Abstract
Trichosporon asahii is an opportunistic pathogenic fungus that causes severe and sometimes fatal infections in immunocompromised patients. Hog1, a mitogen-activated protein kinase, regulates the stress resistance of some pathogenic fungi, however its role in T. asahii has not been investigated. Here, we demonstrated that the hog1 gene-deficient T. asahii mutant is sensitive to high temperature, cell membrane stress, oxidative stress, and antifungal drugs. Growth of the hog1 gene-deficient T. asahii mutant was delayed at 40 °C. The hog1 gene-deficient T. asahii mutant also exhibited sensitivity to sodium dodecyl sulfate, hydrogen peroxide, menadione, methyl methanesulfonate, UV exposure, and antifungal drugs such as amphotericin B under a glucose-rich condition. Under a glucose-restricted condition, the hog1 gene-deficient mutant exhibited sensitivity to NaCl and KCl. The virulence of the hog1 gene-deficient mutant against silkworms was attenuated. Moreover, the viability of the hog1 gene-deficient mutant decreased in the silkworm hemolymph. These phenotypes were restored by re-introducing the hog1 gene into the gene-deficient mutant. Our findings suggest that Hog1 plays a critical role in regulating cellular stress responses in T. asahii.
Similar content being viewed by others
Introduction
The pathogenic fungus Trichosporon asahii is a basidiomycete yeast often isolated from soil, human blood, sputum, skin, feces, and urine1,2,3,4,5,6. T. asahii causes severe fungal infections in immunocompromised patients7,8, and the mortality rate of deep mycoses caused by T. asahii is higher than that of infections caused by Candida albicans, another pathogenic fungus (80% vs. 40%)9. Micafungin, an echinocandin antifungal drug, is used to treat for patients with suspected fungal infections. If infections are caused by T. asahii, severe infections develop because T. asahii is tolerant to micafungin10,11. Amphotericin B- and azole-resistant T. asahii strains have also been isolated from patients12,13. Therefore, infections caused by T. asahii are problematic in clinical settings8.
Hog1-mediated signaling pathway is involved in fungal resistance to several types of stressors14,15,16. The Hog1 protein, a mitogen-activated protein kinase (MAPK), translocates from the cytoplasm to the nucleus in response to environmental stress and acts in conjunction with multiple transcription factors to regulate the expression of genes related to stress resistance15,16. Hog1-mediated stress resistance and gene regulation also affect the virulence of the pathogenic fungus Cryptococcus neoformans17,18,19. The Hog1 protein is also involved in the pathogenicity of C. albicans to the host by increasing its resistance to various stressors and controlling the morphological change from yeast to mycelium20,21,22,23,24. Moreover, stress response systems are related to morphological changes in C. albicans25,26,27,28. However, the role of Hog1 in stress tolerance and virulence of T. asahii remains unknown.
To elucidate the molecular mechanisms underlying T. asahii virulence, a silkworm infection model and a method for constructing a gene-deficient T. asahii mutant were established29,30. The use of an invertebrate silkworm model for infection experiments is highly advantageous because silkworms are less costly to rear in large numbers, and fewer ethical problems are associated with their use31,32. The virulence of T. asahii clinical isolates can be evaluated quantitatively by calculating the half-maximal lethal dose (LD50), which is the dose of a pathogen required to kill half of the animals in a group29. An efficient gene-targeting system was developed to generate gene-deficient mutants of T. asahii30,33. Gene-deficient T. asahii mutants of calcineurin, a calcium-calmodulin-activated phosphatase, exhibit decreased virulence against silkworms34. These findings demonstrate that an evaluation system comprising a silkworm infection model and a gene manipulation system is useful for elucidating the molecular mechanisms of T. asahii infection.
In the present study, we generated a hog1 gene-deficient T. asahii mutant and characterized the phenotypes related to stress resistance and virulence in a silkworm infection model. The hog1 gene-deficient mutant exhibited sensitivity to compounds known to cause membrane damage and to antifungal drugs. Furthermore, the virulence of the hog1 gene-deficient T. asahii mutant against silkworms decreased. Our findings suggest that the virulence and cellular stress responses of T. asahii are controlled by the Hog1 mediated-signaling pathway.
