Abstract
Pelagic predators are effective biological samplers of midtrophic taxa and are especially useful in deep-sea habitats where relatively mobile taxa frequently avoid observation with conventional methods. We examined specimens sampled from the stomachs of longnose lancetfish, Alepisaurus ferox, to describe the diets and foraging behaviors of three common, but poorly known deep-sea fishes: the hammerjaw (Omosudis lowii, n = 79, 0.3–92 g), juvenile common fangtooth (Anoplogaster cornuta, n = 91, 0.6–22 g), and juvenile Al. ferox (n = 138, 0.3–744 g). Diet overlap among the three species was high, with five shared prey families accounting for 63 ± 11% of the total prey mass per species. However, distinct differences in foraging strategies and prey sizes were evident. Resource partitioning was greatest between An. cornuta that specialized on small (mean = 0.13 ± 0.11 g), shallow-living hyperiid amphipods and O. lowii that specialized on large (mean = 0.97 ± 0.45 g), deep-dwelling hatchetfishes. Juvenile Al. ferox foraged on a high diversity of prey from both shallow and deep habitats. We describe the foraging ecologies of three midtrophic fish competitors and demonstrate the potential for biological samplers to improve our understanding of deep-sea food webs.
Similar content being viewed by others
Introduction
Diets, foraging strategies, and migratory behaviors of pelagic animals vary with ontogeny and environmental conditions across trophic levels1,2,3, resulting in complex food webs. Numerous midtrophic consumers vertically migrate from daytime refuges in relatively dark mesopelagic habitats (~ 200–1000 m) to forage in epipelagic nighttime habitats (< 200 m) where primary production and total biomass are highest4,5. Others are non-migratory and rely on migratory prey or the passive flux of carbon from surface production to obtain food at mesopelagic depths6,7. However, many midtrophic fishes and squids are mobile enough to avoid sampling by nets and imaging platforms, limiting observations of their diets and foraging depths. In the deep sea, sampling avoidance has prevented robust quantification of resource partitioning among midtrophic taxa, which remains a persistent gap in our understanding of deep pelagic food webs.
Using a predator as a biological sampler of these mobile taxa can provide much needed insights into an otherwise poorly known component of deep pelagic ecosystems. Predator diet analyses, including stomach contents of fishes and squids, mammal scat, and bird boluses have been used to describe the basic biology of individual prey taxa (e.g., diet and habitat use)8,9,10. When monitored over time, predator diets can also be used to quantify how the composition and size structure of prey assemblages respond to environmental variability11,12. The stomachs of deep-sea fishes often function partly as a storage organ, an adaptation to food-limited habitats that preserves prey mostly undigested in stomachs and makes deep-sea fish predators exceptional candidates as biological samplers of deep-sea food webs13,14.
The longnose lancetfish, Alepisaurus ferox, occurs throughout the tropical and subtropical ocean15,16 and consumes a high diversity of fish, mollusk, and crustacean prey that live throughout the upper 1500 m of the water column17,18,19. Alepisaurus ferox exhibits ontogenetic descent from epipelagic to mesopelagic habitats between larval and adult life stages20,21, and increased foraging depths in individuals > 1.82 kg relative to smaller individuals17. In addition to feeding on poorly sampled taxa, Al. ferox is a useful sampler of deep-sea ecosystems because it is an abundant bycatch species on pelagic deep-set longlines16,22, and can be more readily collected than other deep-sea predators through partnerships with fishers and fisheries monitoring programs. Stomach content analysis of Al. ferox has provided several type specimens of novel species23,24 and has been used to study the feeding habits of mesopelagic taxa found in its stomach25,26.
The hammerjaw, Omosudis lowii, juveniles of the fangtooth, Anoplogaster cornuta (< 80 mm standard length, SL27), and juvenile Al. ferox (< 750 mm SL28) have been observed in relatively high numbers in the stomachs of large Al. ferox collected from the central North Pacific Ocean (CNP)17. All three species are midtrophic predators thought to be relatively common in pelagic ecosystems globally13,29,30, but are mostly able to avoid collection by trawls, with the exception of large commercial high-speed rope trawls that are infrequently used for scientific sampling in deep pelagic habitats31. Anoplogaster cornuta (max. reported size 152 mm SL30) exhibits diel vertical migration and occurs from ~ 135–1050 m27, while O. lowii (max. reported size 270 mm SL, this study) is largely non-migratory and found mostly at ~ 600–1000 m21,31. Very little is known about the feeding habits of these species in the CNP and sparse diet studies from other regions are severely limited in sample size or taxonomic resolution26,32,33. Although the diets of large Al. ferox (max. reported size 2080 mm SL29) are relatively well-described from several ocean basins, individuals < ~ 400 g are also poorly studied, perhaps due to size-specific selection of hooks used on longlines17,34. Omosudis lowii, An. cornuta and juvenile Al. ferox are similarly sized, have overlapping habitats, and likely forage on a shared prey community in the CNP, but little is known about how forage resources are partitioned among them.
Using large Al. ferox as a biological sampler, we present a unique diet dataset to (1) describe the diets of O. lowii, juvenile An. cornuta, and juvenile Al. ferox, (2) quantify diet overlap with respect to taxonomic composition and size structure, and (3) evaluate how differential feeding behaviors and vertical habitat use allow for partitioning of shared resources. We also describe ontogenetic variability in the foraging depth of Al. ferox across body sizes spanning four orders of magnitude. This work demonstrates the importance of depth-informed diet analyses to reveal pelagic food web structure and the potential for biological samplers to expand our understanding of deep-sea ecology.
Methods
Specimen collection
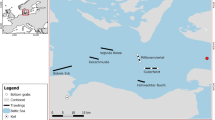
Lancetfish stomachs were collected by federal fisheries observers in the Hawaiʻi-based longline fishery (Hawaiʻi Longline Observer Program, https://www.fisheries.noaa.gov/inport/item/16865) from 2009 to 2020. Observers recorded fork length (FL) to the nearest centimeter as well as the date and location of capture for each specimen. Details of the collection and diet composition of longline-caught individuals, hereafter referred to as “primary lancetfish”, are described in Choy et al.35 and Portner et al.17. Alepisaurus ferox (n = 138), An. cornuta (n = 91), and O. lowii (n = 78) were opportunistically sampled from the stomachs of primary lancetfish for further diet study and are presented here for the first time (Fig. 1, Fig. S1). An additional O. lowii was directly sampled by longline observers and included in the analyses (Table 1). In most cases, specimens were not processed immediately and were re-frozen at − 20 °C. Thawed specimens were weighed (nearest 0.01 g) and measured (SL to the nearest 1 mm) before their stomachs were dissected. Prey were identified to the finest possible taxonomic resolution and assigned a digestion state (following Choy et al.35, 1 = completely intact, 2 = minimally digested, 3 = partially digested, and 4 = heavily digested). Groups of prey with the same taxonomic ID and digestion state in a single stomach (“prey group”) were enumerated and weighed. Stomach content mass was subtracted from whole specimen mass to obtain predator masses used in the analyses. Although we present some of the largest known diet data sets for these deep-sea fishes, sample sizes were not large enough to explore spatial or temporal variability in diets across our study area. The spatial and temporal coverage of sampling was similar among species (Fig. 1, Fig. S1) and stomachs of each predator were pooled across sampling years and locations for all analyses. Unless otherwise specified, analyses were performed with packages in R Statistical Software (version 3.6.336).
