Abstract
The Cryptococcus genus comprises more than 100 species, of which C. neoformans and C. gattii are the leading cause of cryptococcosis. The distribution of C. gattii and C. neoformans species complexes has been extensively studied and widely reported globally. Other species such as Naganishia albida, Papiliotrema laurentii, and Papiliotrema flavescens have been reported as pathogenic yeasts. Since there are no reports of environmental isolation in the Boyacá region (Colombia), this study aimed to isolate and characterize Cryptococcus and Cryptococcus-like yeasts from pigeon feces, Eucalyptus, and olive trees distributed in the municipalities of Tunja and Ricaute Alto. The environmental data was recovered, and the isolations obtained were identified by microscopy, biochemical test, MALDI-TOF MS, URA5-RFLP, and sequencing of the ITS and LSU loci. For the 93 pigeon dropping samples collected in Tunja, 23 yielded to C. neoformans, 3 to N. globosa, 2 N. albida and 1 to P. laurentii. Of the 1188 samples collected from olive trees, 17 (1.43%) positive samples were identified as C. gattii species complex (4), C. neoformans species complex (2), P. laurentii (3), N. albida (2), N. globosa (5) and P. flavescens (1). Likewise, specimens of C. neoformans presented molecular type VNI and molecular type VNII; for C. gattii the molecular types found were VGIII and one VGIV by URA5-RFLP but VGIII by MALDI-TOF and sequencing of the ITS and LSU. Therefore, it can be concluded that the species of Cryptococcus, Naganishia and Papiliotrema genera, are present in the environment of Boyacá, and show a predilection for climate conditions that are typical of this region.
Similar content being viewed by others
Introduction
The most important pathogenic species of the Cryptococcus genus include the Cryptococcus neoformans species complex and the Cryptococcus gattii species complexes, responsible for skin, lung, and, more frequently, central nervous system infections1,2,3. As is the case of cryptococcosis, which worldwide estimates one million positive cases and more than 181.100 deaths from it4, the incidence in Colombia was represented in the last study carried out in 2018 whit an annual incidence of 0.24 × 104 inhabitants and in AIDS patients being 1.1 × 103 inhabitants, considering C. neoformans VNI molecular pattern as the most prevalent (n = 321, 96.1%) and for C. gattii species complex, the most prevalent species was C. deuterogattii (VGII 54.3%), followed by C. bacillisporus (VGIII 32.6%)5 For the department of Boyacá this disease is mostly caused by C.gattii species complex, with an incidence of 3.3%6. The organization of these species concerning to their nomenclature has been established in two hypothesis; the classical that included two species and nine main molecular types, VNI, VNB and VNII (representing C. neoformans var. grubii, serotype A), VNIV (C. neoformans, serotype D) and VNIII (the hybrid of these two species, serotype AD), as well as VGI, VGII, VGIII and VGIV (C. gattii, serotype B or C), and VGII has been subclassified into three associated genotypes: VGIIa, VGIIb, and VGIIc)7. However, differences in genetic and population structure of this yeast are not represented by only two species, and the use the term “species complex”, proposed by Kwon-Chung et al.8, is crucial to represent the diversity of the etiologic agents of Cryptococcosis8. In this regard, the other hypothesis, strongly supported by the phylogenetic studies by Liu et al.9 and Hagen et al.10, 11 suggest the C. neoformans variety grubii serotype A with VNB, VNI and VNII genotypes, C. deneoformans (referred to as C. neoformans var. neoformans with serotype D y genotype VNIV) and a hybrid composed of C. neoformans and C. deneoformans (with serotype AD and genotype VNIII). For the complex of species of C. gattii reorganized as five species: C. gattii with genotype VGI, C. deuterogattii with genotype VGII, C. bacillisporus with genotype VGIII, C. tetragattii with genotype VGIV, and finally C. decagattii with genotype VGIV and the atypical molecular type VGIIIc, initially classified as C. bacillisporus VGIII but after being compared to ISHAM consensus Multilocus sequence typing (MLST) was clustered together with the reference strain C. decagattii. In addition to being hybrids such as C. deneoformans with C. gattii, the hybrid C. neoformans with C. gattii and the hybrid C. neoformans with C. deuterogattii9,10,11,12.
These species present differences in geographic distribution and habitat and have been reported in diverse environments, reflecting the adaptability of these fungi in different parts of the world13. It should be noted that other Cryptococcus-like yeasts, have been reported in human conditions, such as Naganishia liquefaciens (formerly Cryptococcus liquefaciens), which was reported to be the cause of a fungemia in Japan14, Papiliotrema laurentii (formerly Cryptococcus laurentii) related to a case of meningitis15, Naganishia albida (formerly Cryptococcus albidus) reported in a case of fungemia in an immunocompromised child16 and Papiliotrema flavescens (formerly Cryptococcus flavescens) that has been reported in the cerebrospinal fluid of an AIDS patient17. The term “Cryptococcus-like yeasts” denotes a cluster of yeast species that display resemblances in terms of morphological traits with the Cryptococcus genus but these yeast species may not be exclusive to the Cryptococcus genus.
The distribution of the species C. gattii and C. neoformans species complexes has been extensively studied since these generally survive in tropical and subtropical climates. The species C. neoformans has been associated with eucalyptus detritus18 almond tree bark (Terminalia catappa)19 and other species such as Olive trees (Olea europea)20, it can survive and develop thanks to the decomposing organic matter that provides it with the basic nutritional requirements18,19,20,21. Likewise, it is associated with avian excreta, especially pigeons (C. livia); this happens due to the high nitrogen, creatinine, and salts that generate a favorable environment for fungus development. It has been found that feces with low moisture content and low exposure to sunlight are a good reservoir of this species complex22,23,24,25. This species has been recorded in places around the world, such as Malawi26, Nigeria27, China28, South Africa29, Brazil30, United States31, Italy32, Argentina33, 34 and Ecuador35, among others4, 8, 36,37,38,39.
In addition, the main environmental sources of C. gattii species complex is associated to decaying wood from eucalyptus, almond, oak, rubber, olive trees, among others20, 40,41,42, in the same way, it has been found in other environments such as soil, air and water43,44,45.
In the same way different reports associate the presence of C. gattii species complex with regions with temperate climates and periods with higher humidity; however, it is vital to consider the specific climatic conditions of the area since the development of the fungus depends on this46,47,48,49. C. gattii s.l. has been isolated in countries such as Australia50, Africa29, India51, 52, Italy53, United States54 Southern California55, Canada56, Spain9, China28, among other studies39, 40, 47, 52, 57,58,59,60,61.
Environmental isolations of the other Cryptococcus-like yeasts species such as N. liquefaciens (formerly C. liquefacien), P. flavescens (formerly C. flavescens), have been reported in Brazil62, N. albida (formerly C. saitoi) was isolated from the Antarctic soil, indicating that it prevails in cold areas63, 64 and the species of P. laurentii (formerly C. laurentii), Cystofilobasidiales macerans (formerly C. macerans) and N. albida (formerly C. albidus) have been reported in Bogotá city, Colombia65, 66.
Colombia, having a spatial location influenced by the variation of bimodal climatic conditions typical of the tropics, has become, like other countries in the region, a potential area for the spread of these fungi, where species of the C. neoformans complex have been reported in the departments of Cauca, Córdoba, Cundinamarca, Huila, Nariño, Norte de Santander and Valle del Cauca. For the C. gattii complex species, the main reports are in the departments of Norte de Santander and Cundinamarca67.
It should be noted that the department of Boyacá has a high variability of climatic conditions, with sectors where there are 500–1000 mm of average annual rainfall, especially the region of Ricaurte Alto. This bimodal behavior is presented in the west of the department, semi-humid and temperate climates predominate in these sectors, which would positively influence the existence of the fungus in the area49, 68. For this reason, it is of the outmost importance to generate studies for the identification and isolation of Cryptococcus species in this region and encourage further research related to the presence of the fungus and its consequent transmission to human populations.