Results
Conservation of the amino acid sequence of T. asahii Hog1 protein among representative fungi
We performed an amino acid sequence homology analysis with the Hog1 protein of T. asahii based on genomic information from a representative model fungus, Saccharomyces cerevisiae, and representative pathogenic fungi, Aspergillus fumigatus, C. neoformans, and C. albicans. The amino acid sequence of the Hog1 protein of T. asahii was more than 70% identical to those of these fungi (Fig. 1a and Supplementary Table S1). The Hog1 protein of T. asahii contains a protein kinase c-like superfamily domain, which includes an ATP-binding site, polypeptide substrate-binding site, and activation loop (Fig. 1b,c, and Supplementary Table S2).
Estimation of Hog1 protein in T. asahii and conservation of the amino acids among typical fungi. (a) Phylogenetic tree of T. asahii, C. neoformans, S. cerevisiae, C. albicans Hog1, and A. fumigatus SakA was generated according to the maximum likelihood method. (b) Estimation of the functional domain of T. asahii Hog1 and homologs. (c) Conservation of the amino acids of protein kinase c-like superfamily domain in T. asahii Hog1. ATP binding site, polypeptide substrate binding site, and activation loop (A loop) are indicated red, green, and blue lines, respectively.
Generation of the hog1 gene-deficient mutant and the gene-reintroduced strain
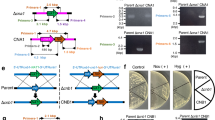
The ku70 gene-deficient strain of T. asahii MPU129 has a high homologous recombination efficiency, making it useful as a parent strain30. Using this strain, we generated the hog1 gene-deficient T. asahii mutant. The targeting DNA fragment used to generate the hog1 gene-deficient mutant contains the NAT1 gene, which leads to nourseothricin resistance (Fig. 2a). Transformants with nourseothricin resistance were obtained by introducing the DNA fragment through electroporation (Fig. 2b). Genomic DNA was extracted from the transformants and replacement of the hog1 gene was confirmed by polymerase chain reaction (PCR) (Fig. 2c,d). Secondary genetic mutations such as point mutations may occur during the generation of gene-deficient mutants. Therefore, we also generated a revertant strain of the hog1 gene-deficient mutant to confirm that the phenotype of the gene-deficient strain is due to a deficiency of the targeted gene. The targeting DNA fragment used to generate the revertant strain of the hog1 gene-deficient mutant contains the hph gene, which leads to hygromycin B resistance (Fig. 2a). Transformants with hygromycin B resistance were obtained by introducing the DNA fragment through electroporation (Fig. 2b). Reintroduction of the hog1 gene was verified by PCR using DNA extracted from the transformants (Fig. 2c,d). These results confirmed the generation of the hog1 gene-deficient T. asahii mutant and the hog1 gene-reintroduced revertant.
Generation of the hog1 gene-deficient T. asahii mutant and its revertant. (a–d) Generation of the hog1 gene-deficient T. asahii mutant and its revertant. (a) Strategy for generating the hog1 gene-deficient mutant (∆hog1) and its revertant (Rev.). Predicted genomes of the hog1 gene-deficient mutant and its revertant are shown. (b) The parent strain (Parent), hog1 gene-deficient mutant (∆hog1), and its revertant (Rev.) were spread on SDA with nourseothricin (Nou) (100 µg/mL) or hygromycin B (Hyg) (100 µg/mL) and incubated at 27 °C for 2 days. (c) Location of the primers for confirming the genome structure of the hog1 gene-deficient candidate by PCR using extracted genome DNA. (d) Confirmation of the genotypes of the hog1 gene-deficient mutant (∆hog1) and its revertant (Rev.) by PCR using extracted genome DNA. Cropped blots were used. Full-length blots are presented in Supplementary Fig. S1.
Role of the hog1 gene in the growth of T. asahii under high temperature
In C. neoformans and S. cerevisiae, the growth of hog1 gene-deficient strains is delayed at 39–40 °C compared to their respective parents14,17,35. We examined whether Hog1 was involved in the high-temperature resistance of T. asahii. Growth of the hog1 gene-deficient T. asahii mutant was delayed at 40 °C (Fig. 3). On the other hand, there was no delay in the growth of the hog1 gene-deficient mutant at 27 °C or 37 °C (Fig. 3). The high-temperature-sensitive phenotype of the hog1 gene-deficient mutants was suppressed in the revertant strain (Fig. 3). These results suggest that hog1 gene-deficient T. asahii mutant is sensitive to high-temperature stress.