Summary of specimen size and collection location across our study area in the central North Pacific Ocean. Longnose lancetfish (Alepisaurus ferox, n = 138), common fangtooth (Anoplogaster cornuta, n = 91), and hammerjaw (Omosudis lowii, n = 79) were collected in the central North Pacific Ocean (a), mostly in waters surrounding the Hawaiian Islands. Heat maps describing the number of stomachs examined per 5° × 5° cell are overlaid on the sampling footprint for all specimens presented in this study (grey, includes primary lancetfish from Portner et al.17). The number of stomachs is given for cells represented by more than 14 stomachs. Specimens ranged in size from 0.32–744.90 g (b), but 91% of all specimens were between 1 and 100 g. Paintings by ACLC.
Diet composition analyses
Diet composition was quantified as the total abundance, mass, and frequency of occurrence for each prey group. The mean proportional abundance (\(\overline{N}\)) and mass (\(\overline{M}\)) per stomach (prey group total/stomach total), as well as the percent frequency of occurrence per predator (%FO) were also quantified. Prior to analyses, \(\overline{M}\) was recalculated at the family level to ensure all prey group identifications were mutually exclusive and reduce the number of zeros in the diet matrix37. Four prey groups considered mutually exclusive from all family-level groups were included at a coarser taxonomic resolution in the family-level analyses. Hyperiid amphipods in the families Brachyscelidae and Lycaeidae were conservatively lumped together into the superfamily Platysceloidea. Crustacean megalopa, fish leptocephali, and deep-sea anglerfishes (Ceratioidei) were not identified further but were each exclusively found in the stomachs of a single predator species and could thus reliably describe diet variability among species. To reduce potential influence of variable predator size distributions on diet comparisons, diet overlap analyses were limited to specimens between 1 and 100 g (91% of specimens). No size restrictions were applied to analyses that explicitly account for predator mass.
Diet overlap among species was quantified using analysis of similarities (ANOSIM) in Primer v.738. Pairwise tests for homogeneity of multivariate dispersion (PERMDISP) and permutational multivariate analysis of variance (PERMANOVA) among species were also performed in Primer v.7. To assess contributions of centroid location and dispersion on observed differences among species, diet overlap was visualized using NMDS in the vegan package (version 2.5-739). In all cases, analyses were performed on a matrix of Morisita-Horn similarities (Cmh40). To reduce the influence of rare taxa on distance metrics (e.g., Legendre & Gallagher41), only prey families contributing more than 1%\(\overline{M}\) for at least one predator species and found in more than one stomach were included in similarity analyses. Unidentified prey groups were also excluded. Family-level sample coverage and diet diversity (Shannon diversity (1D), Hill number of order q = 142) were quantified using the iNEXT package (version 2.0.2043). True diversity was estimated as the asymptotic Shannon diversity (1Dex) and the number of samples required to reach 95% sample coverage are reported to describe the rate of diversity accumulation.
To examine differences in feeding strategies among predators, %FO and prey-specific proportional mass (%\(\overline{M}\)ps) were quantified for the four most important prey groups for each predator. Prey importance was quantified as the index of relative importance ((%\(\overline{N}\) + %\(\overline{M}\))* %FO44). %\(\overline{M}\)ps is a modified proportional mass metric calculated using only stomachs that contained the prey of interest, an analog of the abundance-based metric described in Amundsen et al.45. A 50%\(\overline{M}\)ps threshold can be used to distinguish specialist (> 50) and generalist (< 50) feeding, and when combined with a 50%FO threshold can help describe how prevalent the feeding strategy is among individuals45.
To facilitate comparisons with previous studies, we recalculated \(\overline{M}\) for broad prey types and examined ontogenetic changes in diet contributions. Variability in the \(\overline{M}\) contributions of fish, mollusk, and crustacean prey with predator size were assessed separately for each predator species by fitting generalized additive models (GAMs) specified with beta error distributions and “logit” link functions. All GAMs presented in this study were fit using restricted maximum likelihood estimation in the mgcv package (version 1.8-3846). A single model specified with a beta error distribution describing \(\overline{M}\) of prey in O. lowii was dominated by zeros and ones and would not converge. To improve model performance, we excluded stomachs containing more than one prey type (n = 2, retaining 95% of O. lowii specimens with prey) and fit a GAM specified with a binomial error distribution and “logit” link function.
Prey mass, length, and abundance
For prey groups with digestion states 1–3, individuals were weighed (nearest 0.01 g) and measured (nearest mm; SL for fishes, mantle lengths (ML) for cephalopods, and total lengths for crustaceans). If a prey group contained more than three individuals, a subset representing the minimum, median, and maximum sizes were qualitatively selected and measured. For unmeasured prey items in each stomach, individual mass and length were estimated as the median mass and length of measured individuals from the same prey group. If no individuals of a given prey group were measured, individual mass was estimated by dividing total prey group mass by the number of individuals. GAMs describing family-level length-to-mass relationships for measured individuals were specified with gaussian error distributions and “identity” link functions. Estimated prey lengths for unmeasured individuals were predicted based on estimated prey masses using the family-level GAMs.
The effects of predator species and mass on individual prey mass, total prey mass per stomach, and total number of prey per stomach were examined using multilinear models in R. To meet model assumptions, prey mass, total prey mass, and predator mass were log10-transformed and prey count was log2-transformed prior to model fitting (estimated using ordinary least squares regression). Ninety-five percent confidence intervals and p values were computed using a Wald t-distribution approximation. Model assumptions were checked using standard diagnostic plots in R. The relative explanatory power of predator species and mass in each model was quantified using analysis of covariance (ANCOVA, type III sum of squares) on model outputs in the car package (version 3.0-1347). Statistics describing individual prey mass only include prey in digestive states 1–3. Total prey masses and counts reflect all prey items in a single stomach, regardless of digestion state or level of taxonomic identification.
Foraging depths
To examine variability in foraging depths among predator species and across sizes, the foraging depth of each predator was estimated as the weighted median depth of occurrence of prey in its stomach (Eq. 1). Only prey taxa in families contributing at least 1%\(\overline{M}\) to the overall diet composition for any predator species were included in foraging depth estimations. Median depth of occurrence for each prey taxon was assigned based on reported depths from the literature (Table S1). For taxa that exhibit diel vertical migration, median depths were assigned as the mean of reported daytime and nighttime median depths. For taxa known to exhibit variable habitat depths across ontogeny, length-specific median habitat depths were assigned. For genus- and family-level identifications, median depths of occurrence were averaged from multiple congeners and representative confamilials. The proportional mass of each prey taxa was recalculated using an adjusted total mass per stomach that only included prey for which depth data were available (adj\(\overline{M}\)). Predator foraging depth (Zf) was estimated based on all prey taxa in a single stomach (n) as a function of the median depth of occurrence (z) and adjusted proportional mass (adj\(\overline{M}\)) of each prey taxa (i):
Differences in estimated foraging depths among predator species and across sizes were assessed by fitting a GAM specified with a gaussian error distribution and identity link function.