Various investigations have established that there is a relationship between human infection and exposure to environments where there is the presence of yeast25; therefore, it is essential to understand their distribution in the environment and generate significant contributions that help to deduce the behavior and dynamics of these species that remain in the environment, under specific climatic conditions. Therefore, the objective of this research was to establish the first report of species of the Cryptococcus and Cryptococcus-like yeasts in the department of Boyacá, thus contributing with new knowledge about the environmental distribution of this microorganism that generates relevant data for human health care.
Results
Sample collection
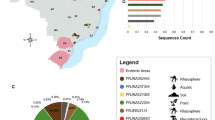
Of the 93 samples recollected from pigeon feces in Tunja, 64 (68.8%) were negative, 23 (24.7%) were positive for C. neoformans and 6 (6.5%) for other Cryptococcus-like yeasts specie; 3 (3,22%) N. globosa, 2 (2.2%) N. albida and 1 (1.07%) P. laurentii, in addition to finding nine species of yeast from other genera. No isolates associated with Cryptococcus species were obtained from the 1211 eucalyptus tree samples taken in Tunja. For the 1188 environmental samples from olive trees collected, 1171 (98.57%) were negative, 4 (0.33%) were positive for C. bacillisporus, 2 (0.17%) for C. neoformans, and 11 (0,93%) for other non-neoformans cryptococcal specie such as P. laurentii (3), N. albida (2), N. globosa (5) and P. flavescens (1), in addition to finding four yeast species of other genera and two bacterial species. The distribution of the species found is presented in Fig. 1.
Climatic and environmental characteristics of the study areas
For the feces sampling of C. livia, variables of solar brightness, relative humidity, evaporation, precipitation, and temperature were taken; where a maximum temperature of 13.8 °C and a minimum of 12.1 °C was evidenced, maximum solar brightness recorded was 180 h per month, relative humidity of 81.8%, observing the highest rainfall in April with 174.7 mm (Supplementary Table 1). The data obtained from the sampling points included variables of maximum, minimum, and average temperature, precipitation, relative humidity, species collected, UV radiation variables, such as direct and indirect sunlight, photosynthetic photon flux density (PPFD), direct and indirect, detailed information in the global data matrix (supplementary table 2).
Within the variables recorded in the sampling associated with olive trees, the maximum temperature recorded was 25 °C and a minimum temperature of 9 °C. In addition, an increase in relative humidity (maximum of 75.8%) was recorded in the municipality of Sutamarchán During July. The most recurrent direct light indices were low, with a value of 0.0394 and indirect light 0.0173.
Microbiological identification
The results of the microbiological identification showed that 45 of 46 presented a capsule. In addition, 35 of the 46 isolates were positive for the urease test, and 42 of the 46 isolates grew at 37ºC, except for three species of N. albida (AM-0277, AM-0286, and AM-0323), and one of N. globosa (AM-0329). All isolates corresponding to C. bacillisporus (AM-0310, AM-0316, AM-0317 and AM-0333) were positive in CGB (L-canavanine-glycine-bromothymol blue) medium, as were three of the four isolates of the species P. laurentii (AM-0313, AM-0314 and AM-0315). The phenoloxidase test was negative for C. bacillisporus AM-316 and AM-317 after ten days of incubation on SSA plates. On the other hand, the C. neoformans AM-310 isolate started pigmentation on the tenth day of incubation; however, it did not have full pigmentation (Fig. 2). Those three isolated negatives for phenoloxidase were urea positive.
MALDI-TOF identification
Sixty-four microorganisms were identified, from which 46 specimens of the genus Cryptococcus were isolated; 11 corresponded to other yeast species and seven to bacteria. Of the stool samples taken from the city of Tunja, 29 (315.18%) positive isolates belonging to the genus Cryptococcus were obtained, of which 23 (24.8%) corresponded to C. neoformans, 3 (3.22%) to N.globosa, 2 (2.2%) N. albida, 1 (1.07%) to P. laurentii. In addition to the findings, other yeasts such as Candida albicans (1), Candida guillermondi (5), Candida parapsilosis (1), Candida tropicalis (1) and Rhodotorula mucilaginosa (1), and some bacterial species such as Bacillus subtillis (4) and Patoea agglomerans (1) were identified. On the other hand, for the samples taken in the Ricaurte Alto region, a lower number of positive isolates was recorded, with a total of 17 (1.43%) positive samples for different species of the genus Cryptococcus, including C. bacillisporus (4), C. neoformans (2), P. laurentii (3), N. albida (2), N. globosa (5) and P. flavescens (1), in addition to finding yeasts such as Rhodotorula mucilaginosa (2) and two species of Pseudomonas; Pseudomonas jesseni (1) and Pseudomonas oryzihabitans (1).
Molecular typification by RFLP of the URA5 gen
The molecular pattern was determined for the isolates identified as species of the C. neoformans and C. gattii species complex. Of the 25 isolates identified by MALDI-TOF MS as C. neoformans, 18 had a VNI molecular pattern, and 7 had a VNII molecular pattern. Likewise, for the four isolates identified as C. gattii species complex, three presented molecular pattern VGIII and one (AM-0317) molecular pattern VGIV contrary to typification by URA5-RFLP isolation AM-0317 was molecular pattern VGIII by sequencing of the ITS and LSU and MALDI-TOF (Fig. 3).
URA5-RFLP profiles obtained after digestion with the restriction enzymes HhaI and Sau96I in reference Cryptococcus spp. strains (lanes 2–9) and environmental isolates (samples AM-0308, AM-0309, AM-0310, AM-0316, AM-0317, AM-0326 and AM-0333). M, DNA size marker 1 kb Opti-DNA Marker Cat#G106. CN, negative control.
Identification by sequencing ITS and LSU gen
Amplification of DNA samples with primers LROR and LR5 resulted in approximately 1,000 bp. The sequences were deposited in GenBank, and accession numbers were obtained (Table 1). Forty- six isolates belonging to the genus Cryptococcus, Naganishia and Papiliotrema were identify; C. neoformans (n = 25), N. globosa (n = 8), N. albida (n = 4), C. bacillisporus (n = 4) (Cryptococcus bacillisporus VGIII), P. laurentii (n = 4), and P. flavescens (n = 1). Also, nine species non-Cryptococcus genus, including Meyerozyma guilliermondii (n = 3) (Kurtzman and Suzuki 2010), (anamorph Candida guilliermondii, Langeron & Guerra 1938), Meyerozyma (Pichia) guilliermondii (n = 1), Candida albicans (n = 1), Candida tropicalis (n = 1), Meyerozyma caribbica (n = 1) and Rhodotorula mucilaginosa (n = 2).
Concatenated ITS and LSU sequences with high bootstrap values generated by neighbor-joining analyses supported the differentiation of the sixclades: 1. N. albida (bootstrap values 100), 2. N. globosa (boot values 100), 3. P. laurentii (bootstrap values 100), 4. P. flavescens (boot values 100) 5. C. bacillisporus (bootstrap 95 values). 6. C. neoformans (boot values 94) (Fig. 4).
Phylogenetic tree of yeast strains identified in this study. Relationships were inferred using the neighbor-joining method in Geneious Prime® 2021.0.3 software. The analysis involved 46 nucleotide sequences and one outgroup with nucleotide sequences of Phaffia rhodozyma CBS5905. All positions containing gaps and missing data were eliminated. Numerical values above the internodes are the percentages of 1000 bootstrap replications. Bootstrap values greater than 50% are indicated. The scale bar of 0.04 represents nucleotide substitutions per position.
Redundancy analysis (RDA) for stool samples of C. livia
It is essential to mention that the data collected in Tunja were organized according to the sampling months, as can be seen in Fig. 5a for the stool samples. It is highlighted that the results indicate that the first three components explain the variability of 93.47% of the data.
Redundancy analysis (RDA). (a) Triplot of the relationship between the presence of species of Cryptococcus, Naganishia and Papiliotrema. per month of sampling and the environmental parameters for the stool samples taken in the city of Tunja. (b) Triplot of the relationship between the presence of Cryptococcus, Naganishia and Papiliotrema species per sampling month and the environmental parameters taken from olive trees in the municipalities of Sáchica, Sutamarchán, and Villa de Leyva.