Temperature sensitivity of the hog1 gene-deficient T. asahii mutant. (a) The T. asahii parent strain (Parent), the hog1 gene-deficient mutant (∆hog1), and the revertant of ∆hog1 (Rev.) were grown on SDA and incubated at 27 °C for 1 day. T. asahii cells were suspended in physiologic saline solution and filtered through a 40-μm cell strainer. Series of tenfold dilution of the fungal suspension were prepared using saline. Five microliters of each cell suspension was spotted on the SDA. Agar plates were incubated at 27 °C, 37 °C, or 40 °C for 24 h. (b) The T. asahii parent strain (Parent), the hog1 gene-deficient mutant (∆hog1), and the revertant of ∆hog1 (Rev.) were inoculated on Sabouraud medium and incubated at 27 °C, 37 °C, or 40 °C. Absorbance of the culture at 630 nm was monitored. Data are shown as means ± standard error of the mean (SEM). Statistically significant differences between groups were evaluated using Tukey’s test. *P < 0.05.
Functions of the hog1 gene in the stress resistance of T. asahii
Next, we examined whether Hog1 contributes to the tolerance to several stresses in T. asahii. NaCl, KCl and sorbitol causes osmotic stress20,21,22,23,24,36,37. Sodium dodecyl sulfate (SDS) damages the cell membrane35,38,39,40. Congo red and caffeine cause cell wall damage38,40. Hydrogen peroxide (H2O2) induces oxidative stress17,19,20,23,41,42. Tunicamycin (TM) and dithiothreitol (DTT) induce endoplasmic reticulum (ER) stress24,35,43. UV exposure and methyl methanesulfonate (MMS) cause DNA damage17,35,44. The number of tolerant cells in the hog1 gene-deficient T. asahii mutant was decreased on Sabouraud dextrose agar (SDA) containing SDS, H2O2, menadione, MMS, or upon UV exposure, but not on SDA containing NaCl, KCl, sorbitol, caffeine, Congo red, TM, or DTT (Fig. 4a–e).
Sensitivity of the hog1 gene-deficient T. asahii mutant against stressors. (a–f) The T. asahii parent strain (Parent), the hog1 gene-deficient mutant (∆hog1), and the revertant of ∆hog1 (Rev.) were grown on SDA and incubated at 27 °C for 1 day. T. asahii cells were suspended in physiologic saline solution. Series of tenfold dilution of the fungal suspension were prepared using saline. Five microliters of each cell suspension was spotted on the SDA containing NaCl (1.2 M), KCl (1.5 M), sorbitol (2 M), SDS (0.008%), caffeine (0.65 mg/mL), Congo red (400 µg/mL), H2O2 (3 mM), menadione (60 µM), tunicamycin (TM, 1 µg/mL), dithiothreitol (DTT, 12 mM), methyl methanesulfonate (MMS, 0.05%), amphotericin-B (AMPH-B, 0.4 µg/mL), fluconazole (18 µg/mL), and voriconazole (VRCZ, 0.2 µg/mL). Cells on SDA were exposed to UV (400 J/m2). Each agar plate of (a–d) or (f) was incubated at 27 °C for 24 h or 48 h, respectively.
Amphotericin B and fluconazole are used to treat T. asahii infections45,46. The hog1 gene-deficient mutants of C. neoformans and C. albicans are sensitive to an antifungal agent the amphotericin B (AMPH-B)19,23,38,42. The hog1 gene-deficient mutant of C. albicans is susceptible to azole antifungals, whereas the hog1 gene-deficient mutant of C. neoformans is resistant19,23,38,42. The number of tolerant cells in the hog1 gene-deficient T. asahii mutant was decreased on SDA containing AMPH-B, fluconazole (FLCZ), or voriconazole (VRCZ) (Fig. 4f). Moreover, we determined the MIC values of antifungal drugs for inhibiting the growth of the parent strain or hog1 gene-deficient T. asahii mutant. Compared to the parental strain, the MIC values of AMPH-B, FLCZ, ITCZ, and VRCZ were lower in the hog1 gene-deficient T. asahii mutant (Table 1). The phenotypes of the hog1 gene-deficient mutant were suppressed in the revertant. These findings suggest that the hog1 gene in T. asahii is involved in resistance to cell membrane damage, oxidative stress, DNA damage, and antifungal drugs, such as AMPH-B and azole antifungals.
