Abstract
A new system based on binary Deep Learning (DL) convolutional neural networks has been developed to recognize specific retinal abnormality signs on Optical Coherence Tomography (OCT) images useful for clinical practice. Images from the local hospital database were retrospectively selected from 2017 to 2022. Images were labeled by two retinal specialists and included central fovea cross-section OCTs. Nine models were developed using the Visual Geometry Group 16 architecture to distinguish healthy versus abnormal retinas and to identify eight different retinal abnormality signs. A total of 21,500 OCT images were screened, and 10,770 central fovea cross-section OCTs were included in the study. The system achieved high accuracy in identifying healthy retinas and specific pathological signs, ranging from 93 to 99%. Accurately detecting abnormal retinal signs from OCT images is crucial for patient care. This study aimed to identify specific signs related to retinal pathologies, aiding ophthalmologists in diagnosis. The high-accuracy system identified healthy retinas and pathological signs, making it a useful diagnostic aid. Labelled OCT images remain a challenge, but our approach reduces dataset creation time and shows DL models’ potential to improve ocular pathology diagnosis and clinical decision-making.
Similar content being viewed by others
Introduction
A large part of clinical data consists of medical images that might contain relevant features that are not visible to the human eye. Thus, there is a growing interest in the development of computer-aided systems for the automated examination of OCT images useful to support ophthalmologists in diagnosis. OCT images are fundamental for the diagnosis of numerous retinal diseases, being able to provide detailed information about all retinal layers.
Deep learning (DL), a method of Machine Learning (ML), is changing the approach to the diagnosis and management of different medical pathologies since advanced DL techniques can detect pathological characteristics1,2. In particular, ML algorithms are powerful tools in the automatic detection and quantification of retinal biomarkers identified on OCT3,4,5. In the last years, different ML models were developed and widely used for the recognition of OCT images acquired on patients with major eye pathologies such as diabetic retinopathy (DR), age-related macular degeneration (AMD), central serous chorioretinopathy (CSC), epiretinal membrane (ERM) and glaucoma6,7,8,9,10,11,12,13,14,15,16.
Regarding OCT images classification, the most used CNN architectures are VGG, ResNet and Inception, and have shown very promising results so far17,18,19,20,21.
Despite the promising results given by the literature on the use of the VGG-16, ResNet-50, and Inception-v3 architectures for the classification of OCT images, the need for large data sets and non-standardized image acquisition techniques limits the applicability of ML in the clinical domain22. Furthermore, a low diffusion of ML-based decision-making in healthcare should be underlined, mainly due to a lack of interpretability of the classification process related to DL-based methods23. In fact, decision-making by VGG-16 as well as by other DL architectures happens in a black-box mode, i.e. without having evidence of the process that led to a certain result.
To overcome some of the challenges of clinical applicability/interpretability and the requirement of large and balanced datasets of the DL, our study focused not on direct pathology classification, but on the identification of retinal abnormality signs by using the DL approach applied to OCT images.
The detection of abnormalities provides direct insight into the presence of one or more signs that can be used by ophthalmologists as a guide in the decision process. Thus, the automatic identification of retinal abnormality signs from OCT images is a fundamental building block in developing a first step of an interpretable decision support system for the diagnosis of retinal pathologies.
Our study aimed to identify the presence of one or more of the following abnormality signs: epiretinal membrane (ERM), intraretinal fluid (IF), subretinal fluid (SF), drusen (D), macular neovascularization (MNV), vitreomacular adhesion (VMA), macular hole (MH) and backscattering (BS).
The identification of singular abnormality signs makes it possible to imitate the deductive process used by the ophthalmologist to diagnose ocular pathologies rather than relying exclusively on the outcome (pathological or not) of a black-box model as that based on DL. This approach also reduces the overall number of images generally necessary to identify different pathologies.