The redundancy analysis established that the main variables that intensify the appearance of species were relative humidity, temperature, and precipitation. Relative humidity (RH) and temperature (T) positively correlated with the appearance of the species N. albida in March. The other environmental variables are positively related to the appearance of this species. Likewise, these environmental variables (HR, T, and P) negatively correlated with the P. laurentii species. As these variables increase, the probability of finding this species in the environment is lower.
For the C. neoformans species, the variables that have a positive correlation and favor its appearance in the environment are solar brightness (BS) with 76.46% and evaporation (EV_mm) with 80.3%, contrary to the negative relationship that was evidenced with the precipitation variable.
Finally, the species N. globosa presented a more significant relationship with the precipitation variable (PR_mm); it is more likely to find this species when precipitation is lower since its correlation with this analysis was 39%.
It should be noted that for the sampling carried out in the city of Tunja, direct and indirect light records were not made, only those mentioned above, which IDEAM provided.
Redundancy analysis (RDA) for olive tree samples
It is essential to mention that to carry out this analysis, data was organized with codes where J was assigned to July, A to August, S to September, O to October, and N to November, as can be shown in Fig. 5b, for the data taken in Sutamarchán, Sáchica and Villa de Leyva.
The redundancy analysis allowed us to establish the main variables that intensify the appearance of Cryptococcus, Naganishia and Papiliotrema species; they were relative humidity and temperature.
The relative humidity (HR) is inversely related to the variables of indirect light (ISite), direct light (Dsite), average temperature (Tprom), and precipitation (Prec). According to the data collected, the relative humidity increases when ISite, Dsite, Tprom, and Prec variables decrease; then, the relative humidity decreases when the mentioned variables increase (Fig. 5b). The species N. albida, P. laurenteii and P. flavescens are found to a greater extent when the relative humidity of the medium is lower. In contrast, C. neoformans and C. gattii species complexes and N. globosa were isolated more frequently when the relative humidity was higher.
Likewise, the maximum temperature is also related to the appearance of species of the genus Cryptococcus, presenting a directly proportional relationship with the species of C. neoformans and C. bacillisporus and, to a lesser extent, N. globosa. These species were found between 20 and 25 °C, reported in July and August in the municipality of Sáchica. It should be noted that the variables of direct and indirect light, precipitation, minimum temperature, and average temperature were inversely related to the presence of all the species above.
Multiple correlations analysis from samples obtained from stool samples
As shown in Fig. 6a, there is a positive correlation (78%) between the isolates of C. neoformans and precipitation; that is, precipitation contributed positively to obtaining isolates of this species; therefore, they are directly found related. Contrary to the negative correlation (30%) evidenced in the variable of solar brightness, that says, as the solar brightness increases, the probability of isolating C. neoformans in the environment decreases. Unlike what was found for N. globosa, which presents a positive correlation (62%), as the solar brightness increases, the probability of finding this species in the environment increases.
Multiple correlation analysis from samples obtained from pigeon feces (a) and samples from the Ricaurte Alto region (b). Color Intensity and cycle size describe the percentage of the relationship of the variables (right side of the figure). The colors on the graph represent positive correlation (Blue) and negative correlation (Red).
On the other hand, for P. laurentii a negative correlation of 80% with relative humidity was determined; that is, it is an inversely proportional relationship; therefore, as relative humidity increases, it is unlikely to find this species in the environment.
Multiple correlations analysis from samples obtained from the Ricaurte Alto region
The variables of direct light (direct Site) and indirect (indirsite) present a positive correlation of 100%, with a relationship percentage of 1, which reflects that they are directly related. Similarly, C. bacillisporus has a slight positive correlation of 40% with relative humidity, which means that it is more likely to find this species when the relative humidity is higher. In the same way, a negative correlation of 30% can be observed between the appearance of C. neoformans and the minimum temperature; this is an inversely proportional relationship. As the temperature increased, it was more likely to isolate C. neoformans from the environment (Fig. 6b).
For the other species of Cryptococcus, it is essential to point out that P. laurentii has a positive correlation of 40% with the average temperature, thus indicating that this particular species is associated with average temperatures that range between 14 and 15 °C, which would indicate an association with cold to temperate places. In the case of the N. albida and N. globosa species, they present an inversely proportional correlation of 50% with the minimum temperature; according to our results, it is more likely to be found at temperatures above 15 °C.
When performing the logistic regression test, it was established that the specie that was most influenced by the environmental variables was N. albida with a p-value of 0.0078.
Discussion
It is essential to point out that the objective of this research was to establish the first report of species of the Cryptococcus and Cryptococcus-like yeasts in the department of Boyacá. As well as to identify through microbiological and molecular tests and differentiate the species of Cryptococcus found. Similarly, establishing an association between environmental variables and the presence of species of Cryptococcus, Naganishia and Papiliotrema spp. In this way, the results permitted establishing the environmental distribution of this yeast in the Boyacá region, thus contributing to human health care.
Several studies have shown that C. neoformans has mainly pigeon feces as an environmental niche. This association is mainly due to the enzyme urease in Cryptococcus species that allows it to assimilate the nitrogen present in the medium. It should be noted that according to our results, P. laurentii isolates were negative for urease, which is also mentioned by Hoog De GS, et al. in (2000), and for the enzyme phenoloxidase in SSA medium, as reported by Pedroso et al.69. Likewise, as reported by Toplis et al.70, when the yeast grows at 37 °C and there is urea deficiency, low levels of melanin production are observed, which can be associated with low or no pigmentation in the SSA medium70. It should be noted that the phenoloxidase test was positive for the vast majority of Cryptococcus spp. isolates, except for strains AM-316, AM-317 and AM-310. The latter started pigmentation in the SSA medium on the tenth day of incubation; this may be due to mutations in the associated genes that prevent melanization71, 72. For the CGB test, all C. bacillisporus were positive, as were three isolates of P. laurentii, which showed a change in the CGB medium, as reported by Tay et al.73.
The isolates identified as C. neoformans complex were primarily obtained from the city of Tunja. The above, related to the presence of pigeons and the average temperature (14 °C), conditions that, according to Quintero, Rosario and Pfeiffer, contribute to generating an environment conducive to the survival of this species18, 42, 74. On the contrary, the species of the C. gatii complex was isolated from tree holes in olive trees, as reported by other authors54, 59, 75,76,77. However, the association of C. gattii species complex with olive trees constitutes the first report in Colombia. On the other hand, only 0.33% of the total samples collected (1188) corresponded to the species of C. gattii, which demonstrates the difficulty of isolating it from environmental sources, as published by Contreras et al.78 and Toro75, who reported very low percentages of C. gattii species complex isolates of 8% and 0.7%75, 78.
Of the species of the genus Cryptococcus different from the C. neoformans and C. gattii complexes, P. laurentii was isolated from pigeon feces and olive trees, showing a predilection for average temperatures typical of the Boyacá territory. P. laurentii has been reported from different tree species79 avian droppings and air samples35, 80. Similarly, this species has been isolated from clinical samples in patients with clinical conditions such as meningitis15, fungemia, and cryptococcosis, considering it a pathogenic species81, 82.
Meanwhile, N. albida has been reported as a pathogenic species for humans15 with a pathogenic behavior similar to C. neoformans83. In our study, this specie was isolated from tree debris and leaves from olive trees in the region of Ricaurte Alto and pigeon feces in Tunja city. In comparison, the environmental record had been reported from tree hollows and excreta in the city of Uberlandia, State of Minas Gerais, Brazil62.
P. flavescens was isolated from tree hollows in the region of Ricaurte alto, in the municipality of Sáchica and associated with temperate temperatures, the same as reported in the study published by Brito et al.62 that isolated this species from Mangifera indica. P. flavescens has also been isolated from the cerebrospinal fluid of a patient with AIDS17.