Ontogeny of predator consumption by lancetfish
To more directly link ontogenetic variability in lancetfish foraging depth to the consumption of Al. ferox, An. cornuta, and O. lowii, diet data from n = 1066 primary lancetfish presented in Portner et al.17 were reanalyzed as described above with some notable differences. Primary lancetfish specimen mass was estimated from FL using a published regression48. Polychaetes in the tribe Alciopini and heteropods in the family Carinariidae are relatively fragile taxa that were common in primary lancetfish stomachs but rarely intact, regardless of digestion state. These prey groups were never individually measured but are best represented by epipelagic taxa and were assigned to a single depth habitat regardless of estimated size (Table S1). For cephalopods in families contributing at least 1%\(\overline{M}\), masses and MLs of individuals identified from beaks were estimated using published regressions following Chen et al.49 and included in adj\(\overline{M}\) calculations.
Differences in foraging depths across predator species and sizes were re-assessed after including primary lancetfish data by fitting a GAM specified with a gaussian error distribution and “identity” link function. To determine how well foraging depths estimated from stomach contents reflect known habitat usage, estimates were compared to reported median depths of occurrence for each predator (Table S1). The estimated foraging depths of all three species, including primary lancetfish, were qualitatively compared to changes in the %FO of all three species in the stomachs of Al. ferox with increasing size.
Results
Diet description and overlap among predators
Of the 308 predator stomachs examined, 116 Al. ferox (84%), 81 An. cornuta (89%), and 40 O. lowii (51%) contained prey. A total of 2035 prey individuals representing 113 unique taxa were identified from 58 families (32 fish, 16 mollusk, 8 crustacean, and 2 other invertebrates). Counts, masses, frequency of occurrence, and proportional metrics (%\(\overline{N}\), %\(\overline{M}\), %FO) of each prey type are given for each predator in Table S2. After removing predators < 1 or > 100 g, unidentified prey groups, and families contributing < 1%\(\overline{M}\), 102 Al. ferox, 73 An. cornuta, and 30 O. lowii were included in diet similarity analyses. Twenty-five prey families contributed at least 1%\(\overline{M}\) (Fig. 2a) and accounted for 88 ± 19% of the total prey mass in each stomach. The most important prey across all predators were hatchetfishes (Sternoptychidae) in the genus Sternoptyx and the hyperiid amphipod, Phrosina semilunata (Phrosinidae). Only five prey taxa were shared among all three predators, but accounted for 50%, 71%, and 67% of the total prey mass in Al. ferox, An. cornuta, and O. lowii stomachs, respectively (Table S2).
Diet overlap is high among predator species, but there is consistent taxonomic resource partitioning. (a–c) Family-level diet composition and overlap among predators 1–100 g for prey families contributing > 1% mean proportional mass (%\(\overline{M}\)). (a) Alepisaurus ferox (n = 102) consumed a high diversity of fish, crustacean, and mollusk families, while Anoplogaster cornuta (n = 73) and Omosudis lowii (n = 30) diets were dominated by crustacean and fish families, respectively. The x-axis is broken into two scales to improve visualization. (b) The first two NMDS axes (stress = 0.096, RMSE = 0.001) and 95% confidence interval ellipses depict relatively high diet overlap between Al. ferox and the other two species, and low overlap between An. cornuta and O. lowii. (c) Percent frequency of occurrence (%FO) and %\(\overline{M}\) recalculated just for stomachs containing the prey family (prey-specific mean proportional mass, %\(\overline{M}\)ps) are given for the four most important prey families for each predator. Dashed lines distinguish specialist (> 50%\(\overline{M}\)ps) from generalist (< 50%\(\overline{M}\)ps) feeding strategies at individual (< 50%FO) and population levels (> 50%FO). Shapes represent the corresponding families in panel (a). (d–h) Partial effects plots from generalized additive models describe changes in the \(\overline{M}\) of prey types with predator size for each species, where axes describe the relationship between a covariate and its parametric contribution (“f(x)”) or the contribution of its smoother (“s(x)”) to the model’s fitted values. Alepisaurus ferox is increasingly piscivorous with size (n = 116) (d–f), while the prey type preferences of An. cornuta (n = 81) (g) and O. lowii (n = 38) (h) did not vary across the sizes examined. Model summaries and partial effects plots for all covariates are given in Table 2 and Fig. S3, respectively.
Alepisaurus ferox diets were the most diverse (1Dex = 17.57 ± 0.90, Table S3, Fig. 2b, Fig. S2) and contained all unique prey types (Table S2). The four most important prey groups (amphipods in the family Phrosinidae and fishes in the families Sternoptychidae, Alepisauridae, and Gempylidae) were consumed with moderate frequency and mostly with low %\(\overline{M}\)ps, reflecting a more generalist feeding strategy (Fig. 2c). Anoplogaster cornuta diets were dominated by crustaceans and exhibited a high degree of specialization on Phrosinidae (65%\(\overline{M}\)ps, 86%FO). Platyscelid and platysceloid hyperiid amphipods were also observed at relatively high frequencies and each stomach contained a high proportion of the total observed diversity (t95% = 12, Table S3). When present, Sternoptychidae comprised most of the stomach content mass for all predators (> 50%\(\overline{M}\)ps) but dominated the diets of O. lowii (91%\(\overline{M}\)ps, 67%FO, Fig. 2c). Omosudis lowii had similar diet diversity to An. cornuta (1Dex = 9.33 ± 3.08 and 7.88 ± 0.37, respectively), but exhibited the lowest rate of family-level prey diversity accumulation (t95% = 113). Diets were significantly different among predators (ANOSIM, Global R = 0.27, p < 0.001). Pairwise differences among predators could be explained by differences in both mean diet composition (PERMANOVA, Table S4) and variance (Fig. 2b, PERMDISP, F = 44.36, p < 0.001, Table S4). Diets of O. lowii were most similar to Al. ferox (Cmh = 22.44) and least similar to An. cornuta (Cmh = 6.71, Fig. 2b).
There was limited variability in the relative mass contributions of broad prey types (mollusk, fish, and crustacean) to diet composition with increasing size for all predators (Table 2). Alepisaurus ferox became increasingly piscivorous with size and exhibited an increase in the \(\overline{M}\) of crustacean prey across intermediate sizes (~ 10–100 g, Fig. 2d–f). Anoplogaster cornuta diet was dominated by crustaceans regardless of size and there was no clear variability in the relative contributions of prey types across the sizes examined (Fig. 2g). Omosudis lowii was strongly piscivorous across all sizes examined. The \(\overline{M}\) of fish prey was two- to three-times larger than the \(\overline{M}\) of mollusk prey on average (Fig. 2h), and there were no changes in the relative contributions of fish and mollusk prey with O. lowii size (Fig. S3).
Size-based diet partitioning and total prey consumption
Individual prey mass varied among predators and increased with predator mass (Fig. 3a; adj. multiple R2 = 0.30, F(5, 1602) = 137.70, p < 2.20 E−16; Table 3, Table S5). Omosudis lowii consumed larger prey than either Al. ferox or An. cornuta across all predator sizes. Trends in prey size differences among predator species were largely consistent across prey types (Fig. S4a–c; Table S6). However, differences in the sizes of the dominant shared prey group, Sternoptychidae, were not statistically clear among predators or across predator sizes for An. cornuta or O. lowii (Fig. S4e, Table S6). Although Al. ferox also consumed larger prey than An. cornuta, the difference in average prey size decreased with increasing predator mass (Fig. 3a), driven by increased crustacean prey size in An. cornuta (Fig. S4a,d).
