due to cross reactions with Asaia sp. in T. dryi, Rhizobium sp. and Sphingomonas sp. in M. koenegii, Sphingomonas sp. and Phyllobacterium sp. in Citrus spp., and uncultured bacterium and Sodalis sp. in T. erytreae. All these microorganisms are associated to plants and/or insects, and share habitat with CaLspp47,66,67,68,69,70; for this reason, it is important to have specific detection methods for CaLspp that do not amplify other microorganisms that share the same niche, causing unspecific cross-reactions.
Since PCR inhibition due to plant or insect tissues may affect sensitivity, different experiments were carried out in the present study to evaluate the selectivity of the new qPCR method. CaLas was detected in the most spiked dilutions of citrus plants, both in plant extract material and DNA purification, obtaining better results using nucleic acid purifications. This fact suggests that citrus tissues may present PCR inhibitors, which can be easily avoided diluting the sample, as recommended by EPPO for other pathogens, such as Xylella fastidiosa71. In the case of CaLsol detection, also the new qPCR protocol was able to detect it even when the DNA was not extracted, directly from potato crude extract samples. The new qPCR method reached higher sensitivity in potato DNA purification than in their corresponding direct samples. This fact evidenced the existence of PCR inhibitor components in potato tissue which could affect the reaction efficiency. For this reason, it is advisable to perform DNA purification for the detection of CaLsol in asymptomatic potato tissues infected with low titers of bacteria. And only analyze direct potato samples when they show symptoms35,36,37,71.
All these data suggest that this new method could detect the target organisms directly from plant material without DNA purification with acceptable efficiency (LOD 3.7–6.9 copies µl−1 in spiked samples). The use of the method without DNA extraction reduces the risk of contamination, saves time and lowers the cost per sample46,72. Therefore, this variable of the described method would be very useful for large scale surveys. Regarding the DNA extractions, it would be useful for more sensitive analysis of plant material, e.g. in disease-free areas and/or asymptomatic samples.
Quantification cycle values (Cq) were used to relatively compare the sensitivity of the new qPCR protocol with previously designed Ananthakrishnan and Bertolini46,48. Firstly, it was studied the sensitivity of the CaLas detection in the citrus samples, obtaining few differences between methods. It should be highlighted that, in most of cases, the new protocol has a sensitivity comparable to other protocols for CaLspp detection, despite the fact that Bertolini et al.46 protocol, as discussed above, is based on the 16S rRNA gene, which has three copies in the CaLspp genomes14,65, giving it an advantage in sensitivity. Secondly, CaLsol sensitivity was evaluated with potato infected samples, in which Ananthakrishnan method48 showed the worst sensitivity in almost all samples tested. Once again, the new protocol was, in most cases, the most sensitive in terms of CaLspp detection. These comparisons provide further data demonstrating the high sensitivity of the new protocol, opening up the possibility of using it as a more accurate RST tool than the current ones.
In conclusion, the new one-step qPCR TaqMan assay is a specific, sensitive and accurate tool, being able to detect a low number of target copies per microliter. The use of this new protocol could improve and reduce the work in plant health laboratories since it allows the screening of the four phytopathogenic bacteria present in this genus and does not present cross reactions or non-specific amplifications with different host plants or insect species.
Methods
Plant and insect material
Collections of plants and insects were carried out in accordance with national and international legislation in force since it did not require any special permit because they were in open access areas. Likewise, experimental research on plant and insect material was carried out in accordance with national and international legislation in force, as no special permits were required. A total of 250 samples from eight different origins and 24 different plant and insect species were used for analytical specificity of the qPCR method developed in the present study. For the validation of the method, 144 samples of CaLsol-, CaLas-, CaLam- and CaLaf-infected plants and insects were selected as positive controls: 45 CaLas-infected samples from Brazil and Costa Rica; one CaLaf-infected sample from INRAE collection; 19 CaLam-infected samples from Brazil and INRAE collection; and 79 CaLsol-infected samples from Finland and Spain. A total of non-infected 42 plant (of 15 different species) and 36 insect samples (from three species) were used as negative controls.
Twenty-six plant and insect samples previously reported as undesired amplifications by Moran et al.47 and the strain BT-1 of Liberibacter crescens were included in the validation.
In addition, 48 carrot samples randomly collected in commercial plots from Villena (Alicante, Spain) were also included.