Involvement of the hog1 gene in the stress resistance of T. asahii under a glucose-restricted condition
The hog1 gene-deficient mutant of T. asahii did not exhibit clear sensitivity to KCl, NaCl, sorbitol, caffeine, Congo red, TM, or DTT under a glucose-rich condition. Hog1 in C. neoformans plays a pivotal role in the regulation of stress responses under a glucose-restricted condition47. Therefore, we investigated whether hog1 gene deficiency in T. asahii causes sensitivity to these compounds under a glucose-restricted condition. Under a glucose-restricted condition, the growth of the hog1 gene-deficient T. asahii mutant was delayed by NaCl and KCl, but not by sorbitol, caffeine, Congo red, TM, or DTT (Fig. 5). The phenotypes of NaCl and KCl sensitivity of the hog1 gene-deficient mutant were suppressed in the revertant (Fig. 5). These findings suggest that the hog1 gene in T. asahii contributes to the stress caused by NaCl and KCl under a glucose-restricted condition.
Effect of glucose on sensitivity of the hog1 gene-deficient mutants against stressors. The T. asahii parent strain (Parent), the hog1 gene-deficient mutant (∆hog1), and the revertant of ∆hog1 (Rev.) were grown on glucose-limited peptone agar and incubated at 27 °C for 1 day. T. asahii cells were suspended in physiologic saline solution. Series of tenfold dilution of the fungal suspension were prepared using saline. Five microliters of each cell suspension was spotted on the glucose-restricted peptone agar containing NaCl (1.2 M), KCl (1.5 M), sorbitol (2 M), caffeine (0.65 mg/mL), Congo red (400 µg/mL), tunicamycin (TM, 1 µg/mL), or dithiothreitol (DTT, 8 mM). Each agar plate was incubated at 27 °C for 48 h.
Role of the hog1 gene in hyphal formation by T. asahii
T. asahii has several morphological forms, including yeast, hyphae (filament form), and arthroconidia (chains of cells and asexual spores)4. In C. albicans, Hog1 is a key factor that regulates hyphal formation20,21,23,25,26,27,28,48. We examined whether hog1 gene deficiency affects hyphal formation in T. asahii. In Sabouraud dextrose medium, the hyphae ratio was higher in the hog1 gene-deficient mutant than in the parental strain (Fig. 6). On the other hand, the yeast and arthroconidia ratio were lower in the hog1 gene-deficient mutant than in the parental strain (Fig. 6). The hyphae per yeast ratios in the parental strain and the hog1 gene-deficient mutant were 0.3 and 2.3, respectively. The phenotype of higher hyphal-forming activity in the hog1 gene-deficient mutant was suppressed in the revertant (Fig. 6). These observations suggest that hog1 gene regulates hyphal formation by T. asahii.
Effect of hog1 gene-deficiency on T. asahii morphology in Sabouraud dextrose medium. The T. asahii parent strain (Parent), the hog1 gene-deficient mutant (∆hog1), and the revertant of ∆hog1 (Rev.) were grown on SDA and incubated at 27 °C for 2 days. T. asahii cells were suspended in physiologic saline solution and filtered through a 40-μm cell strainer. Absorbance at 630 nm of the T. asahii cell suspension was adjusted to 1. The cell suspension (10 µL) was added to Sabouraud dextrose medium (100 µL). The solutions were incubated at 27 °C for 48 h. The incubated solution was placed on glass slides and covered by glass coverslips. (a) Samples were examined with bright light under a microscope. Scale bar, 20 µm. (b) The pictures were randomly obtained. The numbers of 3 cell types: yeast, arthroconidia, and hyphae, were counted.
Hog1-dependency of T. asahii virulence against silkworms
We examined whether a deficiency of the hog1 gene reduced the virulence of T. asahii against silkworms. The survival time of silkworms injected with the hog1 gene-deficient mutant was longer than that of the parental strain (Fig. 7a). The LD50 value of the hog1 gene-deficient mutant was tenfold higher than that of the parent strain (Fig. 7b, and Table 2). This phenotype was suppressed in the revertant (Fig. 7 and Table 2). These findings suggest that the hog1 gene is involved in the virulence of T. asahii against silkworms.