Materials and methods
This retrospective observational study was conducted at the University Eye Clinic of Trieste. All patients enrolled in the study signed an informed consent to use the data. The retrospective study was carried out following the principles of the Declaration of Helsinki, and the research protocol received approval from the Regional Ethics Committee (CEUR) of Friuli Venezia Giulia, Italy (protocol n. 17,094/2022).
Data collection
Completely anonymized OCT scans in A line-scans protocol of 9.0 mm length were retrospectively analyzed. Images were acquired by Spectralis OCT (Heidelberg Engineering, Heidelberg, Germany) with 815 nm laser source, 3.9 μm/pixel axial resolution, 5.7 μm/pixel lateral resolution and 768 × 496 pixel image size.
The study included horizontal and vertical line scans, centered on the fovea, of healthy and pathological eyes, of adults between 18 and 95 years old, acquired from January 2017 to September 2022.
The inclusion criteria for the pathological group were the presence of one or more of the following signs: ERM, IF, SF, D, MNV, VMA, MH and BS. The healthy group consisted of individuals who did not present any retinal abnormal sign on OCT scans. Poor quality images (Spectralis Quality parameter lower than 23) were excluded.
Image labeling and preprocessing
All images were examined and labelled by two experienced retinal specialists (LI,DM). Poor quality images, OCT scans outside the foveal area and images for which an agreement was not reached between the two specialists were excluded from the dataset.
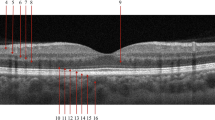
Representative OCT images of each sign are shown in Fig. 1. Each image was cropped in the central area of the scan to 621*445 pixels and then resized to 224*224 pixels, to obtain the default input image size for the VGG-16 convolutional neural networks algorithm. The resizing was accomplished by using a bicubic interpolation.
Datasets population and training process
The labeled images were preprocessed to create 9 predictive binary models. The first model was trained to identify scans belonging to the healthy or pathological group, while the remaining 8 models were used to further identify each of the signs of retinal degeneration. In the first model, the group of all images belonging to healthy eyes and the group of images containing at least one sign was used. In the other eight cases, the group of images including one specific sign and the group containing images lacking that sign was considered. To have a balanced dataset, for each model, the number of images considered was the same for each group. The 10% of images coming from healthy as well as the 10% of images of each sign were randomly selected and used as the test set. The remaining 90% of images was used for the fivefold cross-validation. Table 1 reports the number of images containing one or more abnormal signs.
Modeling
Among the three most used CNN architectures, in this study, we selected VGG-16 because it presents a low number of hidden layers and a small convolution filter (3 × 3), thus requiring a small training data set, probably reducing the network’s tendency to overfit during training.
The selection of VGG has been proven effective in imaging for medical diagnosis. The review of analyzed the trends in the application of deep learning networks in medical image analysis between 2012 and 2020, and found that VGG was among the three most frequently used Convolutional neural network-derived networks applied in medical image analysis24. The architecture was used to diagnose Choroidal Neovascularization (CNV) in retinal OCT images with an accuracy of approximately 97.525. In the classification of diabetic retinopathy, the modified VGG16 has been proposed and outperformed state-of-the-art methods in terms of accuracy and computational resource utilization26. Outside of ophthalmology, VGG16 has also been applied in the classification of breast cancer using mammography images, achieving a test score of 88%27. Additionally, VGG16 has been utilized in brain tumor detection through MRI, achieving a high accuracy of about 96.1% UNet-VGG16 with transfer learning for MRI-based brain tumor segmentation28. In the field of breast histopathology image analysis, VGG16 has been used as a pre-trained model to extract high-level features for breast cancer classification29. Furthermore, a modified version of VGG16, has been proposed for the classification of pneumonia X-ray images, demonstrating superior outcomes compared to other convolutional neural networks30.
As the goal was to obtain nine binary classifiers, we used the modified VGG-16 model depicted in Fig. 2.