Differences in the individual size and number of prey per stomach result in similar total prey mass among predators. Multi-linear regressions describing changes in individual prey size (n = 1608) and the total amount of prey per stomach (n = 237) with increasing predator mass. All metrics increased with predator mass, but although there were differences in the mass of individual prey (a) and total prey count per stomach (b) among predator species, there was no difference in the total prey mass per stomach among species (c) when acounting for predator mass. Y-axes are on the log10-scale in panels (a) and (c) and on a log2-scale in panel (b). Linear models and ANCOVA results for each model are given in Table 3 and model summaries are given in Table S6.
Predator species and mass were both significant predictors of prey counts per stomach (Fig. 3b; adj. multiple R2 = 0.32, F(5, 231) = 23.57, p < 2.20 E−16; Table 3). There was no difference in the number of prey per stomach between Al. ferox and An. cornuta, but both consumed more prey individuals on average than O. lowii (Fig. 3b, Table S6). All predators consumed more prey with increasing size, but the difference in prey counts per stomach between An. cornuta and O. lowii increased with predator size.
The total mass of prey per stomach increased with predator mass (Fig. 3c; adj. multiple R2 = 0.46, F(5, 231) = 41.78, p < 2.20 E−16), but was not clearly different among predator species (Table 3). The interaction effect of predator species and mass on total prey mass is statistically significant but weak (ANCOVA, F = 4.61, p = 0.01), driven by differences in the interaction term for An. cornuta compared to both Al. ferox an O. lowii (lm, t = (− 2.34, − 3.03), p = (0.02, 0.003), respectively, Table S5).
Foraging depths and changes in overlap with predator size
Estimated foraging depths were clearly different among predator species (n = 106 Al. ferox, n = 74 An. cornuta, and n = 36 O. lowii, Table 2) and were quantified using an average 89 ± 8% of the total prey mass per stomach across species. Based on weighted median depths of prey occurrence, the predators foraged in very different depth habitats, mostly either ~ 180 or ~ 675 m (Fig. 4). These depths correlate with the median depths of the two most important prey taxa (Phrosina semilunata and Sternoptyx spp., respectively, Table S1), but also align with the maxima of the bimodal distribution of median depths for all reported prey taxa (Fig. S5).
Foraging depths reflect differential vertical resource use among predators. Foraging depths were estimated for each predator (n = 216) as the weighted median depth of occurrence of all prey in a single stomach that had been identified at least to family and contributed more than 1% mean proportional abundance. Anoplogaster cornuta fed mostly on hyperiid amphipods with relatively shallow median depths of occurence (e.g., Lycaeidae 87.5 m, Platyscelus armatus 185 m, Phrosina semilunata 185.38 m; Table S1), while Omosudis lowii fed mostly on deep-dwelling hatchetfishes (e.g., Sternoptyx diaphana 675 m). Alepisaurus ferox foraged more evenly across the upper 700 m of the water column on shared prey, but also incorporated a higher diversity of prey in their shared habitats (e.g., Gempylus serpens 237 m). Paintings by ACLC.
Omosudis lowii foraged deepest (GAM, intercept = 494.77 m, 95% CI [421.17, 568.37], p < 0.001, Table 2), driven by its consumption of hatchetfishes in the genus Sternoptyx spp., a non-migratory group with a median depth of occurrence of 675 m (Fig. 4). Anoplogaster cornuta had the shallowest mean estimated foraging depth (GAM, intercept = 202.38 m, 95% CI [135.37, 269.39], p < 0.001), consuming large quantities of hyperiid amphipods in six families with median depths of occurrence in the upper 250 m. Alepisaurus ferox fed in both shallow and deep habitats and more broadly throughout the intervening water column (GAM, intercept = 346.12 m, 95% CI [308.47, 383.77], p < 0.001; Fig. 4). Predator size was not a useful predictor of foraging depth for Al. ferox or An. cornuta, but the foraging depth of O. lowii increased with size (GAM, F = 5.51, p = 0.02).
Of the 1066 primary lancetfish containing prey, foraging depths could be estimated for 1004 specimens using an average of 89 ± 21% of the total prey mass per stomach. When these specimens are considered, the average foraging depth of Al. ferox increases with size (Fig. 5a, GAM, F = 28.73, p < 2.2E−16, Table 2). Overlap in foraging depths between Al. ferox and O. lowii also increased with size and there was no discernable difference in foraging depths between them once large Al. ferox (> ~ 4 kg) were included. The average foraging depths estimated from stomach contents are very similar to the median depths of occurrence reported for all three predators (Fig. 5b, Table S1). Estimated foraging depths for most An. cornuta match reported nighttime median depths of occurrence (160–375 m)27. Eleven percent of An. cornuta stomachs contained Sternoptyx spp., which were likely consumed closer to the predator’s reported daytime median depths of occurrence (650–950 m).
Vertical habitat overlap among predator species varies with size, and predator consumption by Alepisaurus ferox increases with foraging depth. The foraging depths of Anoplogaster cornuta and Al. ferox did not vary with mass, while the foraging depth of Omosudis lowii increased with mass across the size of specimens examined in this study (a). When primary lancetfish from Portner et al.17 were included [>99% of specimens larger than 100 g, indicated by vertical line and shading in panels (a) and (c)], overlap in foraging depths between Al. ferox and O. lowii increased with Al. ferox mass. Regression lines in panel (a) were fit with generalized additive models (GAM) for each predator species. Partial effects plots for the full GAMs with and without primary lancetfish as described in Table 2 are given in Fig. S3. The median reported habitat depths of each predator (b) during day (“d”) and night (“n”), represented as bars extending from the right-hand y axis of panel (a), were very similar to the foraging depths estimated in this study. Only An. cornuta is known to undergo diel vertical migration. Regressions fit with generalized linear models using a binomial error distribution describe increased frequency of occurrence of all three predator species in the stomach of Al. ferox with specimen mass (c).
The %FO of Al. ferox, An. cornuta, and O. lowii prey were also quantified for the 1004 primary lancetfish with estimated foraging depths. As prey, the frequency of occurrence of all three species increased with Al. ferox size. The incidence of cannibalism increased from 5%FO in the smallest individuals to 35%FO in the largest, and cannibalism was more frequent than the consumption of An. cornuta or O. lowii across all sizes of Al. ferox (Fig. 5c). However, the %FO of O. lowii in the stomachs of Al. ferox was near zero for specimens < 100 g and increased rapidly in specimens > 1 kg as overlap in foraging depths between the two species increased.
Discussion
Stomach content analysis of Al. ferox is well-suited to address a practical gap in our sampling of mobile midtrophic taxa in pelagic ecosystems, providing specimens of poorly sampled fauna from shallow and deep-sea habitats in suitable condition for diet analyses. Additionally, there is a rich literature describing the depth habitats of small organisms that comprise the forage base for larger midtrophic taxa found in the stomachs of Al. ferox. By using observations of prey vertical habitats to infer predator foraging depths, we describe how variability in foraging ecology and resource overlap among three mesopelagic fishes is mediated by ontogenetic variability in habitat use. Although less specific than observations made with depth-discrete trawls for describing vertical habitats, this work demonstrates that an understanding of prey ecology can be leveraged to elucidate basic information about habitat use and resource partitioning in poorly known taxa through studies of their diets.