N. globosa was isolated from pigeon feces and olive trees, in a range of 9 °C–25 °C, contrary to reported by Butinar and collaborators84; who reported this specie from the Arctic soil (5 °C), specifically in the coastal glaciers Conwaybreen, Kongsvegen, and Austria Love ́nbreen and the interior glacier Austre Brøggerb-reen84. Likewise, Cornnell et al. (2008) and Singh et al.64 identified strains of N. globosa from glacial ice cores63, 64. These results allow recognizing the adaptation of N. globosa to different sources and its affinity to environmental variables such as low precipitation, average temperature, and low relative humidity, demonstrating new conditions that allow its development in our results.
Regarding the analysis of the relationship between the environmental variables and the species found from pigeon feces, it can be associated that the lower the brightness of the sun, the greater the probability of finding the species C. neoformans. The Tunja city registered solar brightness was low, varying between 180 h/month to 78 h/month, a characteristic associated with many C. neoformans isolates (n = 8). While in the region from Ricaurte Alto, only one sample of this species was isolated in the month with the lowest incidence of direct and indirect light, as reported by Ellis et al.85 and Rosario et al.48. Other variables that favor the presence of this species are; low temperatures and precipitation, together with an increase in humidity, as reported by other authors18, 42, 85. The same happens with N. albida, which presents the same characteristics with a greater predilection for average temperature and a positive relationship when relative humidity increases. While for P. laurentii it was established in this study that as all the variables decrease, the chance of finding it in the environment increases, contrary to what was reported by Pedroso et al.80, where, despite not specifying the variables studied, they relate it to a tropical climate79, 80.
For the isolates obtained from olive trees, as a first report, it was established that the species N. albida, P. laurenteii, and P. flavescens are isolated mainly in environments with low relative humidity. In this regard, the variable relative humidity was negatively correlated for the species mentioned above. On the other hand, a positive correlation was established for the maximum recorded temperature variable and C. neoformans and C. gattii species complex, equal to that reported for Bogotá city by Castañeda et al.65.
Furthermore, it is essential to note that the molecular patterns for the species of C. neoformans and C. gattii species complexes isolated in this study coincide with those reported for other departments of Colombia. The VNI molecular pattern is mainly reported in environmental isolations, followed by the VNII molecular pattern86,87,88,89. For the C. gattii complex the VGIII molecular pattern is reported more frequently, followed by VGII and VGI77, 87, 88. Additionally, the RFLP analysis identified the AM-0317 strain as VGIV but was classified as VGIII by MALDI-TOF and ITS/LSU sequences, the incorrectly grouping to VGIV can be the result of a point mutation in the RFLP restriction site. As reported by Trilles et al.90 and Firacative et al.91, a single nucleotide mutation resulting in the misidentification of isolates as VGIV, subsequently classified as molecular type VGIII by MLST analysis90, 91. In this respect, further phylogenetic analysis based on MLST and Whole Genome Sequencing (WGS) is required to establish the molecular structure of this strain.
Finally, molecular techniques using ITS and LSU allowed us to identify all isolates to the species level. However, it was observed that strains AM-0277, AM-0286, AM-0312, and AM-0323 identified as N. albida by LSU and ITS sequencing presented inconsistent results with MALDI-TOF which resulted in N. liquefaciens. Likewise, C. neoformans AM-0299 was identified as N. globosa by MALDI-TOF. Also, N. globosa (AM-0322) and C. neoformans AM-0325 were identified as N. liquefaciens by MALDI-TOF. Therefore, improving MALDI-TOF MS spectra libraries and implementing another characterization method to identify the species-level isolates is necessary.
Conclusions
The findings of this study constitute the first report of C. neoformans and C. gattii species complexes, P. laurentii, N. liquefaciens, N. globosa, and P. flavescens in the department of Boyacá.
The data obtained from the microbiological identification were similar to those obtained by the molecular identification. The MALDI-TOF MS identification presented a correct recognition of the C. neoformans and C. gattii species complexes, including the molecular type. Most of the results obtained by URA5-RFLP were consistent with the other techniques employed, with only one atypical RFLP pattern identified as VGIV. Additionally, 89% of the non-neoformans/non-gattii Cryptococcus species, were correctly identify. In the same way, ribosomal subunit DNA sequencing allows to differentiate C. neoformans and C. gattii, however other Cryptococcus species requires more than one set of DNA markers.
In the Ricaurte Alto region, the environmental variables related to a higher recovery of Cryptococcus; Naganishia and Papiliotrema species were medium to high temperatures and the relative humidity of the environment.
For the sampling of C. livia feces carried out in Tunja, it is concluded that the environmental. conditions that favored the recovery of C. neoformans were low sunshine and increased precipitation.
For the species P. laurentii and P. flavescens, the first report of association with environmental conditions that favor their recovery, was made. In addition, the species N. globosa was reported to have a new favorable environment for its development.
Methods
Study area
The first stage of the study was carried out in the city of Tunja, capital of the department of Boyacá—Colombia, for seven months, from January to July 2018. The sample collection was done in six points of Tunja city (República Forest, San Ricardo Forest, Escuela Normal Superior, Forest of the Universidad Pedagógica y Tecnológica de Colombia, Sugamux park and Plaza de Bolívar), which were selected for presenting an abundant avian population (Fig. 7). For the second stage, samples were collected for five months, from June to November 2019. The study area included the upper province of the department of Boyacá, made up of three municipalities, such as Villa de Leyva, located 40 km west of Tunja; Sáchica, located 32 km west of Tunja and Sutamarchán, which is 44 km from Tunja92. The selected sectors corresponded to areas with the presence of olive trees, such as; the central parks of Sáchica, Sutamarchán, the Nariño Park in Villa de Leyva, since they are a potential source of contamination and human influence, and the olive grove in the municipality of Sutamarchán, which is considered one of the oldest trees in the region (Fig. 7).
Sample collection
A total of 93 samples of pigeon feces and 1211 from eucalyptus trees in Tunja and 1188 environmental samples from olive trees were collected. The sampling was random for each site, using biosecurity measures, to avoid the inhalation of viable spores. At the tree collection points, samples of bark, leaves, soil, cavities, and debris were taken, and stool samples were collected in different parts of the city. The samples were deposited in hermetically sealed bags, transferred and processed in the Biological Sciences Laboratory of the University of Boyacá.
Eucalyptus globulus and Olea europea, the species associated with the samples obtained in this study, are not endangered. The sample collection is non-lethal for the plants because it includes non-living parts of the plant such as bark, fallen leaves, soil, cavities, and debris36.
The sample collection complied with relevant institutional, national, and international guidelines and legislation. The permission to collect biological samples was granted by Autoridad Nacional de Licencias Ambientales—ANLA (Resolution No 01300 de 2019).
Environmental data
In the sampling points associated with trees, environmental data were collected through what was reported by the Institute of Hydrology, Meteorology, and Environmental Studies of Colombia. Canopy cover was evaluated using the WinCanopy equipment (canopy structure and solar radiation, with cover and light variables), which allowed characterizing the habitat where Cryptococcus species isolated from trees are found.
Microbiological identification
During a maximum of 24 h after collecting the samples, processing of the samples was carried out following the protocol described by Escandon et al. (2010), using the extraction technique with phosphate-buffered saline (PBS)77. Briefly, 1 g of each pigeon dropping sample and 5 g of soil, bark, leaves, cavities, and debris were suspended in 50 ml of PBS 1X with 0.2 g of chloramphenicol, followed by manual shaking for 5 min. After resting 30 min, 100 µl of the supernatant was sown in sunflower seed agar (SSA) medium plates (Khan et al. 2004). The plates were then incubated at 25 °C and checked daily for brown colonies for five days. Each phenol oxidase-positive/brown colony was sub-cultured on Sabouraud agar for purification and phenotypical characterization.
All single isolated colonies, not only pigmented colonies, were tested for the presence of capsules by India ink examination, urease production on urea agar93 and thermotolerance at 37 °C on Sabouraud agar. The species C. neoformans and C. gattii were differentiated on canavanine-glycine-bromothymol blue (CGB) medium94, 95. Additionally, the colonies that did not pigment in SSA agar despite presenting a capsule, positive urea, and growth at 37 °C, were seeded again in SSA agar and incubated for ten days with daily observation. C. albicans ATCC 10,231 and C. neoformans ATCC 32,045 were used as negative and positive controls of the phenol oxidase test.