Each of the nine binary models was developed using transfer learning and fine-tuning techniques on the pre-trained model (VGG-16). To build the model, the top (rightmost in Fig. 2) layers of the VGG-16 were replaced by a custom layer, and the sigmoid dense layer was used for classification, while the previous layers were kept frozen. However, since dense layers take 1D vectors as input, while the output of previous layers is 3D tensors, a flattened layer converting data into a 1-dimensional array was used.
The new layers can learn patterns from previously learnt convolutional layers because a very small learning rate is utilized (Adaptive Moment Estimation Algorithm (ADAM) with a learning rate of 0.0001)31.
By applying this approach, the retinal abnormality signs could be recognized even if the pre-trained VGG-16 were not trained using our images. For training each model, the images were resized and augmented using typical data augmentation techniques. Once this was done, we flowed them in batches of 32 into the model and started the training. Each model was trained through two steps: in the first step, the model was trained with frozen convolution layers for adjusting the top layers (transfer learning). In the second step, the early stopping technique was used if after eight epochs there is no improvement in the accuracy measured on the validation set.
Each model was trained for a maximum of 70 epochs: 40 epochs for the transfer learning phase and 30 epochs for fine-tuning, always using batches of 32 elements. The number of maximum epochs was determined empirically in preliminary trials after recording the number of steps the model needed to converge. The early Stopping technique was applied to monitor the accuracy of the models for each epoch on the validation datasets and to terminate the process when the performances did not further improve. At the end of the training, the model with the best performance on the validation set was selected and tested on the test sets.
Models were trained using Python version 3.10 and Keras, a high-level API of Tensorflow 2, on a computer equipped with Ryzen 7 2700 processor, NVIDIA RTX 3070ti graphic card and 16 GB DDR4 ram.
Evaluation metrics
Confusion matrices were generated to understand the detail of the misinterpretations and to evaluate the performance of the model by computing the following metrics: accuracy, sensitivity and specificity, and area under the ROC curve (AUC). Cohen's Kappa indexes were obtained to examine the agreement between the systems with the ground truth on the assignment of categories of labelled variables. All analyses were carried out through the Python library scikit-learn32.
Model visualization (GRAD-CAM)
To understand the CNN predictions, Gradient-weighted class activation mapping (Grad-CAM heatmap) for each CNN model was used. Grad-CAMs were implemented before the last fully connected layer of VGG16 and allowed to highlight the regions most involved in the decision made by the model. The regions of interest or crucial features within the input data that influenced the model's decision were visually identified by generating heatmaps. Insights into the reasoning behind the model's predictions were gained with the help of this approach. Examples of Grad-CAM heat maps are shown in Fig. 3.
Results
A total of 21,500 completely anonymized OCT scans of 11,245 patients (5258 Male and 5987 Female) with a mean age of 71.2 ± 16.5 were screened. These images were collected randomly from Heidelberg Spectralis OCT database. After this initial selection 10,770 images were included in the study. Of those, 3120 did not show any pathological sign and were marked as normal and 7650 were labelled as pathological, specifying the detected abnormality sign/s. Images presenting more than one sign were counted multiple times, thus 1587 ERM, 2086 IF, 762 SF, 1698 D, 819 MNV, 2186 VMA, 489 MH and 985 BS images, for a total of 10,612 images presenting one or more signs, were utilized.
Nine CNN models were created and trained to recognize an image as normal (no pathological signs) vs. pathological (presence of pathological signs), as well as to differentiate each pathological sign from the others. An example of a typical increase in the accuracy metric as well as the decrease in loss during the training phases is shown in Fig. 4.
Nine confusion matrices were calculated both on the validation and the test set. The results are reported in Tables 2 and 3. In each matrix, the rows represent the instances in the actual classes while the columns represent the instances in the predicted classes. Tables 4 and 5 show the accuracy, sensitivity, specificity, kappa value, and AUC for each of the nine CNN models calculated on the test and the validation set, respectively.