Resource partitioning among Alepisaurus, Anoplogaster, and Omosudis
As is the case for many deep-sea fauna, reports on the foraging ecology of An. cornuta and O. lowii are limited to few diet observations and estimates of trophic position (TP) and relative feeding depths inferred from stable carbon and nitrogen isotope data. Stomach content analysis provides direct observations of trophic linkages, but the data represent a snapshot of diet and low sample sizes can result in incomplete or mischaracterized feeding habits. Stable isotope analysis (SIA) reflects more integrated feeding signals and can provide useful descriptions of general feeding guilds (e.g., micronektivores that consume fauna ~ 20–200 mm, zooplanktivores that primarily consume fauna < 20 mm), especially when samples are limited, but provide a low-resolution picture of food web structure. When many samples are available, individual diet snapshots are integrated into a finer-resolution picture of feeding habits that can be used to contextualize SIA and provide an overall broader understanding resource partitioning.
Juvenile Al. ferox, juvenile An. cornuta, and O. lowii have distinct overlap in forage resources that is partitioned through differential vertical habitat use and feeding strategies. All three predators consume prey with median depth habitats that correlate with two distinct depths of peak biomass in the CNP observed as epipelagic (~ 0–250 m) and mesopelagic (~ 400–750 m) acoustic scattering layers50,51. Anoplogaster cornuta consumed large quantities of small, mostly epipelagic crustaceans while O. lowii specialized on relatively large mesopelagic fishes, but infrequently contained more than one or two prey items in its stomach. Alepisaurus ferox exhibited a generalist strategy, consuming diverse intermediate-sized prey that occur throughout the water column. Although we carefully estimated foraging depths, we did not explicitly observe the depths at which each prey was captured and our method does not account for the likely consumption of prey away from their core distributions. However, our estimates generally match the known vertical habitats of all three predator species and suggest depth-stratified foraging as an important mechanism of prey partitioning.
Of the three predators, An. cornuta is the only species reported to perform regular diel vertical migration. Although its mesopelagic daytime habitat overlaps with the non-migratory O. lowii, An. cornuta mostly consumed prey with median depths similar to its relatively shallow nighttime distribution. Conversely, O. lowii only occasionally foraged on prey that predominantly occur outside of its core mesopelagic range. Some individuals fed on epipelagic prey (e.g., Heteroteuthis hawaiiensis, Sepiolidae) and the largest O. lowii in this study (270 mm SL) was directly captured on a longline (max hook depth ~ 250 m52). Alepisaurus ferox is not known to migrate on a diel cycle but is observed throughout epipelagic and mesopelagic habitats21. Competition with An. cornuta and O. lowii for shared prey would be reduced by foraging more evenly across the shared water column.
Predators of similar sizes had similar masses of prey in their stomachs, but almost half of the O. lowii stomachs were empty. Low average prey numbers and high vacuity indices are common in deep-sea fishes, consistent with infrequent feeding and low metabolic rates relative to their shallow-dwelling or migratory counterparts5,53,54. In deep-sea habitats, where prey densities are low, sit-and-wait foraging is the predominate feeding strategy used by fishes that consume micronekton44. Remotely operated vehicle observations show that both O. lowii (pers. observation EJP) and Al. ferox55 position vertically in the water column, oriented with their head up. This posture is exhibited by other mesopelagic sit-and-wait predators with forward- or lateral-facing eyes and is thought to facilitate prey detection56,57. Omosudis lowii, Al. ferox, and other fishes that consume micronekton also have large teeth and gapes, which are adaptations thought to increase predation success rates in food-limited habitats44. Fishes that consume zooplankton, including juvenile An. cornuta, generally have smaller teeth and actively pursue prey54,58. Diel migration is more energetically expensive than employing sit-and-wait strategies at depth, but there are clear benefits to feeding on higher density prey in the epipelagic5,59,60. Access to higher density epipelagic prey could explain the low vacuity indices of Al. ferox and An. cornuta stomachs (< 16% empty). The diverse diet of A. ferox reflects not just generalist feeding, but also flexibility in the behaviors employed to capture prey.
Juvenile Anoplogaster specialize on hyperiid amphipods
Anoplogaster cornuta is typically considered to be a generalist4,54. However, we describe a more specialist feeding strategy in juvenile An. cornuta, feeding predominantly on hyperiid amphipods. Many stomachs were full of epipelagic crustaceans, similar to those “generally greatly extended by quantities of larval crustaceans” described in Mead26 (n = 14, 13.7–88.0 mm SL), but in a few cases instead contained only mesopelagic fish. Persistent supplementation of epipelagic forage with mesopelagic prey supports the hypothesis put forth by Romero-Romero et al.7 to explain increases in stable nitrogen isotope composition (δ15N values) with foraging depth for migratory animals relative to their epipelagic, non-migratory counterparts.
Anoplogaster cornuta exhibits ontogenetic descent between juvenile (< 24 mm SL) and larger subadult individuals (> 77 mm SL)27. Diet descriptions of individuals > 80 mm SL are limited; a total of five individuals from three combined reports suggest An. cornuta becomes more piscivorous as an adult33,61,62. The ontogeny of its dentition and gill raker morphology tracks the proposed transition away from a zooplanktivorous diet; the large, eponymous fangs begin to develop in the upper jaw at 53 mm SL and gill rakers develop into short spikes better-suited for retention of larger prey58. Studies using δ15N to estimate trophic position (TP) of sub-adult and adult An. cornuta consistently report a TP of ~ 3.5, reflecting a mixed diet of micronekton and zooplankton6,62,63. Richards et al.33 observed an ontogenetic increase in TP from ~ 3 to ~ 4 across individuals 84–148 mm SL in the Gulf of Mexico (GOM) that may reflect a transition from crustacean zooplankton to micronektonic fish diets.
Omosudis maintains a consistent feeding strategy with size
Diets of O. lowii were dominated by non-migratory Sternoptyx spp., with sporadic consumption of lanternfishes (Myctophidae), fire squids (Pyroteuthidae), and enope squids (Enoploteuthidae) that migrate between upper mesopelagic and epipelagic habitats. Our findings are consistent with observations from the North Atlantic of O. lowii feeding almost exclusively on fishes and squids32,61, with Sternoptyx spp. being the most common fish prey reported by Rofen13. The smallest post-larvae of O. lowii are observed in the epipelagic, but juveniles 5–9 mm SL rapidly descend to the adult depth range > 600 m13,31. We observed a positive relationship between foraging depth and O. lowii size, but the relationship is weak, driven by shallow feeding in a few of the smallest individuals. Examination of additional small individuals is necessary to resolve whether the observed ontogenetic descent in foraging depth at small sizes is robust. Increased sample size would also improve our assessment of overall diet diversity, especially with respect to uncommon prey. However, the rate of diversity accumulation was very low over a broad region and time period (Fig. S2, Table S3), suggesting that our analysis likely captures the dominant prey and feeding strategy of O. lowii despite a low sample size relative to the other predator species.
In the GOM, Richards et al.33 observed no variability in δ15N values with O. lowii body size (36–260 mm SL), which could be influenced by prey size, identity, and habitat depth. Although we observed an increase in average prey size, we did not observe variability in the average size of the dominant prey species (Sternoptyx spp., 0.97 ± 0.45 g, 26.82 ± 5.42 mm SL), nor variability in the relative contributions of cephalopods and fish prey with size of O. lowii (43–270 mm SL). Thus, consistency in δ15N values across sizes reported by Richards et al.33 likely reflects limited ontogenetic variability in foraging ecology.