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF)
To obtain protein profiles for each yeast isolate, these were seeded in Sabouraud agar and incubated for 24 h at 37 °C. An inoculum of each pure culture was deposited in a well of a metal plate for analysis (Bruker Daltonics®), formic acid was added twice, allowing it to dry between each application. Then, 1μL of the matrix was added (2.5 mg/mL of α-cyano-4-hydroxycinnamic acid HCCA Bruker Daltonics®) in 50% acetonitrile, 2.5% trifluoroacetic acid, and 47.5% HPLC Water-Sigma). The mixture was allowed to dry at room temperature. The spectra for each isolate were obtained after 240 laser shots in six different regions within the well by a Microflex spectrometer LT MALDI-TOF MS (Bruker Daltonics) and analyzed using Bruker Flex software and MALDI Biotyper RTC 3.0 (Bruker Daltonics®). The BTS standard (Bacterial Test Standard) Escherichia coli DH5 from alpha peptide was used as a calibration standard. All the spectra were analyzed from 2,000 to 20,000 Da and then compared with the BDAL database provided by Bruker. All protein profiles were used to make a correlation dendrogram using RTC Biotyper 3.0 software96.
An extraction process was carried out for the isolates that did not present protein profiles, which is briefly described below; 300 μL of HPLC water was added to a 1, 5 ml tube, after which several isolated colonies were transferred to the tube, and vortexed, 300 μL of ethanol was added and turned to pass by vortex, it was centrifuged for 2 min at 1400 rpm, the ethanol was decanted, and it was centrifuged again with the same conditions. The excess ethanol was stirred with the pipette, and 10 μL to 50 μL of formic acid was added, depending on the formed pellet, and vortexed, 10 μL to 50 μL of acetonitrile was added, it is essential to note that the amounts of formic acid and acetonitrile must be in identical volumes, it was centrifuged again for 2 min at 1400 rpm. 0.85 μL of the supernatant was pipetted into a well of the MALDI-TOF plate. Avoid touching the pellet at the bottom and allow it to dry. The sample was covered with 0.85 μL of the matrix and dry.
Molecular identification
DNA extraction
The Wizard® Genomic DNA Purification Kit protocol from Promega was used with some modifications. The isolates belonging to the genus Cryptococcus were plated 48 h in advance on yeast extract, peptone, dextrose (YEPD) agar, at a temperature of 37 °C, then 0.9 to 0.12 g of the culture was transferred to a 1.5 ml Eppendorf tube containing 293 µL of EDTA (50 mM). Subsequently, 0.10 g of glass beads were added and subjected to vortex agitation for 10 min, the supernatant was transferred to a clean tube, and the procedure indicated by the extraction kit, described for yeasts. The quantification of DNA was made with QuantiFluor® dsDNA System (Promega).
Polymorphisms of the URA5 gene by RLFP
Molecular type was determined by RFLP analysis of the URA5 gene. This URA5 gene was amplified with the two primers URA5 (5'ATGTCCTCCCAAGCCCTCG ACTCCG 3') and SJ01 (5'TTAAGACCTCTGAACAC-CGTACTC 3'). PCR amplification of the URA5 gene was performed as described by Meyer et al.38, which was carried out with one cycle of 94 °C for the initial denaturation of 2 min, for 35 cycles in a thermocycler brand (AXYGEN MAXGYGENE) as follows: 94 °C for denaturation for 45 s, 1 min annealing at 61 °C and 2 min extension at 72 °C, followed by a final extension cycle for 10 min at 72 °C. The amplification products were visualized on 1.5% agarose gels in 1X TBE buffer stained with SafeViem™ Classic Cat. G108 0.3 mg/ml, at 100 V for 1 h. Subsequently, 30 μl of each PCR product was digested twice with Sau96I (10 U/μL) and HhaI (20 U/μL) for three hours and separated by 3% agarose gel electrophoresis at 90 V for five hours. The RFLP patterns were visually assigned by comparing them with the patterns obtained from the standard strains (VNI-VNIV and VGI-VGIV) provided by the Colombian National Institute of Health38, 97.
PCR amplification and Sequencing of rLSU region
Two nuclear loci were amplified, the long subunit of ribosomal RNA (LROR: 5' -ACCCGCTGAACTTAAGC-3' LR5: 5' -ATC CTG AGG GAA ACT TC-3') and nuclear ribosomal internal transcribed spacer region (ITS1: 5' -CTTGGTCATTTAGAGGAAGTAA-3' ITS4: 5' -GGAAGTAAAAGTCGTAACAAGG-3'). Amplification followed the procedure reported by Gardes and Bruns98 and Vilgalys and Sun99 with some modifications. The reactions were carried out in 30 µL. The PCR mix contained primers at a final concentration of 0.5 µM, 1 µL of genomic DNA (2 ng/µL), and 12.5 µL of 2X PCR MasterMix (Applied Biological Materials Inc. (Abm)). The amplification program consisted of one initial cycle of 3 min at 94 °C, followed by 35 cycles comprising denaturation (1 min at 94 °C), annealing (30 s at 56 °C), and extension (1 min at 72 °C), and then a final extension (7 min at 72 °C). The amplified products were analyzed on 1% agarose gels stained with SYBR® Green (Applied Biological Materials Inc. (Abm)).
The PCR products obtained were purified and sequenced using the Sanger platform. The sequences obtained were edited using Geneious Prime® 2021.0.3 software; subsequently, with the consensus sequences, BLAST was performed in the GenBank database and MycoBank databases to determine the species or genus of each isolate. The identification presented in this article corresponds to the better similarity and overlap percent. The assembled sequences were submitted to the GenBank Database.
The isolates were preserved in glycerol 10% at − 80 °C and deposited in the Culture Collection of Fungi and Microorganisms of the University of Boyacá.
Statistical analysis
Statistical analyzes were performed with the R studio version 4.1.1 program to find the relationship between environmental variables and the incidence of the fungus in the region, firstly, with descriptive statistics for PCA and RDA and basic statistics and probabilities to perform correlations and multivalent statistics, in this case, the logistic regression (to determine the variable of more weight) for the samples taken from olive trees. Multiple correlation was done to with the use of a matrix of 64 data and eight variables, among which are positive isolates (postcryto) for Cryptococcus, maximum temperature (Temmax), minimum (Temmin) and average (Tempro), relative humidity (RH), precipitation, direct light (Directside), and indirect (indirectside)100. Rstudio software, with packages (FactoMineR, factoextra, readxl, performanceAnalitytics, ggplot2, MVN) was done for this analysis.
Logistic regression was used to predict the probability of finding Cryptococcus spp. positive in association with the environmental variables studied, which are maximum (Temmax), minimum (Temmin) and average (Tempro) temperature, relative humidity (RH), precipitation, direct light (Directside), and indirect (indirecside), for this the analysis was carried out using the Rstudio program, with the packages (corrplot, ggplot2) (Chitarroni 2002)101.
RDA redundancy analysis was done to evaluate the presence of Cryptococcus species during the sampling months, taking into account the aforementioned environmental variables.
Data availability
All data generated or analysed during this study are included in this published article. The sequence data have been deposited in GeneBank with the accession codes list in a Table 1.
Abbreviations
- AIDS:
-
Acquired immunodeficiency syndrome
- ATCC:
-
American type collection culture
- BS:
-
Solar brightness
- CGB:
-
Canavanine-glycine-bromthymol blue agar
- EDTA:
-
Ethylenediaminetetraacetic acid
- IDEAM:
-
Instituto de Hidrología, Meteorología y Estudios Ambientales
- ITS:
-
Internal transcribed spacer
- LSU:
-
Large subunit
- MALDI-TOF:
-
Matrix-assisted laser desorption/ionization time-of-flight
- MLST:
-
Multilocus sequence typing
- PBS:
-
Phosphate-buffered saline
- PPFD:
-
Photosynthetic photon flux density
- RDA:
-
Redundancy analysis
- RFLP:
-
Restriction fragment length polymorphism
- RH:
-
Relative humidity
- SSA:
-
Sunflower seed agar
- UBCHM:
-
Universidad de Boyacá Colección de Hongos y Microorganismos
- WGS:
-
Whole genome sequencing
- YEPD:
-
Yeast extract, peptone, dextrose agar
References
Kwon-Chung, K. J. & Bennett, J. E. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120(1), 123–130 (1984).