Figure 3 shows an example of the heat maps of each pathological sign highlighting the correct localization and identification obtained by the algorithm. The system was also capable of recognizing multiple signs present in a single OCT image, as shown in the example of Grad-cam heatmaps in Fig. 5 in which the image presents three different signs. In some cases, CNNs have misclassified OCT images. This has happened only a few times as all the networks achieved very high accuracies, as highlighted before. Figure 6 shows some cases where the CNNs have produced incorrect heatmaps.
Cases in which neural networks have misclassified OCT images, providing incorrect heatmaps. (A) N classified as VMA. (B) D and BS as MNV (C) D classified as N. (D) IRF and BS classified as N. (E) MNV, SRF and IRF classified as MH. (F) SRF and VMA localized in the wrong position (G) IRF and SRF classified as IRF (H) IRF classified as MH (I) IRF classified as VMA.
Discussion
Nowadays OCT is an essential exam to diagnose several retinal pathologies such as DR, AMD, ERM, and MH along with other techniques, such as fundus photography and fluorescein angiography33,34,35,36,37.
Several authors developed DL systems to detect DR and diabetic macular oedema by using OCT6,38,39,40. Ting successfully trained a DL system to recognize DR, achieving remarkable results with an AUC of 0.958, a sensitivity of 100%, and a specificity of 91.1%6. In 2017, Lee and coworkers developed a DL system for the automated segmentation of macular oedema and showed an excellent performance in comparison with retina experts4. Kermany developed a CNN capable of distinguishing normal from diabetic retinopathy on 62,489 OCT images, with an impressive accuracy of 98.2%, a sensitivity of 96.8%, and a specificity of 99.6%41. In a study published in 2017, Schlegl and coworkers developed a fully automated method to detect and quantify intraretinal cystoid fluid (IRC) with an AUC of 0.9440. Abràmoff et al. created a CNN capable of recognizing DR on OCT images with an AUC of 0.98, a sensitivity of 96.8% and a specificity of 87.0%8.
An AI system proposed by Burlina and coworkers was capable of detecting AMD from OCT images with a good performance and an accuracy between nearly 92% and 95% in different groups42. Similarly, Ting et al. developed a DL system that recognizes AMD in a multiethnic population with diabetes with an AUC of 0.93, a sensitivity of 93.2% and a specificity of 88.7%6. Moreover, Kermany and coworkers obtained an accuracy of 96.6%, a sensitivity of 97.8% and a specificity of 97.4% to diagnose AMD from OCT images41. CNNs were also trained to recognize specific biomarkers for the prediction and progression of AMD disease43,44,45,46,47,48,49. Despite the availability of a large number of studies, their applicability in clinical practice is limited when considering real-world hospital conditions. Yanagihara et al.22 showed that one of the challenges among the others is the limited interpretability of a DL model and the non-standardized datasets, which imposes that each hospital creates its dataset.
All the studies mentioned above utilized binary classification methods to distinguish between pathological and normal OCT images and made a black-box diagnosis based on a single OCT image per patient. However, clinical diagnoses rely on identifying abnormalities across a series of OCT images taken from the same patient, as a single image may not capture all the necessary information. One possible way to address this issue is to focus on classifying signs of retinal abnormality rather than the pathologies themselves. Not many studies reported the recognition of signs. Son et al. created a system that accurately detects 15 abnormal retinal findings and diagnoses 8 major eye diseases using macula-centered fundus images. They introduced the concept of counterfactual attribution ratio (CAR) to illuminate the system's diagnostic reasoning, showing how each abnormal finding contributes to its prediction. CAR allows for quantitative and qualitative interpretation, interactive adjustments, and confirms the model's ability to identify findings and diseases similar to ophthalmologists50. Lu et al. proposed a DL system capable to discriminate normal images, cystoid macular oedema, serous macular detachment, ERM, and MH with an accuracy of 97%, 84%, 94%, 96% and 98%, respectively51. Rajagopalan et al. classified choroidal neovascularization (CNV), drusen and diabetic macular oedema (DME) with an accuracy of 97%, a sensitivity of 93%, and a specificity of 98%52. In another study, Kurmann implemented a machine learning method capable of recognizing various conditions in OCT B-scan images, including subretinal fluid (SRF), intraretinal fluid (IRF), intraretinal cysts (IRC), hyperreflective foci (HF), drusen, reticular pseudodrusen (RPD), epiretinal membrane (ERM), geographic atrophy (GA), outer retinal atrophy (ORA), and fibrovascular pigment epithelial detachment (FPED). They developed the DL system using 23,030 OCT B-scan images, achieving remarkable results53.