Ontogenetic descent in Alepisaurus
Diets of juvenile Al. ferox were similar to larger individuals from the central North Pacific; 95% of all prey identified here were also reported in Portner et al.17 and novel prey types were mostly small fishes and crustaceans. Habitat depth generally increases with size across deep-sea taxa (e.g., Pearcy et al.64, Young65), but we observed no clear variability in the estimated foraging depth of individuals ~ 1–500 g, even as prey size increased with predator size. It is also possible that our methods of estimating foraging depth by prey size class could be refined to better capture finer scale variability in foraging depths that might reflect more continuous ontogenetic descent with size in some prey species. For individuals ~ 0.5 – 8 kg, we observed a positive trend in mean foraging depth. This ontogenetic increase in estimated foraging depth is correlated with diet variability between individuals greater and less than 1.82 kg17 and increases in δ15N values spanning two TPs for individuals across similar size ranges in the central and western Pacific Ocean63,66. Little is known about the life history of Al. ferox, but a histological study by Gibbs28 described 17 individuals 43–109 cm FL (0.19–2.50 kg) as “immature”. Further examination of a single “large” specimen from the same collection described mature, inactive ovaries containing Stage 3 ova67,68. The onset of rapid ontogenetic descent in Al. ferox at ~ 0.5 kg could reflect a stepwise change in depth habitat between life stages more similar to that observed in some cephalopod species65.
Competitors in shared feeding grounds become prey
Variability in the consumption of each predator by Al. ferox can be explained by ontogenetic differences in the degree of diet and habitat overlap. Alepisaurus ferox has high intraspecific resource and habitat overlap, and cannibalism is most frequent across all sizes, followed by the consumption of An. cornuta and O. lowii. Given its prevalence in the diets of numerous pelagic predators69 and high catch rates in longline fisheries16, it is likely that Al. ferox also has higher total biomass than An. cornuta and O. lowii in the CNP. Juvenile Al. ferox and An. cornuta have intermediate overlap in foraging depths, but their comparatively low diet overlap increases with size as the size spectra of their prey converge. Anoplogaster cornuta > 80 mm SL were absent from Al. ferox stomachs. This size correlates with an ontogenetic change in coloration from the silvery gray of juveniles to the black or dark brown of adults26,30, as well as the putative transition to piscivory. Associated ontogenetic changes in the degree of competition or detectability may explain the absence of adult An. cornuta. Despite having the highest diet overlap, Al. ferox does not begin to consume O. lowii at appreciable frequencies until their mean foraging depths also overlap.
The vacuity indices of An. Cornuta and O. lowii are much lower here than previously reported for individuals sampled with nets32,33. Higher prey incidence in these stomachs could indicate that the predator was consumed at or near the time of feeding. The dominant crustacean (Phrosina semilunata) and fish (Sternoptyx spp.) prey taxa among all three predators are known to form relatively dense aggregations or schools70,71,72,73 and were sometimes found in exceptionally high numbers in Al. ferox stomachs (P. semilunata, max n = 86; Sternoptyx spp., max n = 124). Even if the absolute concentrations of these aggregations in the CNP are low compared to higher productivity regions, locally dense prey patches may ‘aggregate’ pelagic predators throughout the water column74,75,76. Considering the low vacuity indices, increased consumption of An. cornuta and O. lowii with increased overlap in diet and foraging depth suggests the act of feeding on prey aggregations might increase the rates at which large Al. ferox encounters these three deep-sea fishes.
Future directions
This work highlights our ability to harness pelagic predators as biological samplers to address critical gaps in our understanding of deep-sea food web structure. Expanding this type of work to other pelagic predators that are captured by longlines (e.g., snake mackerels and barracudinas) would increase the diversity of prey taxa that could be sampled. Collaboration with fisheries observer programs allows for much higher spatial and temporal resolution sampling than is possible with ship-based scientific exploration. Long term monitoring of pelagic predator diets would greatly facilitate fundamental biogeographic descriptions of common deep-sea taxa and their foraging ecologies, as well as large-scale studies of the responses of pelagic prey communities to environmental perturbations.
Data availability
The data required to replicate the analyses presented in this study are available as supplemental material. See “Read me” sheet in Supplementary Data File S1 for a description of the data provided for each figure/analysis.
References
Gastauer, S., Nickels, C. F. & Ohman, M. D. Body size- and season-dependent diel vertical migration of mesozooplankton resolved acoustically in the San Diego Trough. Limnol. Oceanogr. 67, 300–313. https://doi.org/10.1002/lno.11993 (2022).
Staby, A., Srisomwong, J. & Rosland, R. Variation in DVM behaviour of juvenile and adult pearlside (Maurolicus muelleri) linked to feeding strategies and related predation risk. Fish. Oceanogr. 22, 90–101. https://doi.org/10.1111/fog.12012 (2013).
Portner, E. J., Snodgrass, O. & Dewar, H. Pacific bluefin tuna, Thunnus orientalis, exhibits a flexible feeding ecology in the Southern California Bight. PLoS ONE 17, e0272048. https://doi.org/10.1371/journal.pone.0272048 (2022).
Drazen, J. C. & Sutton, T. T. Dining in the deep: The feeding ecology of deep-sea fishes. Annu. Rev. Mar. Sci. 9, 1–26. https://doi.org/10.1146/annurev-marine-010816-060543 (2017).
Pearre, S. Eat and run? The hunger/satiation hypothesis in vertical migration: History, evidence and consequences. Biol. Rev. Camb. Philos. Soc. 78, 1–79. https://doi.org/10.1017/S146479310200595X (2003).
Gloeckler, K. et al. Stable isotope analysis of micronekton around Hawaii reveals suspended particles are an important nutritional source in the lower mesopelagic and upper bathypelagic zones: Suspended particles as a mesopelagic food source. Limnol. Oceanogr. 63, 1168–1180. https://doi.org/10.1002/lno.10762 (2018).
Romero-Romero, S., Choy, C. A., Hannides, C. C. S., Popp, B. N. & Drazen, J. C. Differences in the trophic ecology of micronekton driven by diel vertical migration. Limnol. Oceanogr. 64, 1473–1483. https://doi.org/10.1002/lno.11128 (2019).
Abreu, J. et al. Long-term changes in habitat and trophic level of Southern Ocean squid in relation to environmental conditions. Sci. Rep. 10, 15215. https://doi.org/10.1038/s41598-020-72103-6 (2020).
Cherel, Y. & Hobson, K. A. Stable isotopes, beaks and predators: A new tool to study the trophic ecology of cephalopods, including giant and colossal squids. Proc. R. Soc. B Biol. Sci. 272, 1601–1607 (2005).
Negri, A. et al. The cephalopod prey of the Weddell seal, Leptonychotes weddellii, a biological sampler of the Antarctic marine ecosystem. Polar Biol. 39, 561–564. https://doi.org/10.1007/s00300-015-1794-9 (2016).
Olson, R. J. et al. Decadal diet shift in yellowfin tuna Thunnus albacares suggests broad-scale food web changes in the eastern tropical Pacific Ocean. Mar. Ecol. Prog. Ser. 497, 157–178 (2014).
Portner, E. J., Markaida, U., Robinson, C. J. & Gilly, W. F. Trophic ecology of Humboldt squid, Dosidicus gigas, in conjunction with body size and climatic variability in the Gulf of California, Mexico. Limnol. Oceanogr. 65, 732–748. https://doi.org/10.1002/lno.11343 (2020).
Rofen, R. R. Family Omosudidae. in Orders Iniomi and Lyomeri 462–481 (Yale University Press, 1966). https://doi.org/10.2307/j.ctvbcd095.16.