Maziarz, J. EK y Perfect, Cryptococcosis. Intraocular Inflamm. 30, 1277–1283. https://doi.org/10.1007/978-3-540-75387-2_123 (2016).
Neilson, et al. Cryptococcus neoformans: pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect. Immun. 20(1), 262–266 (1978).
Rajasingham, R. et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet. Infect. Dis. 17(8), 873–881 (2017).
Escandón, P., Lizarazo, J., Agudelo, C. I. & Castañeda, E. Cryptococcosis in Colombia: Compilation and analysis of data from laboratory-based surveillance. J. Fungi. 4(1), 32 (2018).
Escandón, P., De Bedout, C., Lizarazo, J. & Agudelo, C. I. Cryptococcosis in Colombia: Results of the national surveillance program for the years 2006–2010. Biomedica 32, 386–392 (2012).
Meyer, W. et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol 47(6), 561–570. https://doi.org/10.1080/13693780902953886 (2009).
Kwon-Chung, K. J. et al. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. MSphere. 2(1), 10–128 (2017).
Liu, X. Z. et al. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Stud. Mycol. 81, 1–26. https://doi.org/10.1016/j.simyco.2015.08.001 (2015).
Hagen, F. et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 78, 16–48. https://doi.org/10.1016/j.fgb.2015.02.009 (2015).
Hagen, F. Importance of resolving fungal nomenclature: the case of multiple pathogenic species in the Cryptococcus Genus. Msphere. 2(4), e00238-e317. https://doi.org/10.1128/msphere.00238-17 (2017).
Rodrigues, J. Pathogenic diversity amongst serotype C VGIII and VGIV Cryptococcus gattii isolates. Sci. Rep. 5(1), 11717. https://doi.org/10.1038/srep11717 (2015).
Chambers, C., MacDougall, L., Li, M. & Galanis, E. Tourism and specific risk areas for Cryptococcus gattii, Vancouver Island, Canada. Emerg. Infect. Dis. 14(11), 1781–1783. https://doi.org/10.3201/eid1411.080532 (2008).
Takemura, H. et al. The first reported case of central venous catheter-related fungemia caused by Cryptococcus liquefaciens. J. Infect. Chemother. 21(5), 392–394. https://doi.org/10.1016/j.jiac.2014.11.007 (2015).
Kordossis, T. et al. First report of Cryptococcus laurentii meningitis and a fatal case of Cryptococcus albidus cryptococcaemia in AIDS patients. Med. Mycol. 36(5), 335–339. https://doi.org/10.1046/j.1365-280x.1998.00166.x (1998).
Choe, Y. J. et al. Cryptococcus albidus Fungemia in an Immunosuppressed Child: Case report and systematic literature review. J. Pediatric. Infect. Dis. Soc. 9(1), 100–105. https://doi.org/10.1093/jpids/piz039 (2019).
Ikeda, R. & Maeda, T. Structural studies of the capsular polysaccharide of a non-neoformans Cryptococcus species identified as Cryptococcus laurentii, which was reclassified as Cryptococcus flavescens, from a patient with AIDS. Carbohydr. Res. 339(3), 503–509. https://doi.org/10.1016/j.carres.2003.11.015 (2004).
Quintero, E., Castañeda, E. & Ruiz, A. Environmental distribution of Cryptococcus neoformans in the department of Cundinamarca-Colombia. [Distribución ambiental de Cryptococcus neoformans en el departamento de Cundinamarca-Colombia.]. Rev. Iberoam. Micol. 22(2), 93–98 (2005).
Virviescas, C. et al. Molecular characterization of Cryptococcus neoformans recovered from pigeon droppings in Rivera and Neiva. Colombia. Revista MVZ Córdoba. 20, 6991–6997 (2018).
Ergin, Ç. Cryptococcus neoformans recovered from olive trees (Olea europaea) in Turkey reveal allopatry with African and South American lineages. Front. Cell. Infect. Microbiol. 8(9), 384 (2019).
Xue, C., Tada, Y., Dong, X. & Heitman, J. The human fungal pathogen Cryptococcus can complete Its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1(4), 263–273. https://doi.org/10.1016/j.chom.2007.05.005 (2007).
Bauwens, L., Swinne, D., De Vroey, C. & De Meurichy, W. Isolation of Cryptococcus neoformans var. neoformans in the aviaries of the Antwerp Zoological Gardens. Mykosen 29(7), 291–294 (1986).
Cabañes, F. J. Mycoses and zoonoses: Cryptococcus spp. Rev Iberoam Micol 25(1), S1–S3 (2008).
Huamán, A. et al. Cryptococcus neoformans in pigeon feces (Columba livia) in Metropolitan Lima. [Cryptococcus neoformans en heces de palomas (Columba livia) en Lima Metropolitana.]. Revista Médica Herediana 29(2), 85–89 (2018).
Vallejo Timarán, D. A., Benavides Melo, C. J., Chaves Velásquez, C. A., Morillo Caicedo, M. I. & Castillo Ceballos, A. M. Aislamiento de Cryptococcus neoformans en heces de palomas (Columba livia) en el casco urbano del municipio de Pasto, Colombia. Biosalud 15(1), 62–71. https://doi.org/10.17151/biosa.2016.15.1.7 (2016).
Archibald, L. K. et al. Antifungal Susceptibilities of Cryptococcus neoformans. Emerg. Infect. Dis. 10(1), 143–145 (2004).
Nnadi, N. E. et al. Caractérisation moléculaire de souches de Cryptococcus neoformans VNII isolés dans l’environnement à Jos, état du Plateau, Nigeria. J. Mycol. Med. 26(4), 306–311. https://doi.org/10.1016/j.mycmed.2016.04.001 (2016).
Dou, H. et al. Molecular characterization of Cryptococcus neoformans isolated from the environment in Beijing, China. Med. Mycol. 55(7), 737–747 (2017).
Mseddi, F. et al. First environmental isolations of Cryptococcus neoformans and Cryptococcus gattii in tunisia and review of published studies on environmental isolations in Africa. Mycopathologia 171(5), 355–360 (2011).
Nishikawa, M. M. et al. Serotyping of 467 Cryptococcus neoformans isolates from clinical and environmental sources in Brazil: Analysis of host and regional patterns. J Clin Microbiol 41(1), 73–77 (2003).
Kidd, S. E. A. rare genotype of cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proceed. Nat. Acad. Sci. 101(49), 17258–17263 (2006).
Montagna, M. T. Molecular characterization of Cryptococcus neoformans and Cryptococcus gattii from environmental sources and genetic comparison with clinical isolates in Apulia. Italy. Environ. Res. 160, 347–352 (2018).
Cattana, M. E. et al. Native trees of the northeast argentine: Natural hosts of the Cryptococcus neoformans-Cryptococcus gattii species complex. Rev. Iberoam. Micol. 31(3), 188–192 (2014).
Refojo, N. et al. Isolation of Cryptococcus neoformans and Cryptococcus gattii from trunk hollows of living trees in Buenos Aires City. Argent. Med. Mycol. 47(2), 177–184 (2009).
Pedroso, R. S., Lavrador, M. A. S., Ferreira, J. C., Candido, R. C. & Maffei, C. M. L. Cryptococcus neoformans var. grubii - Pathogenicity of environmental isolates correlated to virulence factors, susceptibility to fluconazole and molecular profile. Mem. Inst. Oswaldo. Cruz. 105(8), 993–1000. https://doi.org/10.1590/S0074-02762010000800008 (2010).
Callejas, A., Ordoñez, N., Rodriguez, M. C. & Castañeda, E. First isolation of Cryptococcus neoformans var. gattii, serotype C, from the environment in Colombia. Med. Mycol. 36(April), 341–344 (1998).