Our DL models like those of Kurmann were trained on small datasets (which could be more easily acquired within a single hospital) and were designed to detect a variety of retinal abnormalities in multiple input images from the same patient. By identifying individual abnormality signs, they replicate the deductive process followed by the ophthalmologist in diagnosing ocular pathologies, rather than solely relying on the results generated by a black-box DL learning model. The clinical procedure, where doctors have access to multiple images and use them to assess the presence of all nine signs, was aimed to be replicated. A comprehensive understanding of the process of sign identification was sought by observing the classifier output, including class probabilities, and analyzing the heatmaps. This method also minimizes the total number of images typically required to distinguish between different pathologies. Whereas the creation of a CNN model specific to a particular pathology requires many images associated with that pathology, the identification of a sign could be accomplished by using images that are common to different pathologies. Therefore, our approach drastically reduced the overall time necessary for image collection. The VGG-16 is based on a relatively simple CNN design consisting of a series of stacked convolutional layers followed by max pooling and then fully connected layers at the end. This simple architecture means that VGG-16 has a smaller number of parameters compared to ResNet and Inception, which have more complex architectures with skip connections, residual blocks, and inception modules that enable them to learn more complex features.
Lee et al. demonstrated that CNN can be successfully used to distinguish normal OCT images from patients with AMD20. The authors extracted 2.6 million OCT images from normal subjects and AMD patients. Of these, 80,839 images were selected to train a CNN model, while 20,163 images were used to validate it. The architecture chosen was a modified version of the VGG-16 network. ROC curves were created at the image level, macular level and patient level, and the AUCs achieved were 92.78%, 93.83%, and 97.45%, respectively. Choi et al. trained and validated three CNNs to classify normal, high myopia, and other retinal disease groups based on OCT images21. The authors adopted three specific architectures (VGG-16, ResNet-50, and Inception-v3) as a backbone and developed models to perform image classification. The best AUCs of the three CNNS models were 99.9% for VGG-16, 100.0% for ResNet-50 and 96.1% for Inception-v3.
Despite using simpler architecture, comparably to the previous works, our models achieved a high level of accuracy on both the training and test sets, ranging from 93 to 99%, for identifying healthy retinas and eight specific pathological signs. The similar model performance on both the validation and the test sets, suggests that our nine models were robust, did not overfit during the training and learnt to capture the underlying patterns related to retinal abnormality signs so that they could classify well also unseen data. Finally, the relatively high performance of our models, demonstrated by the results, underlines the potential capacity of these models to identify single or multiple signs in OCT images.
We acknowledge that VGG16 is not the most recent architecture and might not have the highest accuracy, and that some approaches might achieve some more, however, even such achieving the clinically relevant results for retinal deterioration sign detections. We believe also that utilizing VGG's general-purpose nature and the abundance of tutorials and implementations available is its advantage. Apart from that, the time it takes for the models to classify the image depends mostly on the characteristics of the computer used. In our settings (with our computer), the time to classify the uploaded image in the system was 2.2 s. The real-time classifier performance holds significance, it is not the primary factor determining its clinical relevance. While achieving extremely high accuracy (e.g., close to 100%) may be unrealistic or impractical for some medical imaging tasks, it is important to focus on achieving clinically relevant accuracy levels54,55.
These levels may vary depending on the specific medical task, the potential impact on patient care, and the specific use-case scenario56.