The presence of ‘Ca. Liberibacter’ spp. in control samples was confirmed using the real time PCR detection protocols described by Li et al.44 and Bertolini et al.46, in the case of HLB associated CaLspp, and Li et al.54 and Teresani et al.25, in the case of CaLsol.
Sample preparation
Plant crude extracts were prepared by crushing leaves, seeds and tuber potato from each plant sample in extraction bags (Bioreba, Switzerland) with 10 mM phosphate buffered saline (PBS) (8.0 g NaCl; 2.9 g Na2HPO4·12 H2O; 0.2 g KH2PO4; 0.2 g KCl; pH 7.2) at 1/5 (w/v) ratio. DNA was purified from 400 µl of these crude extracts following the CTAB protocol recommended by EPPO35,36,37. Total DNA was quantified by using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, USA).
Regarding insect crude extracts, single specimens were squashed on Whatman paper (GE Healthcare, Europe), according to the procedure developed by Bertolini et al.46. Then, membranes were resuspended with 100 µl of Tween 20 at 0.05%, and the suspension directly analyzed by qPCR.
Both plant and insect direct samples consisted of 1/10 dilutions of crude extracts in PBS. All total purified DNA and direct samples were kept at −20 °C until use.
Primer and probe design
In order to find the most conserved region for the design of primers and probe, nine whole genome sequences, from species of ‘Ca. Liberibacter’ from different hosts and origins, were aligned using the software package MAUVE73: CaLsol (GenBank accession number: NC_014774), CaLas (NC_012985, NC_020549, NZ_AP014595, NZ_CP010804, NZ_CP019958 and NZ_CP029348), CaLam (NC_022793) and CaLaf (NZ_CP00402). The best alignment identity percentages (AIP) in the most conserved region were used. To improve the design of the highly conserved region selected, a second alignment with 106 sequences of this region of Liberibacter species available in NCBI data bases was performed using Clustal W74, implemented in MEGA 7 software75.
Amplification conditions by conventional PCR and quantitative real-time PCR
Conventional PCR amplifications were carried out to test the primers, using 2 U Taq DNA polymerase (Biotools, Spain), 1 µM of each primer, 0.1 mM of each dNTPs, 1.5 mM MgCl2 and 5 µl of DNA template (50 ng), in a total reaction volume of 25 µl. Conventional PCR conditions consisted of a first denaturalization step at 94 °C for 3 min, followed by 40 cycles of amplification (94 °C for 30 s, 63 °C for 30 s and 72 °C for 45 s) and a final extension step at 72 °C for 10 min. A Veriti Dx Thermal Cycler (Thermo Fisher Scientific, USA) was used. The amplification products obtained were visualized through 1.5% (w/v) agarose gel electrophoresis in 0.5X TAE buffer (40 mM Tris pH 7.6, 20 mM CHsCOOH, 1 mM EDTA) with Good View™ staining (SBS Genetech Co., Ltd., China).
Quantitative real-time PCR (qPCR) reaction mix optimized using GoTaq® DNA Probe qPCR Master Mix (Promega Corporation, USA) consisted of 500 nM of each primer (CaL_rpoB-F and CaL_rpoB-R), 80 nM of TaqMan probe (CaL_rpoB-P) and 5 μl of DNA template (50 ng) or crude extract as template in a total volume of 25 µl. To normalize fluorescent signals between wells, 30 µM of CXR reference dye was added to the Master Mix. The conditions of qPCR assay consist of an initial denaturalization step at 95 °C for 10 min, followed by 45 cycles of amplification (95 °C for 15 s and 60 °C for 1 min). A StepOne Plus qPCR thermocycler (Applied Biosystems, USA) and a LightCycler 480 thermocycler (Roche, USA) were used.
Default threshold lines set by the thermocycler software were adjusted slightly above the background noise, according to the manufacturers, to obtain the cycle quantification value (Cq). All samples were analyzed in triplicate.
A selection of 23 PCR products, obtained from different bacterial hosts originating from Brazil, Costa Rica, Finland, Spain, and USA (Table 3), were purified using mi-PCR Purification Kit (Metabion International AG, Germany), and sequenced through Sanger sequencing methods by Macrogen sequencing service (Macrogen Inc., Spain) in both directions (forward and reverse complimentary DNA strands). Sequences were analyzed using MEGA 7 software75.