Laurenson, I. F. et al. Cryptococcus neoformans in Papua New Guinea: a common pathogen but an elusive source. J. Med. Vet. Mycol. 35(6), 437–440 (1997).
Meyer, W. et al. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis 9(2), 189–195. https://doi.org/10.3201/eid0902.020246 (2003).
Randhawa, H. S. et al. Distribution of Cryptococcus gattii and Cryptococcus neoformans in decayed trunk wood of Syzygium cumini trees in north-western India. Med Mycol 44(7), 623–630 (2006).
A. Casadevall and J. R. Perfect, Cryptococcus neoformans, vol. 595. Citeseer, 1998.
Mattsson, R., Haemig, P. D. & Olsen, B. Feral pigeons as carriers of Cryptococcus laurentii, Cryptococcus uniguttulatus and Debaryomyces hansenii. Med. Mycol. 37(5), 367–369 (1999).
Rosario, I., Acosta, B. & Colom, F. The pigeon and other birds as reservoirs for Cryptococcus spp. [La paloma y otras aves como reservorio de Cryptococcus spp.]. Rev. Iberoam. Micol. 25(1), S13–S18 (2008).
Bartlett, K. H., Kidd, S. E. & Kronstad, J. W. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Curr. Infect. Dis. Rep. 10(1), 58–65 (2008).
Byrnes, E. J. III. et al. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the pacific northwest in the United States. J. Infect. Dis. 199(7), 1081–1086. https://doi.org/10.1086/597306 (2009).
Upton, A. et al. First contemporary case of human infection with Cryptococcus gattii in puget sound: Evidence for spread of the Vancouver Island outbreak. J Clin Microbiol 45(9), 3086–3088 (2007).
Cogliati, M. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: an atlas of the molecular types. Hindawi Publ. Corp. Sci. 2013, 1–23. https://doi.org/10.1155/2013/675213 (2013).
Ellis, D. & Pfeiffer, T. Natural Habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 8(3), 321–325 (1990).
P. Escandón, E. Quintero, D. Granados, S. Huérfano, A. Ruiz, and E. Castañeda, “Aislamiento de Cryptococcus gattii serotipo B a partir de detritos de Eucalyptus spp. en Colombia.,” Biomédica, vol. 25, no. 3, p. 390, 2005, doi: https://doi.org/10.7705/biomedica.v25i3.1363.
Kidd, S. E. et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A 101(49), 17258–17263 (2004).
Campbell, L. T. et al. Clinical and environmental isolates of Cryptococcus gattii from Australia that retain sexual fecundity. Eukaryot. Cell. 4(8), 1410–1419 (2006).
Chowdhary, A., Randhawa, H. S., Prakash, A. & Meis, J. F. Environmental prevalence of Cryptococcus neoformans and Cryptococcus gattii in India: An update. Crit. Rev. Microbiol. 38(2011), 1–16 (2012).
Girish Kumar, C. P., Prabu, D., Mitani, H., Mikami, Y. & Menon, T. Environmental isolation of Cryptococcus neoformans and Cryptococcus gattii from living trees in Guindy National Park, Chennai. South India. Mycoses. 53(3), 262–264 (2010).
Cogliati, M. et al. Environmental distribution of Cryptococcus neoformans and Cryptococcus gattii around the Mediterranean basin. FEMS Yeast. Res. 16(4), 045 (2016).
Datta, K. et al. Spread of Cryptococcus gattii into Pacific Northwest Region of the United States. Emerg. Infect. Dis. 15(8), 1185–1191 (2009).
Hurst, S. et al. Molecular typing of clinical and environmental isolates of Cryptococcus gattii species complex from southern California, United States. Mycoses 62(11), 1029–1034 (2019).
Springer, D. J. & Chaturvedi, V. Projecting global occurrence of Cryptococcus gattii. Emerg. Infect. Dis. 16(1), 14–20 (2010).
Andrade-Silva, L. et al. Susceptibility profile of clinical and environmental isolates of Cryptococcus neoformans and Cryptococcus gattii in Uberaba, Minas Gerais, Brazil. Med. Mycol. 51(6), 635–640 (2013).
Brito-Santos, F. et al. Environmental isolation of Cryptococcus gattii VGII from indoor dust from typical wooden houses in the deep Amazonas of the rio Negro basin. PLoS ONE 10(2), 1–11 (2015).
Davel, G. et al. Primer aislamiento ambiental de Cryptococcus neoformans var. gattii en Argentina. Rev. Argent. Microbiol. 35(2), 110–112 (2003).
Kassi, F. K. et al. Comparative typing analyses of clinical and environmental strains of the Cryptococcus neoformans/Cryptococcus gattii species complex from ivory coast. J. Med. Microbiol. 67(1), 87–96 (2018).
Sorrell, T. C. Cryptococcus neoformans variety gattii. Med. Mycol. 39(2), 155–168 (2001).
Brito, M. D. O. et al. Isolation of Cryptococcus species from the external environments of hospital and academic areas. J. Infect. Dev. Ctries 13(6), 545–553 (2019).
Connell, L. et al. Diversity of soil yeasts isolated from South Victoria Land, Antarctica. Microb. Ecol. 56(3), 448–459. https://doi.org/10.1007/s00248-008-9363-1 (2008).
Singh, P., Tsuji, M., Singh, S. M., Roy, U. & Hoshino, T. Taxonomic characterization, adaptation strategies and biotechnological potential of cryophilic yeasts from ice cores of Midre Lovénbreen glacier, Svalbard, Arctic. Cryobiology 66(2), 167–175. https://doi.org/10.1016/j.cryobiol.2013.01.002 (2013).
A. Castañeda and E. Castañeda, “Isolation of Cryptococcus species associated with Eucalyptus in a park in Bogotá,” Biomédica, vol. 21, n, pp. 75–78, 2001.
Duarte, A., Ordoñez, N. & Castañeda, E. Association of yeasts of the genus Cryptococcus with Eucalyptus species in Santafe de Bogota. [Asociación de levaduras del género Cryptococcus con especies de Eucalyptus en Santafé de Bogotá.]. Rev. Inst. Med. Trop. Sao Paulo 36(2), 125–130 (1994).
Serna-Espinosa, B. N., Guzmán-Sanabria, D., Forero-Castro, M., Escandón, P. & Sánchez-Quitian, Z. A. Environmental status of Cryptococcus neoformans and Cryptococcus gattii in Colombia. J. Fungi. 7(6), 410. https://doi.org/10.3390/jof7060410 (2021).
M. Bello, “Guía turistica de Boyacá,” Ministerio de Industria y Comercio, p. 76, 2011, [Online]. Available: http://www.colombia.travel/es/descargas/guias_turisticas/GUIA_BOYACA-web.pdf
Pedroso, R. D., Costa, K. R., Ferreira, J. C. & Candido, R. C. Avaliação da produção de melanina por espécies de Cryptococcus em quatro diferentes meios de cultura. Revista da Soc. Brasileira de Med. Trop. 40, 566–568 (2007).
Toplis, B. et al. The virulence factor urease and its unexplored role in the metabolism of Cryptococcus neoformans. FEMS Yeast Res. 4, foaa031 (2020).
Chrissian, C. et al. Melanin deposition in two Cryptococcus species depends on cell-wall composition and flexibility. J. Biol. Chem. 295(7), 1815–1828. https://doi.org/10.1074/jbc.RA119.011949 (2020).
Kwon-Chung, K. J., Polacheck, I. & Popkin, T. J. Melanin-Lacking Mutants of Cryptococcus neoformans and Their Virulence for Mice. J. Bacteriol. 150(3), 1414–1421 (1982).
Tay, S. T., Na, S. L. & Tajuddin, T. H. Natural occurrence and growth reaction on canavanine-glycine-bromothymol blue agar of non-neoformans Cryptococcus spp. in Malaysia. Mycoses 51(6), 515–519. https://doi.org/10.1111/j.1439-0507.2008.01516.x (2008).
Pfeiffer, T. J. & Ellis, D. H. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. J. Med. Vet. Mycol. 30, 407–408 (1992).