Attenuated virulence of the hog1 gene-deficient T. asahii mutant against silkworms. (a) The T. asahii parent strain (Parent; 1.3 × 103 cells/larva), the hog1 gene-deficient mutant (∆hog1; 1.6 × 103 cells/larva), and the revertant from ∆hog1 (Rev.; 1.1 × 103 cells/larva) were injected into the silkworm hemolymph and the silkworms were incubated at 37 °C. Silkworm survival was monitored for 96 h. The significance of differences between the parent strain group and the hog1 gene-deficient mutant group was calculated by the log-rank test based on the curves by the Kaplan–Meier method. P < 0.05 was considered significant. n = 10/group. (b) Number of surviving silkworms at 37 °C was determined at 72 h after administration of the fungal cells (1 × 102 to 5.5 × 104 cells/larva) into the silkworm hemolymph. Surviving and dead silkworms are indicated as 1 and 0, respectively. n = 4/group. Curves were drawn from combined data of 2 independent experiments by a simple logistic regression model. The result of the half-maximal lethal time (LT50), which is the time required to kill half of the animals in a group, were shown in the Supplementary Fig. S2.
Decreased viability of the hog1 gene-deficient T. asahii mutant in silkworm hemolymph
We examined the effect of hog1 gene deficiency on T. asahii cell viability in harvested silkworm hemolymph in vitro. In the parent strain, the number of viable cells in the silkworm hemolymph increased after incubation (Fig. 8a). On the other hand, the viable cell number of the hog1 gene-deficient mutant decreased in the silkworm hemolymph after 96 h of incubation (Fig. 8a). The viable cell number of the hog1 gene-deficient mutant in the silkworm hemolymph after 96 h of incubation was lower than that of the parental strain (Fig. 8b). In the silkworm hemolymph, the hyphae ratio was higher in the hog1 gene-deficient mutant than in the parental strain (Fig. 9). The phenotypes of higher hyphal-forming activity in the hog1 gene-deficient mutant were suppressed in the revertant (Figs. 8 and 9). This observation suggests that the hog1 gene is involved in the morphological changes of T. asahii in the silkworm hemolymph and tolerance against the factors in silkworm hemolymph.
Decrease in number of viable cells of the hog1 gene-deficient T. asahii mutant in harvested silkworm hemolymph. The T. asahii parent strain (Parent), the hog1 gene-deficient mutant (∆hog1), and the revertant of ∆hog1 (Rev.) were grown on SDA and incubated at 27 °C for 1 day. T. asahii cells were suspended in physiologic saline solution and filtered through a 40-μm cell strainer. Absorbance at 630 nm of the T. asahii cell suspension was adjusted to 1–1.5. The cell suspension (10 µL) was added to harvested silkworm hemolymph (90 µL) and the solutions were incubated at 37 °C for 4 days. The T. asahii viable cells were determined by the CFU method. (a) Time course experiment. n = 3/group. Statistically significant differences between 0 and 4 days were evaluated using Student’s t-test. *P < 0.05. (b) T. asahii viable cells were determined at 4 days after inoculation. Statistically significant differences between groups were evaluated using Tukey’s test. *P < 0.05.
Role of hog1 gene on T. asahii morphological change in silkworm hemolymph. The T. asahii parent strain (Parent), the hog1 gene-deficient mutant (∆hog1), and the revertant of ∆hog1 (Rev.) were grown on SDA and incubated at 27 °C for 2 days. T. asahii cells were suspended in physiologic saline solution and filtered through a 40-μm cell strainer. Absorbance at 630 nm of the T. asahii cell suspension was adjusted to 1. The cell suspension (10 µL) was added to hemolymph (100 µL). The solutions were incubated at 27 °C for 48 h. The incubated solution was placed on glass slides and covered by glass coverslips. (a) Samples were examined with bright light under a microscope. Scale bar, 20 µm. (b) The pictures were randomly obtained. The numbers of 3 cell types: yeast, arthroconidia, and hyphae, were counted.
Discussion
In the present study, we generated a hog1 gene-deficient T. asahii mutant to examine its role of the hog1 gene on stress tolerance and virulence. The hog1 gene in T. asahii plays a vital role in stress tolerance to high-temperature, oxidative agents, antifungal drugs, and factors in the silkworm hemolymph. Our findings suggest that the stress tolerance of T. asahii is regulated by the Hog1-mediated signaling pathway and that the stress tolerance of T. asahii might contribute to its virulence.
Hog1 is a MAPK that is widely conserved in fungi16,49. In fungi such as S. cerevisiae and C. neoformans, the Hog1-mediated signaling pathway is involved in resistance to various stressors15,16. A gene encoding a protein with more than 70% homology to the amino acid sequence of Hog1 in S. cerevisiae and C. neoformans was identified in the genome of T. asahii. Furthermore, Hog1 protein in T. asahii contains a phosphorylation domain. Therefore, we assumed that Hog1 is also involved in the kinase cascade related to the stress responses in T. asahii.