Markedly, our approach could allow ophthalmologists to analyze each OCT image separately, as not all signs might be discernible in every image. Furthermore, since the system could identify individual signs rather than being restricted to single retinal pathologies, it could serve as a diagnostic aid for a much wider range of pathologies presenting a different combination of these signs. On the other hand, the classification of singular signs might be considered a drawback, as it still requires the intervention of the ophthalmologist to identify a pathology as required in automated screening applications.
Conclusions
The development of DL models that can accurately and automatically detect abnormal retinal signs from OCT images has significant implications for patient care. Although many studies have focused on the classification of ocular pathologies, our study aimed to identify individual signs related to a pathology, which allows the ophthalmologist more room to provide additional interpretation to reach a correct diagnosis. Our system achieved high accuracy in identifying healthy retinas as well as specific pathological signs making it a useful diagnostic aid for a wide range of pathologies. The Grad-Cam visualization enhanced the interpretability of our CNN's results, allowing ophthalmologists to assess the model's efficacy. While the need for a considerable amount of labelled OCT images to train the model remains a challenge, our approach reduced the time required to create separate datasets for each retinal pathology. In our study, we utilized the VGG16 architecture. Despite its accessibility, there are potential drawbacks associated with it. Exploring the feasibility of employing newer deep learning architectures and comparing their performance could enhance the integration of machine learning into the diagnostic process. Overall, our study demonstrated the potential of DL models in improving the diagnosis of ocular pathologies and supporting clinical decision-making.
Data availability
The datasets generated and analyzed during the study are not publicly available due to privacy constraints. The data may however be available from the University of Trieste subject to local and national ethical approvals. In addition, we have made the models and relevant code available upon request. Any requests should be sent to the corresponding author.
References
Jiang, F. et al. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2(4), 230–243 (2017).
Korot, E. et al. Code-free deep learning for multi-modality medical image classification. Nat. Mach. Intell. 3, 288–298 (2021).
Liefers, B. et al. Quantification of key retinal features in early and late age-related macular degeneration using deep learning. Am. J. Ophthalmol. 226, 1–12 (2021).
Lee, C. S. et al. Deep-learning based, automated segmentation of macular edema in optical coherence tomography. Biomed. Opt. Express. 8(7), 3440 (2017).
Schmidt-Erfurth, U. et al. AI-based monitoring of retinal fluid in disease activity and under therapy. Prog. Retin. Eye Res. 86, 100972 (2022).
Ting, D. S. W. et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA–J. Am. Med. Assoc. 318(22), 2211–2223 (2017).
Gulshan, V. et al. Development and validation of a deep learning algorithm for the detection of diabetic retinopathy in retinal fundus photographs. JAMA–J. Am. Med. Assoc. 316(22), 2402–2410 (2016).
Abràmoff, M. D. et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Investig. Ophthalmol. Vis. Sci. 57(13), 5200–5206 (2016).
Gargeya, R. & Leng, T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology 124(7), 962–969 (2017).
Li, Z. et al. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology 125(8), 1199–1206 (2018).
Burlina, P. M. et al. Automated grading of age-related macular degeneration from colour fundus images using deep convolutional neural networks. JAMA Ophthalmol. 135(11), 1170–1176 (2017).
Grassmann, F. et al. A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology 125(9), 1410–1420 (2018).
Yousefi, S. et al. Detection of longitudinal visual field progression in glaucoma using machine learning. Am. J. Ophthalmol. 193, 71–79 (2018).
Lo, Y. C. et al. Epiretinal membrane detection at the ophthalmologist level using deep learning of optical coherence tomography. Sci. Rep. 10(1), 8424 (2020).
Kim, S. H., Ahn, H., Yang, S., Soo Kim, S. & Lee, J. H. Deep learning-based prediction of outcomes following noncomplicated epiretinal membrane surgery. Retina 42(8), 1465–1471 (2022).
Crincoli, E. et al. New artificial intelligence analysis for prediction of long-term visual improvement after epiretinal membrane surgery. Retina 43(2), 173–181 (2023).