Toro Zúñiga, V. & Brevis, A. P. Presumptive isolation and characterization of Cryptococcus neoformans and Cryptococcus gattii from trees in the region of O’Higgins and Maule Chile. Boletín Micológico. 30(2), 6–15 (2015).
Castañeda, A., Huérfano, S., Rodríguez, M. C. & Castañeda, E. Recovery of Cryptococcus neoformans var. gattii serotype C from almond tree debris. [Recuperación de Cryptococcus neoformans var. gattii serotipo C a partir de detritos de almendros]. Biomedica 21(1), 70–74 (2001).
Escandón, P., Sánchez, A., Firacative, C. & Castañeda, E. Isolation of Cryptococcus gattii molecular type VGIII, from Corymbia ficifolia detritus in Colombia. Med Mycol 48(4), 675–678 (2010).
Contreras Martínez, O. I., Aycardi Morinelli, M. P., Alarcón Furnieles, J. L. & Jaraba Ramos, A. M. Identificación presuntiva de Cryptococcus gattii aislado de Terminalia catappa en Montería, Córdoba Colombia. Revista Cubana de Med Trop. 63(2), 117–122 (2011).
Andrade-Silva, L., Ferreira-Paim, K., Silva-Vergara, M. L. & Pedrosa, A. L. Molecular characterization and evaluation of virulence factors of Cryptococcus laurentii and Cryptococcus neoformans strains isolated from external hospital areas. Fungal Biol 114(5–6), 438–445. https://doi.org/10.1016/j.funbio.2010.03.005 (2010).
Pedroso, R. S., Ferreira, J. C. & Candido, R. C. The isolation and characterization of virulence factors of Cryptococcus spp. from saprophytic sources in the city of Ribeirão Preto, São Paulo, Brazil. Microbiol. Res. 164(2), 221–227. https://doi.org/10.1016/j.micres.2007.01.002 (2009).
Cheng, M. F., Chiou, C. C., Liu, Y. C., Wang, H. Z. & Hsieh, K. S. Cryptococcus laurentii fungemia in a premature neonate. J. Clin. Microbiol. 39(4), 1608–1611. https://doi.org/10.1128/JCM.39.4.1608-1611.2001 (2001).
Burbano Pérez, S., Gómez Querales, N., González, A., Tummino, C. & Asquineyer, Y. Compromiso pulmonar por Cryptoccus laurentii en paciente inmunocomprometido. Revista Am. de Med. Respir. 20(2), 185–188 (2020).
Araújo, G. R. et al. The environmental yeast Cryptococcus liquefaciens produces capsular and secreted polysaccharides with similar pathogenic properties to those of C Neoformans. Sci. Rep. 7(1), 46768. https://doi.org/10.1038/srep46768 (2017).
Butinar, L., Spencer-Martins, I. & Gunde-Cimerman, N. Yeasts in high Arctic glaciers: The discovery of a new habitat for eukaryotic microorganisms Antonie van Leeuwenhoek. Int. J. General Mol. Microbiol. 91(3), 277–289. https://doi.org/10.1007/s10482-006-9117-3 (2007).
Ellis, D. & Pfeiffer, T. Cryptococcosis and the ecology of Cryptococcus neoformans. Jpn. J. Med. Mycol. 35(2), 111–122. https://doi.org/10.3314/jjmm.35.111 (1994).
Anacona, C., Vásquez-A, L. R. & Escandón, P. First isolation and molecular characterization of Cryptococcus neoformans var. grubii in excreta of birds in the urban perimeter of the Municipality of Popayán. Colomb. Rev. Iberoam. de Micol. 35(3), 123–129 (2018).
Angarita-Sánchez, A., Cárdenas-Sierra, D., Parra-Giraldo, C., Diaz-Carvajal, C. & Escandon-Hernandez, P. Recovery of environmental Cryptococcus neoformans and Cryptococcus gattii and their association with clinical isolates in Cúcuta, Colombia. [Recuperación de Cryptococcus neoformans y Cryptococcus gattii ambientales y su asociación con aislados clínicos en Cúcuta, Colombia.]. MVZ Córdoba 24(1), 7137–7144 (2019).
Firacative, C., Torres, G., Escandón, P. & Rodríguez, M. C. First environmental isolation of Cryptococcus gattii serotype B, from Cúcuta, Colombia. Biomedica 31(1), 118–123 (2011).
Noguera, M. C., Escandón, P. & Castañeda, E. Cryptococcosis in Atlántico, Colombia: An approximation of the prevalence of this mycosis and the distribution of the etiological agent in the environment. Rev. Soc. Bras. Med Trop 48(5), 580–586 (2015).
Trilles, L. et al. Identification of the major molecular types of Cryptococcus neoformans and C gattii by hyperbranched rolling circle amplification. PLoS ONE 9(4), e94648. https://doi.org/10.1371/journal.pone.0094648 (2014).
Firacative, C. et al. MLST and whole-genome-based population analysis of Cryptococcus gattii VGIII Links clinical, veterinary and environmental strains, and Reveals divergent serotype specific sub-populations and distant ancestors. PLoS Negl. Trop. Dis. 10(8), e0004861. https://doi.org/10.1371/journal.pntd.0004861 (2016).
Google Maps., “[Coordenadas Ricaurte Alto Boyacá, Colombia en Google maps].” https://www.google.com.co/maps/@4.6315748,-74.0699088,11.79z?hl=en
Torres-Rodríguez, J. M., Alvarado-Ramírez, E. & Gutiérrez-Gallego, R. Diferencias en la actividad de la enzima ureasa entre Cryptococcus neoformans y Cryptococcus gattii. Rev. Iberoam. Micol. 25(1), 27–31 (2008).
Min, K. H. & Kwon-Chung, K. J. The biochemical basis for the distinction between the two Cryptococcus neoformans varieties with CGB medium. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 261(4), 471–480. https://doi.org/10.1016/S0176-6724(86)80079-7 (1986).
Pérez, C. et al. Mantenimiento de Cryptococcus sp. con el método de Castellani. Rev. de la Soc. Venez. de Microbiol. 2, 153–157 (2003).
Croxatto, A., Prod’hom, G. & Greub, G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 36(2), 380–407. https://doi.org/10.1111/j.1574-6976.2011.00298.x (2012).
Escandón, P., Sánchez, A., Martínez, M., Meyer, W. & Castañeda, E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res 6(4), 625–635 (2006).
Gardes, M. & Bruns, T. D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118 (1993).
Vilgalys, R. Phylogenetic implications of generic concepts in fungal taxonomy: the impact of molecular systematic studies. Mycol. Helv. 6, 73–91 (1994).
Vallejo, P. M. Correlación y regresión, simple y múltiple (Universidad Pontificia Comillas, 2012).
H. Chitarroni, “El análisis de correlación y regresión lineal entre variables cuantitativas.” [Online]. Available: http://www.salvador.edu.ar/csoc/idicso, 2002.
Acknowledgements
The authors thank Jeimmy Valbuena, Javier Sierra, and Diego Moreno for their help collecting samples. To Pablo Andrés Gil for their collaboration in the construction of the map. Also, to Universidad Pedagógica y Tecnológica de Colombia (UPTC), Pontificia Universidad Javeriana, Instituto Nacional de Salud, and Universidad de Boyacá for the technical and financial support. And the anonymous reviewers who participated in the critical review of the manuscript.
Author information
Authors and Affiliations
Contributions
Study design, M.F.C., Z.A.S.Q.; development and methodology, B.N.S.E., M.F.C., Z.A.S.Q., C.M.P.G.; collection of data, B.N.S.E., Z.A.S.Q.; formal analysis, B.N.S.E., M.F.C., P.E., Z.A.S.Q.; writing—original draft preparation, B.N.S.E., M.F.C., Z.A.S.Q.; writing—review and editing, M.F.C., P.E., Z.A.S.Q., B.N.S.E. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Serna-Espinosa, BN., Forero-Castro, M., Morales-Puentes, M. et al. First report of environmental isolation of Cryptococcus and Cryptococcus-like yeasts from Boyacá, Colombia. Sci Rep 13, 15755 (2023). https://doi.org/10.1038/s41598-023-41994-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-41994-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.