Validation of the qPCR designed
Validation of the new real-time qPCR protocol developed in this study was performed according to PM 7/98 EPPO guidelines49. The following parameters were evaluated: analytical specificity for inclusivity (detection of CaLspp strains from different origins, hosts and genetic diversity) and for exclusivity (negative detection of relevant non-targets that might be present in the same matrix), selectivity (evaluation of different host species), limit of detection (LOD) and standard curve, analytical sensitivity (maximum dilution of target DNA detected) and repeatability and reproducibility (evaluation of the test performance consistency on different replicates by different operators and with different equipments).
Analytical specificity
It was evaluated for inclusivity and exclusivity with target and non-target samples, respectively. As described above, a total of 250 plant and insect samples from different geographical origins and hosts, including some samples reported previously as undesired amplifications in Morán et al.47, were evaluated using three repetitions per sample (Table 3).
LOD and standard curve
For qPCR standard curve generation, synthetic double stranded DNA commercially named gBlock (Integrated DNA Technologies, USA) with the sequence of a partial region (156 bp) of the rpoB gene from CaLsol whole genome (NC_014774), containing the PCR target designed in this study, was used.
Analytical sensitivity
It was determined testing six serial dilutions (from 1/2 to 1/40), and their corresponding ten-fold dilutions in PBS of spiked crude extract samples of C. lemon, C. reticulata, and S. tuberosum. Thus, non-infected Citrus spp. extracts were spiked with DNA isolated from CaLas-infected citrus samples CC116 and CC144 (Table 3); and non-infected S. tuberosum extracts were spiked with CaLsol-infected potato crude extract sample PA (Table 3). For all cases, three replicates per sample were used, and the absolute quantification was done as described in the above section (LOD and standard curve). The presence of CaLspp bacteria in tissues was confirmed by qPCR25,44,46,54, and quantified by generating standard curves as described above with the new real-time qPCR protocol.
Repeatability and reproducibility
It was evaluated in the context of a Test Performance Study (TPS) organized in 2020 by the Netherlands Institute for Vectors, Invasive plants and Plant health (NIVIP): TPS for the molecular detection of regulated ‘Ca. Liberibacter’ species in C. sinensis and C. reticulata using conventional and real-time PCR tests (TPS 2020HLB). Eight laboratories from different countries (Italy, Belgium, Slovenia, Netherlands, France, Brazil, Spain, and Japan) participated in the TPS. Twenty samples were tested according to the agreement of qualitative reported results of biological duplicates, i.e. positive or negative, under repeatability and reproducibility conditions, per TPS participant or between TPS participants, respectively, according to EPPO standard PM7/76 (5)76.
Comparison with different real-time PCR protocols
On the one hand, the sensitivity of the new qPCR protocol was compared with two real-time PCR protocols for ‘Ca Liberibacter’ spp. detection previously described, based on TaqMan probes, which have as target the 16S rRNA and rpoB housekeeping genes (Bertolini et al.46 and Ananthakrishnan et al.48, respectively). For this comparison, ten-fold serial dilutions (from 10–1 to 10–4) from the samples CC123 (C. sinensis cv. Valencia naturally infected with Calas) and PB (S. tuberosum, naturally infected with CaLsol) were prepared in healthy plant material. On the other hand, the sensitivity of the new qPCR protocol was also compared with two real-time PCR protocols for CaLsol specific detection previously described (Li et al.54; Teresani et al.25) with psyllids. For it, ten psyllid samples of B. trigonica naturally infected with CaLsol were used. Direct samples and DNA purifications were analyzed with three repetitions by the real-time PCR protocols. Average Cq values and standard deviations were used for comparison of the relative sensitivity between the three real-time PCR methods.
Data availability
All the datasets and material generated and analyzed during the current study are available from the corresponding author on reasonable request. The sequences datasets generated and/or analyzed during the current study are available in the GenBank repository (OM831078; OM831079; OM831080; OM831081; OM831082; OM831083; OM831084; OM831085; OM831086; OM831087; OM831088; OM831089; OM831090; OM831091; OM831092; OM831093; OM831094; OM831095; OM831096; OM831097; OM831098; OM831099 and OM831100).
References
Jagoueix, S., Bové, J. M. & Garnier, M. The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of the Proteobacteria. Int. J. Syst. Bacteriol. 44, 379–386 (1994).