Simonyan, K., Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition.; Computer Science > Computer Vision and Pattern Recognition (2015).
He, K., Zhang, X., Ren, S., Sun, J. Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016-December. 770–778 (2016).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016-December. 2818–2826 (2016).
Lee, C. S., Baughman, D. M. & Lee, A. Y. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol. Retin. 1(4), 322–327 (2017).
Choi, K. J. et al. Deep learning models for screening of high myopia using optical coherence tomography. Sci. Rep. 11(1), 21663 (2021).
Yanagihara, R. T., Lee, C. S., Ting, D. S. W. & Lee, A. Y. Methodological challenges of deep learning in optical coherence tomography for retinal diseases: A review. Transl. Vis. Sci. Technol. 9(2), 11 (2020).
Singh, A., Mohammed, A.R., Zelek, J., Lakshminarayanan, V. Interpretation of deep learning using attributions: application to ophthalmic diagnosis. In: Proc. SPIE 11511, Applications of Machine Learning. 11511: 9 (2020).
Wang, L. et al. Trends in the application of deep learning networks in medical image analysis: Evolution between 2012 and 2020. Eur. J. Radiol. 146, 110069 (2022).
Abirami, M. S., Vennila, B., Suganthi, K., Sarthak, K. & Anuja, V. Detection of choroidal neovascularization (CNV) in retina OCT images using VGG16 and DenseNet CNN. Wireless Pers. Commun. 127, 2569–2583 (2022).
Khan, Z. et al. Diabetic retinopathy detection using VGG-NIN a deep learning architecture. IEEE Access 9, 61408–61416 (2021).
Sashikanta, P., Sujit, K. D. & Srikanta, P. A novel transfer learning technique for detecting breast cancer mammograms using VGG16 bottleneck feature. ECS Trans. 107, 733–746 (2022).
Anindya Apriliyanti, P. et al. UNet-VGG16 with transfer learning for MRI-based brain tumor segmentation. TELKOMNIKA 18(3), 1310–1318 (2020).
D. Albashish, R., Al-Sayyed, A., Abdullah, M. H. R., Nedaa, A. A. Deep CNN model based on VGG16 for breast cancer classification. International Conference on Information Technology (ICIT). 805–810 (2021).
Jiang, Z. P., Yi-Yang, L., Zhen-En, S. & Ko-Wei, H. An improved VGG16 model for pneumonia image classification. Appl. Sci. 23, 11185 (2021).
Kingma, D.P., Ba, J. Adam: A method for stochastic optimization. December 2014 (2014).
Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 12(85), 2825–2830 (2011).
Spaide, R. F., Fujimoto, J. G., Waheed, N. K., Sadda, S. R. & Staurenghi, G. Optical coherence tomography angiography. Prog. Retin Eye Res. 64, 1–55 (2018).
Laíns, I. et al. Retinal applications of swept source optical coherence tomography (OCT) and optical coherence tomography angiography (OCTA). Prog. Retin. Eye Res. 84, 100951 (2021).
Schneider, E. W. & Fowler, S. C. Optical coherence tomography angiography in the management of age-related macular degeneration. Curr. Opin. Ophthalmol. 29(3), 217–225 (2018).
Corvi, F. et al. Optical coherence tomography angiography for detection of macular neovascularization associated with atrophy in age-related macular degeneration. Graefe’s Arch. Clin. Exp. Ophthalmol. 259(2), 291–299 (2021).
Lindtjørn, B., Krohn, J. & Forsaa, V. A. Optical coherence tomography features and risk of macular hole formation in the fellow eye. BMC Ophthalmol. 21, 351 (2021).
Abbas, Q., Fondon, I., Sarmiento, A., Jiménez, S. & Alemany, P. Automatic recognition of severity level for diagnosis of diabetic retinopathy using deep visual features. Med. Biol. Eng. Comput. 55(11), 1959–1974 (2017).