The hog1 gene is required for the normal growth of T. asahii under a high-temperature condition (40 °C). Severe growth inhibition of the hog1 gene-deficient T. asahii mutant was observed at 40 °C compared with that at 27 °C and 37 °C. Therefore, the high-temperature stress response in T. asahii is mediated by Hog1. In C. neoformans and S. cerevisiae, the Hog1-mediated signaling pathway is required for growth at 40 °C14,17,35. S. cerevisiae Hog1 activates by high temperature and is required for immediate recovery from high temperature conditions14. Moreover, high temperature stress simultaneously activates Hog1 and the cell wall integrity pathway in S. cerevisiae50. In Candida albicans, Hog1 regulates the cell wall integrity pathway51. Deletion of the CEL1 gene, which encodes a lytic polysaccharide monooxygenase involved in cell wall integrity in Cryptococcus neoformans, causes susceptibility to high temperature stress52. Therefore, we assumed that Hog1 in T. asahii may be activated by high temperature stress and regulates the cell wall integrity pathway for high temperature stress tolerance.
Hog1 is required for several different types of stress responses in T. asahii. Hog1 in S. cerevisiae and C. albicans, and SakA in A. fumigatus play important roles in osmotic stress responses20,21,22,23,24,36,37. In C. neoformans, Hog1 is required for the resistance to stress caused by NaCl and KCl under a glucose-restricted condition19,47. In T. asahii, Hog1 is not involved in osmotic stress responses under a glucose-rich condition. On the other hand, Hog1 in T. asahii is related to resistance to NaCl and KCl under a glucose-restricted condition. The phenotypes of T. asahii are similar to those of C. neoformans. Upstream of the Hog1 MAPK signaling pathway in S. cerevisiae are two branching cascades consisting of Sho1 or Sln1, which phosphorylate Pbs2 MAPK kinase kinase14,15. The gene encoding the Sln1 homolog of S. cerevisiae is not present in the C. neoformans genome, however, and histidine kinase Tco2, instead of Sln1, acts as an osmotic sensor15,19,53. The histidine kinase in T. asahii has 46% amino acid sequence homology with C. neoformans Tco2. Moreover, no protein in T. asahii has at least 30% amino acid sequence homology with the osmotic sensor Sho1 in S. cerevisiae. Therefore, T. asahii may have a Tco2-mediated response mechanism to osmotic stress similar to that of C. neoformans. In C. neoformans, Ena1, a Na+/ATPase regulated by Hog1, exports cations such as sodium and potassium ions47,54. The ena1 gene-deficient C. neoformans mutant exhibits similar growth as the wild-type strain under stress induced by NaCl and KCl in glucose-rich conditions, but shows susceptibility under low-glucose conditions47,54. In T. asahii, Hog1 may regulate the stress tolerance mechanism for sodium and potassium ions mediated by Ena1 under low-glucose conditions. Further studies are needed to clarify the relationships between Hog1 and Tco2 or Ena1 in the cation-stress response of T. asahii. Hog1 in C. neoformans and SakA, a homolog of Hog1, in A. fumigatus show susceptibility to compounds that cause damage to the cell membrane35,39,40. SakA in A. fumigatus is required for the cell wall integrity signaling pathway and contributes to resistance against cell wall damage39. On the other hand, because the Hog1 signaling pathway and the cell wall integrity signaling pathway are not associated in C. neoformans, the hog1 gene-deficient mutant does not show marked susceptibility to cell wall stress38,40. In T. asahii, Hog1 contributes to tolerance to cell membrane damage, but not to cell wall damage. Therefore, the function of Hog1 in T. asahii for the maintenance of cell membranes and cell walls is similar to that in C. neoformans. In C. neoformans, C. albicans, and S. cerevisiae, Hog1 plays crucial roles in the resistance to oxidative stress and ER stress17,20,23,24,35,41,42,43. The hog1 gene-deficient T. asahii mutant showed sensitivity to oxidative stress, but not to ER stress. These observations suggest that Hog1 in T. asahii is an essential factor for oxidative stress resistance, and T. asahii possesses a mechanism involving Hog1-mediated oxidative stress resistance similar to other fungi such as C. neoformans. Hog1 in C. neoformans regulates the expression of oxidative stress-related factors such as catalase and mitochondrial manganese superoxide dismutase54. The increased sensitivity to H2O2 and menadione observed in the hog1 gene-deficient T. asahii mutant may be due to a decrease in the activity of enzymes involved in the elimination of reactive oxygen species. Identification of the responsible enzymes regulated by Hog1 to eliminate reactive oxygen species in T. asahii will be an important future research aim. Hog1 in C. neoformans and S. cerevisiae contributes to resistance to DNA damage17,35,44. The sensitivity of the hog1 gene-deficient T. asahii mutant against MMS and UV suggests that Hog1 is an essential factor for resistance to DNA mutations in T. asahii. We assumed that Hog1 in T. asahii is phosphorylated upon exposure to these stressors. The development of a specific antibody against T. asahii Hog1 is important to reveal the stress response mechanisms by phosphorylation.