Wassersug, R. J. & Johnson, R. K. A remarkable pyloric caecum in the evermannellid genus Coccorella with notes on gut structure and function in alepisauroid fishes (Pisces, Myctophiformes). J. Zool. Lond. 179, 273–289. https://doi.org/10.1111/j.1469-7998.1976.tb02296.x (1976).
Reinhardt, J. F. et al. Catch rate and at-vessel mortality of circle hooks versus J-hooks in pelagic longline fisheries: A global meta-analysis. Fish Fish. 19, 413–430. https://doi.org/10.1111/faf.12260 (2018).
Woodworth-Jefcoats, P. A., Polovina, J. J. & Drazen, J. C. Synergy among oceanographic variability, fishery expansion, and longline catch composition in the central North Pacific Ocean. Fish. Bull. 116, 228–239 (2018).
Portner, E. J., Polovina, J. J. & Choy, C. A. Patterns in micronekton diversity across the North Pacific Subtropical Gyre observed from the diet of longnose lancetfish (Alepisaurus ferox). Deep-Sea Res. Part I 125, 40–51. https://doi.org/10.1016/j.dsr.2017.04.013 (2017).
Bañón, R., Roura, Á., García-Fernández, C., Alonso-Fernández, A. & de Carlos, A. Coastal habitat evidences and biological data of Alepisaurus ferox (Aulopiform; Alepisauridae) from northwestern Iberian Peninsula. Mar. Biodivers. 52, 22. https://doi.org/10.1007/s12526-022-01261-9 (2022).
Potier, M. et al. Forage fauna in the diet of three large pelagic fishes (lancetfish, swordfish and yellowfin tuna) in the western equatorial Indian Ocean. Fish. Res. 83, 60–72. https://doi.org/10.1016/j.fishres.2006.08.020 (2007).
Loeb, V. Vertical distribution and development of larval fishes in the North Pacific central gyre during summer. Fish. Bull. 77, 777–793 (1979).
Mundy, B. M. Checklist of the Fishes of the Hawaiian Archipelago (2005).
Pan, B. et al. Study on the catch, bycatch and discard of Chinese pelagic longline fisheries in the Atlantic Ocean. Aquac. Fish. https://doi.org/10.1016/j.aaf.2022.03.002 (2022).
Okutani, T. Two new species of the squid genus Onykia from the tropical Indian Ocean (Cephalopoda, Onychoteuthidae). Bull. Natl. Mus. Nat. Sci. Ser. Zool. 7, 155–163 (1981).
Richards, W. J. Stemonosudis rothschildi, a new paralepidid fish from the central Pacific. Calif Fish Game 53, 35–39 (1967).
Hopkins, T. L. & Baird, R. C. Diet of the hatchetfish Sternoptyx diaphana. Mar. Biol. 21, 34–46. https://doi.org/10.1007/BF00351190 (1973).
Maul, G. E. Monografia dos peixes do Museu Municipal do Funchal: Ordem Berycomorphi. Bol. Mus. Munic. Funchal 7, 5–41 (1954).
Clarke, T. A. & Wagner, P. J. Vertical distribution and other aspects of the ecology of certain mesopelagic fishes taken near Hawaii. Fish. Bull. 74, 635–645 (1976).
Gibbs, R. H. Alepisaurus brevirostris, a new species of lancetfish from the western North Atlantic. Breviora 123, 1–14 (1960).
Gibbs, R. H. et al. Family Alepisauridae. in Orders Iniomi and Lyomeri 482–497 (Yale University Press, 1966). https://doi.org/10.2307/j.ctvbcd095.17.
Woods, L. P. et al. Order Berycomorphi (Beryciformes). in Orders Heteromi (Notacanthiformes), Berycomorphi (Beryciformes), Xenoberyces (Stephanoberyciformes), Anacanthini (Gadiformes) 263–396 (Yale University Press, 1973). https://doi.org/10.2307/j.ctvbcd0bn.7.
Slayden, N. Age and Growth of Predatory Mesopelagic Fishes in a Low-Latitude Oceanic Ecosystem (Nova Southeastern University, 2020).
Appelbaum, S. Studies on food organisms of pelagic fishes as revealed by the 1979 North Atlantic Eel Expedition. Helgol. Meeresunters 35, 357–367 (1982).
Richards, T. M., Gipson, E. E., Cook, A., Sutton, T. T. & Wells, R. J. D. Trophic ecology of meso- and bathypelagic predatory fishes in the Gulf of Mexico. ICES J. Mar. Sci. 76, 662–672. https://doi.org/10.1093/icesjms/fsy074 (2019).
Curran, D. & Beverly, S. Effects of 16/0 circle hooks on pelagic fish catches in three South Pacific albacore longline fisheries. Bull. Mar. Sci. 88, 485–497. https://doi.org/10.5343/bms.2011.1060 (2012).
Choy, C., Portner, E., Iwane, M. & Drazen, J. C. Diets of five important predatory mesopelagic fishes of the central North Pacific. Mar. Ecol. Prog. Ser. 492, 169–184. https://doi.org/10.3354/meps10518 (2013).
R Core Team. R: A Language and Environment for Statistical Computing (2020).
Legendre, P. & Legendre, L. Numerical Ecology 2nd edn, 1998 (Elsevier, 1998).
Anderson, M. J., Gorley, R. N. & Clarke, K. R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods (PRIMER-E, 2008).
Oksanen, J. et al. Vegan: Community Ecology Package (2020).
Horn, H. S. Measurement of ‘overlap’ in comparative ecological studies. Am. Nat. 100, 419–424 (1966).
Legendre, P. & Gallagher, E. D. Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280. https://doi.org/10.1007/s004420100716 (2001).
Chao, A. et al. Rarefaction and extrapolation with Hill numbers: A framework for sampling and estimation in species diversity studies. Ecol. Monogr. 84, 45–67. https://doi.org/10.1890/13-0133.1 (2014).
Hsieh, T. C., Ma, K. H. & Chao, A. iNEXT: Interpolation and Extrapolation for Species Diversity (2020).
Pinkas, L., Oliphant, M. S. & Iverson, I. L. K. Food habits of Albacore, Bluefin Tuna, and Bonito in California waters. Calif. Dep. Fish Game Fish Bull. 152, 1–105 (1971).
Amundsen, P.-A., Gabler, H.-M. & Staldvik, F. J. A new approach to graphical analysis of feeding strategy from stomach contents data—Modification of the Costello (1990) method. J. Fish Biol. 48, 607–614. https://doi.org/10.1111/j.1095-8649.1996.tb01455.x (1996).
Wood, S. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. Ser. B 73, 3–36. https://www.jstor.org/stable/41057423 (2011).
Fox, J. & Weisberg, S. An R Companion to Applied Regression (Sage, 2019).
Uchiyama, J. & Kazama, T. Updated weight-on-length relationships for pelagic fishes caught in the Central North Pacific Ocean and bottomfishes from the Northwestern Hawaiian Islands. Fish. Sci. 1–34 (2003).
Chen, R. S., Portner, E. J. & Choy, C. A. Gelatinous cephalopods as important prey for a deep-sea fish predator. Mar. Biol. 169, 155. https://doi.org/10.1007/s00227-022-04116-w (2022).
Domokos, R. Seamount effects on micronekton at a subtropical central Pacific seamount. Deep Sea Res. Part Oceanogr. Res. Pap. 186, 103829. https://doi.org/10.1016/j.dsr.2022.103829 (2022).