Duan, Y. et al. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. MDPI. 22, 1011–1020 (2009).
Bové, J. M. A destructive, newly-emerging, century-old disease of citrus. Plant Pathol. 88, 7–37 (2006).
Gottwald, T. R. Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48, 119–139 (2010).
Capoor, S. P., Rao, D. G. & Viswanath, S. M. Greening disease of citrus in the deccan trap country and its relationship with the vector, Diaphorina citri Kuwayama. In IOCV Conference Proceedings (1957–2010) vol. 6 (California Digital Library (CDL), 1974).
Xu, C. F., Xia, Y. H., Li, K. B. & Ke, C. Further study of the transmission of citrus huanglongbing by a psyllid, Diaphorina citri Kuwayama. IOCV Conference Proceedings (1957–2010) 10, 10 (1988).
Haapalainen, M. Biology and epidemics of ‘Candidatus Liberibacter’ species, psyllid-transmitted plant-pathogenic bacteria. Ann. Appl. Biol. 165, 172–198 (2014).
Parker, J. K. et al. Viability of ‘Candidatus Liberibacter asiaticus’ prolonged by addition of citrus juice to culture medium. Phytopathology 104, 15–26 (2014).
Ha, P. T. et al. Host-free biofilm culture of ‘Candidatus Liberibacter asiaticus,’ the bacterium associated with Huanglongbing. Biofilm. 1, 100005 (2019).
Fagen, J. R. et al. Liberibacter crescens gen. nov., sp. nov., the first cultured member of the genus Liberibacter. Int. J. Syst. Evol. Microbiol. 64, 2461–2466 (2014).
Teixeira, D. C. et al. A new Liberibacter species, ‘Candidatus Liberibacter americanus’, associated with citrus huanglongbing (greening disease) in São Paulo State, Brazil. Int. J. Syst. Evol. Microbiol. 55, 1857–1862 (2005).
Lopes, S. A. et al. Liberibacters associated with citrus huanglongbing in Brazil: ‘Candidatus Liberibacter asiaticus’ is heat tolerant ‘Ca L. americanus’ is heat sensitive. Plant Dis. 93, 257–262 (2009).
Wang, N. & Trivedi, P. Citrus huanglongbing: A newly relevant disease presents unprecedented challenges. Phytopathology 103, 652–665 (2013).
Kim, J. S. & Wang, N. Characterization of copy numbers of 16S rDNA and 16S rRNA of ‘Candidatus Liberibacter asiaticus’ and the implication in detection in planta using quantitative PCR. BMC Res Notes 2, 37 (2009).
EPPO Global Database. https://gd.eppo.int/reporting/article-7262 (2022).
Capoor, S. P., Rao, D. G. & Viswanath, S. M. Diaphoria citri Kuway., a vector of the greening disease of citrus in India. ICAR 37, 572–579 (1967).
Martinez, A. & Wallace, J. Citrus leaf-mottle-yellows disease in the Philippines and transmission of the causal virus by a Psyllid, Diaphorina citri. Plant Dis. 51, 692–695 (1967).
McClean, A. & Oberholzer, P. Citrus psylla, a vector of the greening disease of sweet orange-research note. S. Afr. J. Plant Agric. 8, 297–298 (1965).
Siverio, F. et al. Survey of huanglongbing associated with ‘Candidatus Liberibacter’ species in Spain: analyses of citrus plants and Trioza erytreae. Phytopathol. Mediterr. 56, 98–110 (2017).
Ruíz-Rivero, O. et al. Insights into the origin of the invasive populations of Trioza erytreae in Europe using microsatellite markers and mtDNA barcoding approaches. Sci. Rep. 2021(11), 1–15 (2021).
EPPO Reporting Service No 02 - 2022 Num. article: 2022/032. https://gd.eppo.int/reporting/article-7262.
Liefting, L. W. et al. A new Candidatus Liberibacter species associated with diseases of solanaceous crops. Plant Dis. 93, 208–214. https://doi.org/10.1094/PDIS-93-3-0208 (2009).
Nelson, W. R., Fisher, T. W. & Munyaneza, J. E. Haplotypes of ‘Candidatus Liberibacter solanacearum’ suggest long-standing separation. Eur. J. Plant Pathol. 130, 5–12 (2011).