Li, F., Chen, H., Liu, Z., Zhang, X. & Wu, Z. Fully automated detection of retinal disorders by image-based deep learning. Graefe’s Arch. Clin. Exp. Ophthalmol. 257(3), 495–505 (2019).
Schlegl, T. et al. Fully automated detection and quantification of macular fluid in OCT using deep learning. Ophthalmology 125(4), 549–558 (2018).
Kermany, D. S. et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172(5), 1122-1131.e9 (2018).
Burlina, P., Freund, D.E., Joshi, N., Wolfson, Y., Bressler, N.M. Detection of age-related macular degeneration via deep learning. In: Proceedings - International Symposium on Biomedical Imaging. Vol 2016-June. Czech Republic; 184–188 (2016).
Russakoff, D. B., Lamin, A., Oakley, J. D., Dubis, A. M. & Sivaprasad, S. Deep learning for prediction of AMD progression: A pilot study. Investig. Ophthalmol. Vis. Sci. 60(2), 712–722 (2019).
Thakoor, K. A. et al. A multimodal deep learning system to distinguish late stages of AMD and to compare expert vs. AI ocular biomarkers. Sci. Rep. 12(1), 2585 (2022).
Saha, S. et al. Automated detection and classification of early AMD biomarkers using deep learning. Sci. Rep. 9(1), 10990 (2019).
Vyas, A., Raman, S., Surya, J., Sen, S. & Raman, R. The need for artificial intelligence based risk factor analysis for age-related macular degeneration: A review. Diagnostics 13(1), 130 (2023).
Samagaio, G. et al. Automatic macular edema identification and characterization using OCT images. Comput. Methods Programs Biomed. 163, 47–63 (2018).
Saha, S. et al. Automated detection and classification of early AMD biomarkers using deep learning. Sci. Rep. 9, 10990 (2019).
Thakoor, K. A. et al. A multimodal deep learning system to distinguish late stages of AMD and to compare expert vs. AI ocular biomarkers. Sci. Rep. 12, 2585 (2022).
Son, J. et al. An interpretable and interactive deep learning algorithm for a clinically applicable retinal fundus diagnosis system by modelling finding-disease relationship. Sci. Rep. 13, 5934 (2023).
Lu, W. et al. Deep learning-based automated classification of multi-categorical abnormalities from optical coherence tomography images. Transl. Vis. Sci. Technol. 7(6), 41 (2018).
Rajagopalan, N., N V, Josephraj, A.N., E S. Diagnosis of retinal disorders from optical coherence tomography images using CNN. PLoS One. 16(7): e0254180 (2021).
Kurmann, T. et al. Expert-level automated biomarker identification in optical coherence tomography scans. Sci. Rep. 9, 13605 (2019).
Kennedy, A. G. Imaging, Representation and diagnostic uncertainty. In: Lalumera, E., Fanti, S. Philosophy of Advanced Medical Imaging. Springer Briefs in Ethics. Springer, Cham. (2020).
Aggarwal, R. et al. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 4(1), 65 (2021).
Varoquaux, G. & Cheplygina, V. Machine learning for medical imaging: Methodological failures and recommendations for the future. NPJ Digit. Med. 5(1), 48 (2022).
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
I.L. designed the experiment, conducted the experiment, provided materials, wrote the article; B.L. conducted the experiment, analyzed/interpreted data, provided materials, wrote the article; M.A. analyzed/interpreted data, wrote the article, proofed/revised article; G.R. proofed/revised article; M.D. conducted the experiment, proofed/revised article; A.A. designed the experiment, analyzed/interpreted data, proofed/revised article; T.D. provided materials, proofed/revised article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leandro, I., Lorenzo, B., Aleksandar, M. et al. OCT-based deep-learning models for the identification of retinal key signs. Sci Rep 13, 14628 (2023). https://doi.org/10.1038/s41598-023-41362-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-41362-4
This article is cited by
-
OCTDL: Optical Coherence Tomography Dataset for Image-Based Deep Learning Methods
Scientific Data (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.