Hazen, E. L. & Johnston, D. W. Meridional patterns in the deep scattering layers and top predator distribution in the central equatorial Pacific. Fish. Oceanogr. 19, 427–433. https://doi.org/10.1111/j.1365-2419.2010.00561.x (2010).
Bigelow, K., Musyl, M. K., Poisson, F. & Kleiber, P. Pelagic longline gear depth and shoaling. Fish. Res. 77, 173–183. https://doi.org/10.1016/j.fishres.2005.10.010 (2006).
Koslow, J. A. Energetic and life-history patterns of deep-sea benthic, benthopelagic and seamount-associated fish. J. Fish Biol. 49, 54–74. https://doi.org/10.1111/j.1095-8649.1996.tb06067.x (1996).
Priede, I. G. Deep-Sea Fishes: Biology, Diversity, Ecology and Fisheries. (Cambridge University Press, 2017).
NOAA OER. Deep-Sea Symphony: Exploring the Musicians Seamounts. Dive 11. https://www.ncei.noaa.gov/waf/okeanos-rov-cruises/ex1708/#tab-11 (2017).
Kupchik, M. J., Benfield, M. C. & Sutton, T. T. The first in situ encounter of Gigantura chuni (Giganturidae: Giganturoidei: Aulopiformes: Cyclosquamata: Teleostei), with a preliminary investigation of pair-bonding. Copeia 106, 641–645. https://doi.org/10.1643/CE-18-034 (2018).
Miller, M. J. et al. Vertical body orientation by a snipe eel (Nemichthyidae, Anguilliformes) in the deep mesopelagic zone along the West Mariana Ridge. Mar. Freshw. Behav. Physiol. 47, 265–272. https://doi.org/10.1080/10236244.2014.926128 (2014).
Kotlyar, A. N. Classification and distribution of fishes of the family Anoplogasteridae (Beryciformes). J. Ichthyol. 26, 133–152 (1986).
Hays, G. C. A review of the adaptive significance and ecosystem consequences of zooplankton diel vertical migrations. Hydrobiologia 503, 163–170. https://doi.org/10.1023/B:HYDR.0000008476.23617.b0 (2003).
Kiørboe, T. How zooplankton feed: Mechanisms, traits and trade-offs. Biol. Rev. 86, 311–339. https://doi.org/10.1111/j.1469-185X.2010.00148.x (2011).
Hopkins, T. L., Sutton, T. T. & Lancraft, T. M. The trophic structure and predation impact of a low latitude midwater fish assemblage. Prog. Oceanogr. 38, 205–239. https://doi.org/10.1016/S0079-6611(97)00003-7 (1996).
Parzanini, C., Parrish, C. C., Hamel, J.-F. & Mercier, A. Trophic ecology of a deep-sea fish assemblage in the Northwest Atlantic. Mar. Biol. 164, 206. https://doi.org/10.1007/s00227-017-3236-4 (2017).
Choy, C. A., Popp, B. N., Hannides, C. C. S. & Drazen, J. C. Trophic structure and food resources of epipelagic and mesopelagic fishes in the North Pacific Subtropical Gyre ecosystem inferred from nitrogen isotopic compositions: Trophic structure of pelagic fishes. Limnol. Oceanogr. 60, 1156–1171. https://doi.org/10.1002/lno.10085 (2015).
Pearcy, W. G., Krygier, E. E., Mesecar, R. & Ramsey, F. Vertical distribution and migration of oceanic micronekton off Oregon. Deep Sea Res. 24, 223–245. https://doi.org/10.1016/S0146-6291(77)80002-7 (1977).
Young, R. Vertical distribution and photosensitive vesicles of pelagic cephalopods from Hawaiian waters. Fish. Bull. 76, 583–615 (1978).
Ohshimo, S., Hiraoka, Y., Sato, T. & Nakatsuka, S. Feeding habits of bigeye tuna (Thunnus obesus) in the North Pacific from 2011 to 2013. Mar. Freshw. Res. https://doi.org/10.1071/MF17058 (2018).
Smith, C. L. The patterns of sexuality and the classification of serranid fishes. Am. Mus. Novit. 1–20 (1965).
Smith, C. L. & Atz, E. H. Hermaphroditism in the mesopelagic fishes Omosudis lowei and Alepisaurus ferox. Copeia 1, 41–44. https://doi.org/10.2307/1442355 (1973).
Moteki, M., Arai, M., Tsuchiya, K. & Okamoto, H. Composition of piscine prey in the diet of large pelagic fish in the eastern tropical Pacific Ocean. Fish. Sci. 67, 1063–1074. https://doi.org/10.1046/j.1444-2906.2001.00362.x (2001).
Gibbs, R. H. & Krueger, W. H. Biology of midwater fishes of the Bermuda Ocean Acre. Smithson. Contrib. Zool. 452, 1–187. https://doi.org/10.5479/si.00810282.452 (1987).
Ridge-Cooney, V. L. Aspects of the Ecology of the Pelagic Hatchetfishes, Family Sternoptychidae, in the Pacific Ocean Near Hawaii (University of Hawaii at Manoa, 1987).
Thurston, M. The vertical distribution and diurnal migration of the Crustacea Amphipoda collected during the Sond Cruise, 1965: II. The Hyperiidea and general discussion. J. Mar. Biol. Assoc. UK 56, 383–470. https://doi.org/10.1017/S0025315400018981 (1976).
Vinogradov, M., Volkov, A. & Semenova, T. Hyperiid Amphipods (Amphipoda, Hyperiidea) of the World Oceans (Smithsonian Instituiton Libraries, 1996).
Benoit-Bird, K. J. et al. Prey patch patterns predict habitat use by top marine predators with diverse foraging strategies. PLoS ONE 8, e53348–e53348. https://doi.org/10.1371/journal.pone.0053348 (2013).
Bost, C. A. et al. The importance of oceanographic fronts to marine birds and mammals of the southern oceans. J. Mar. Syst. 78, 363–376. https://doi.org/10.1016/j.jmarsys.2008.11.022 (2009).
Sutherland, W. J. Aggregation and the `ideal free’ distribution. J. Anim. Ecol. 52, 821. https://doi.org/10.2307/4456 (1983).
Acknowledgements
This work would not have been possible without the efforts of federal Fisheries Observers at the Pacific Islands Regional Office. Cindy Klepadlo, Bruce Mundy, and Dick Young generously lent their expertise to prey identifications. Jessica Perelman and Jennifer Wong-Ala supported the acquisition of target predators. Rachel Chen assisted with stomach dissections. Barbara Muhling and Ryan Rykaczewski provided thoughtful feedback on the analyses and manuscript. EJP was supported by a National Science Foundation Postdoctoral Fellowship in Biology (Award # 2011031). CAC was supported by funds from the Maxwell/Hanrahan Foundation and an Alfred P. Sloan Research Fellowship in Earth System Science.
Author information
Authors and Affiliations
Contributions
E.J.P., P.W.J., and C.A.C. designed the study. All authors collected the data. E.J.P. and T.M.L. analyzed the data. E.J.P. wrote the manuscript. All authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Portner, E.J., Mowatt-Larssen, T., Carretero, A.CL. et al. Harnessing a mesopelagic predator as a biological sampler reveals taxonomic and vertical resource partitioning among three poorly known deep-sea fishes. Sci Rep 13, 16078 (2023). https://doi.org/10.1038/s41598-023-41298-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-41298-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.