Nelson, W. R. et al. A new haplotype of ‘Candidatus Liberibacter solanacearum’ identified in the Mediterranean region. Eur. J. Plant Pathol. 135, 633–639 (2013).
Teresani, G. R. et al. Association of ‘Candidatus Liberibacter solanacearum’ with a vegetative disorder of celery in Spain and development of a real-time PCR method for its detection. Phytopathology 104, 804–811 (2014).
Alfaro-Fernández, A., Hernández-Llopis, D. & Font, M. I. Haplotypes of ‘Candidatus Liberibacter solanacearum’ identified in Umbeliferous crops in Spain. Eur. J. Plant Pathol. 149, 127–131 (2017).
Crosslin, J. M., Munyaneza, J. E., Brown, J. K. & Liefting, L. W. Plant management network. PHP https://doi.org/10.1094/PHP-2010 (2010).
Munyaneza, J. E. Zebra chip disease of potato: Biology, epidemiology, and management. Am. J. Potato Res. 89, 329–350 (2012).
Munyaneza, J. E., Lemmetty, A., Nissinen, A. I., Sengoda, V. G. & Fisher, T. W. Molecular detection of aster yellows phytoplasma and ‘Candidatus Liberibacter solanacearum’ in carrots affected by the psyllid Trioza apicalis. Plant Pathol. 93, 697–700 (2011).
Alfaro-Fernández, A. et al. First Report of ‘Candidatus Liberibacter solanacearum’ in carrot in mainland Spain. Plant Dis. 96, 582–582 (2012).
Alfaro-Fernández, A., Siverio, F., Cebrián, M. C., Villaescusa, F. J. & Font, M. I. ‘Candidatus Liberibacter solanacearum’ associated with bactericera trigonica-affected carrots in the canary islands. Plant Dis. 96, 581–581 (2012).
Pierson, E. A. et al. ‘Candidatus Liberibacter’ pathosystems at the forefront of agricultural and biological research challenges. Phytopathology 112, 7–10 (2022).
Munyaneza, J. E. et al. Association of ‘Candidatus Liberibacter solanacearum’ with the psyllid, Trioza apicalis (Hemiptera: Triozidae) in Europe. J. Econ. Entomol. 103, 1060–1070 (2010).
Tahzima, R. et al. First report of ‘Candidatus Liberibacter solanacearum’ on carrot in Africa. Plant Dis. 98, 1426 (2014).
PM 7/121 (1). Candidatus Liberibacter africanus’, ‘Candidatus Liberibacter americanus’ and ‘Candidatus Liberibacter asiaticus. EPPO Bull. 44, 376–389 (2014).
PM 7/121 (2). Candidatus Liberibacter africanus’, ‘Candidatus Liberibacter americanus’ and ‘Candidatus Liberibacter asiaticus. EPPO Bull. 51, 267–282 (2021).
PM 7/143 (1). Candidatus Liberibacter solanacearum. EPPO Bull. 50, 49–68 (2020).
Jagoueix, S., Bové, J. M. & Garnier, M. PCR detection of the two ‘Candidatus’ Liberibacter species associated with greening disease of citrus. Mol. Cell Probes. 10, 43–50 (1996).
Hansen, A. K., Trumble, J. T., Stouthamer, R. & Paine, T. D. A new huanglongbing species, ‘Candidatus Liberibacter psyllaurous’, found to infect tomato and potato, is vectored by the psyllid Bactericera cockerelli (Sulc). Appl. Environ. Microbiol. 74, 5862–5865 (2008).
Liefting, L. W., Weir, B. S., Pennycook, S. R. & Clover, G. R. G. ‘Candidatus Liberibacter solanacearum’, associated with plants in the family Solanaceae. Int. J. Syst. Evol. Microbiol. 59, 2274–2276 (2009).
Ravindran, A., Levy, J., Pierson, E. & Gross, D. C. Development of primers for improved PCR detection of the potato zebra chip pathogen, ‘Candidatus Liberibacter solanacearum’. Plant Dis. 95, 1542–1546 (2011).
Iftikhar, Y., Rauf, S., Shahzad, U. & Zahid, M. A. Huanglongbing: Pathogen detection system for integrated disease management: A review. J. Saudi Soc. Agric. Sci. 15, 1–11 (2016).
Cellier, G. et al. Comparison of the performance of the main real-time and conventional PCR detection tests for ‘Candidatus Liberibacter’ spp, plant pathogenic bacteria causing the Huanglongbing disease in Citrus spp. Eur. J. Plant Pathol. 157, 919–941 (2020).
Li, W., Hartung, J. S. & Levy, L. Quantitative real-time PCR for detection and identification of 'Candidatus Liberibacter’ species associated with citrus huanglongbing. J. Microbiol. Methods. 66, 104–115 (2006).
Hocquellet, A., Toorawa, P., Bové, J. M. & Garnier, M. Detection and identification of the two ‘Candidatus Liberibacter’ species associated with citrus huanglongbing by PCR amplification of ribosomal protein genes of the beta operon. Mol. Cell Probes. 13, 373–379 (1999).
Bertolini, E. et al. Tissue-print and squash real-time PCR for direct detection of ‘Candidatus Liberibacter’ species in citrus plants and psyllid vectors. Plant Pathol. 63, 1149–1158 (2014).
Morán, F. et al. The Challenge of Environmental samples for PCR detection of phytopathogenic bacteria: A case study of citrus huanglongbing disease. Agronomy 11, 10 (2021).
Ananthakrishnan, G. et al. Development of primers and probes for genus and species specific detection of ‘Candidatus Liberibacter’ species by real-time PCR. Plant Dis. 97, 1235–1243 (2013).
PM 7/98 (5). Specific requirements for laboratories preparing accreditation for a plant pest diagnostic activity. EPPO Bulletin. 51, 468–498 (2021).
Lin, H. et al. Complete genome sequence of ‘Candidatus Liberibacter africanus’, a bacterium associated with citrus huanglongbing. Genome Announc. 3, e00733-15 (2015).
Wulff, N. A. et al. The complete genome sequence of ‘Candidatus Liberibacter americanus’, associated with citrus huanglongbing. MDPI 27, 163–176 (2014).
Zheng, Z., Deng, X. & Chen, J. Whole-genome sequence of Candidatus Liberibacter asiaticus’ from Guangdong, China. Genome Announ. 2, e00273 (2014).
Lin, H. et al. The complete genome sequence of ‘Candidatus Liberibacter solanacearum,’ the bacterium associated with potato zebra chip disease. PLoS ONE 6, e19135 (2011).
Li, W. et al. Multiplex real-time PCR for detection, identification and quantification of ‘Candidatus Liberibacter solanacearum´ in potato plants with zebra chip. J. Microbiol. Methods 78, 59–65 (2009).
Orce, I. G. et al. Novel set of real-time PCR primers for simultaneous detection of Liberibacter species associated with citrus huanglongbing. Sci. Agric. 72, 252–259 (2015).
Adékambi, T., Shinnick, T. M., Raoult, D. & Drancourt, M. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int. J. Syst. Evol. Microbiol. 58, 1807–1814 (2008).
Borukhov, S. & Nudler, E. RNA polymerase holoenzyme: structure, function and biological implications. Curr. Opin. Microbiol. 6, 93–100 (2003).
Culbreath, K. D., Simmon, K. E. & Petti, C. A. Application of identification of bacteria by DNA target sequencing in a clinical microbiology laboratory. Mol Microbiol https://doi.org/10.1128/9781555819071.CH2 (2016).
Wen, A. et al. Detection, distribution, and genetic variability of ‘Candidatus Liberibacter’ species associated with zebra complex disease of potato in north America. Plant Dis. 93, 1102–1115 (2009).
Fujikawa, T. & Iwanami, T. Sensitive and robust detection of citrus greening (huanglongbing) bacterium ‘Candidatus Liberibacter asiaticus’ by DNA amplification with new 16S rDNA-specific primers. Mol. Cell Probes. 26, 194–197 (2012).
Beard, S. S. & Scott, I. A. W. A rapid method for the detection and quantification of the vector-borne bacterium ‘Candidatus Liberibacter solanacearum’ in the tomato potato psyllid, Bactericera cockerelli. Entomol. Exp. Appl. 147, 196–200 (2013).
Nissinen, A. I., Haapalainen, M., Jauhiainen, L., Lindman, M. & Pirhonen, M. Different symptoms in carrots caused by male and female carrot psyllid feeding and infection by ‘Candidatus Liberibacter solanacearum’. Plant Pathol. 63, 812–820 (2014).
Park, J. W. et al. A new diagnostic real-time PCR method for huanglongbing detection in citrus root tissue. J. Gen. Plant Pathol. 84, 359–367 (2018).
Clarridge, J. E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17, 840–862 (2004).
Teixeira, D. C. et al. Distribution and quantification of ‘Candidatus Liberibacter americanus’, agent of huanglongbing disease of citrus in São Paulo State, Brasil, in leaves of an affected sweet orange tree as determined by PCR. Mol. Cell Probes. 22, 139–150 (2008).
Schäfer, A. et al. Hemicellulose-degrading bacteria and yeasts from the termite gut. J. Appl. Bacteriol. 80, 471–478 (1996).
Yamada, Y. et al. Asaia bogorensis gen. nov., sp. Nov., an unusual acetic acid bacterium in the alpha-Proteobacteria. Int. J. Syst. Evol. Microbiol. 50, 823–829 (2000).
Favia, G. et al. Bacteria of the genus Asaia: a potential paratransgenic weapon against malaria. Adv. Exp. Med. Biol. 627, 49–59 (2008).
Eleftherianos, L., Atri, J., Accetta, J. & Castillo, J. C. Endosymbiotic bacteria in insects: Guardians of the immune system. Front. Physiol. 4, 46 (2013).
Rasowo, B. A. et al. Diversity and phylogenetic analysis of endosymbionts from Trioza erytreae (Del Guercio) and its parasitoids in Kenya. J. Appl. Entomol. 145, 104–116 (2021).
PM 7/24 (4). Xylella fastidiosa. EPPO Bull. 49, 175–227 (2019).
Barbé, S., Bertolini, E., Roselló, M., Llop, P. & López, M. M. Conventional and real-time PCRs for detection of Erwinia piriflorinigrans allow its distinction from the fire blight pathogen, Erwinia amylovora. AEM 80, 2390–2398 (2014).
Darling, A. C. E., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
PM 7/76 (5). Use of EPPO diagnostic standards. EPPO Bull. 48, 373–377 (2018).
Acknowledgements
The authors would like to thank for the Grants IVIA/52202D–Sostenible–funded by Instituto Valenciano de Investigaciones Agrarias, Generalitat Valenciana, LIFE18 CCA/ES/001109 (Development of sustainable control strategies for citric under threat of climate change & preventing entry of HLB in EU -LIFE Vida for Citrus-) and PID2021-124145OR-C21 (Breeding, selection and evaluation of new citrus materials for more sustainable plantations in the face of emerging threats due to global change -FORTECITRUS-). This research was funded by INIA E-RTA2014-00008-C4-01 and INIA E-RTA2015-00005-C06-01 from the Spanish Government. MQGC is recipient of PhD grant 2017-2020 from Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria. We would like to thank University of Costa Rica-Tropical Disease Research Unit (San José, Costa Rica), Institut National de la Recheche Agronomique, INRAE (Bordeaux) for the CaLaf, CaLas and CaLam positives samples, Anne Nissinen (Institute of Natural Resources of Finland, Luke) for samples of T. apicalis, Servicio de Sanidad Vegetal de la Dirección General de Agricultura del Gobierno de Canarias for allowing the use of its laboratory equipment. Also Dr. L. De La Fuente and E. Naranjo, Auburn University (USA), for kindly providing Liberibacter crescens strain BT-1. And Tim Warbroek and Maria Bergsma-Vlami from Netherlands Institute for Vectors, Invasive plants and Plant health (NIVIP), co-ordinator of the European Union Reference Laboratory for pests of plants on bacteria, for allowing us to cite the results of the TPS regarding the protocol developed in the present study.
Author information
Authors and Affiliations
Contributions
Conceptualization: E.M.-N. and F.M. Methodology: M.Q.; S.B. and F.M. Validation: M.Q.; S.B. and F.M. Formal analysis: M.Q, S.B, F.M. and E.M.-N. Resources: E.M.-N. and F.S. Writing-original draft preparation: M.Q.; S.B. and F.M. Writing-review and editing: M.Q.; S.B.; F.M.; E.B.; F.S. and E.M.-N.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Chaves, M.QG., Morán, F., Barbé, S. et al. A new and accurate qPCR protocol to detect plant pathogenic bacteria of the genus ‘Candidatus Liberibacter’ in plants and insects. Sci Rep 13, 3338 (2023). https://doi.org/10.1038/s41598-023-30345-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-30345-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.