T. asahii exhibits Hog1-mediated resistance to antifungal drugs. The hog1 gene-deficient mutant in C. albicans shows sensitivities to AMPH-B and azole antifungal drugs23. In C. neoformans, the hog1 gene-deficient mutant shows sensitivity to AMPH-B, but not to azole antifungal drugs19,35,38. Hog1 in T. asahii is involved in resistance against antifungal drugs. The combined use of Hog1 inhibitors and AMPH-B or azole antifungal agents might be effective in treating T. asahii infections.
C. albicans exhibits 2 forms of yeast and hyphae, and the hyphal elongation is negatively regulated by Hog120,21,23,25,26,27,28,48. Because the hog1 gene-deficient T. asahii mutant forms more hyphae than the parent strain, Hog1 might regulate hyphal formation in T. asahii. We hypothesized that Hog1 in T. asahii controls the expression of genes related to hyphal formation. Hyphal formation in T. asahii is induced by magnesium55. In C. albicans, magnesium deficiency activates Hog156. Therefore, magnesium plays an important role in regulating Hog1 activation in fungi. In T. asahii, magnesium may suppress the Hog1 pathway and promote hyphal formation that is similar to the phenotype of the hog1 gene-deficient mutant. Identifying the important genes involved in hyphal formation regulated by Hog1 is an important future challenge toward deepening our understanding of the morphological aspects of T. asahii.
The virulence of T. asahii against silkworms was reduced by hog1 gene deficiency. Moreover, the number of viable cells in the hog1 gene-deficient mutant decreased in the silkworm hemolymph. We speculate that hog1 gene deficiency causes several stress sensitivities in host environments and results in a virulent phenotype in T. asahii. Therefore, the identification of host factors that induce Hog1-mediated stress responses is an important topic for future research.
Hog1 inhibitors may attenuate T. asahii virulence. However, these inhibitors may affect humans because Hog1 is highly conserved among eukaryotes. Therefore, the development of fungal Hog1-specific inhibitors is required. In calcineurin, which is highly conserved in eukaryotes like Hog1, a fungal-specific calcineurin inhibitor have been synthesized by focusing on the subtle structural differences between human calcineurin and fungal calcineurin57. We assumed that it might be possible to construct selective inhibitory compounds for fungal Hog1 based on structural differences, similar to the development of inhibitory compounds for calcineurin.
In conclusion, T. asahii controls stress response and virulence via Hog1. Hog1-mediated stress responses might contribute to antifungal drug resistance and virulence of T. asahii by facilitating its adaptation to the host environments. These findings suggest that Hog1 inhibitors, as anti-infectious agents in combination with antifungal drugs, may be useful in treating T. asahii infection.
Methods
Reagents
Nourseothricin was purchased from Jena Bioscience (Dortmund, Germany). Cefotaxime sodium, D-glucose, agar, NaCl, KCl, sorbitol, H2O2, DTT, MMS, and AMPH-B were purchased from Fujifilm Wako Pure Chemical Industries (Osaka, Japan). Hygromycin B, caffeine, FLCZ, VRCZ, and N-phenylthiourea were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Congo Red and menadione were purchased from Sigma-Aldrich (St. Louis, MO, USA). G418 was purchased from Enzo Life Science, Inc. (Farmingdale, NY, USA). Hipolypeptone was purchased from Nihon Pharmaceutical Co., Ltd. (Tokyo, Japan). Tunicamycin (TM) was purchased from Cayman Chemical Company (Ann Arbor, MI, USA). Sodium dodecyl sulfate (SDS) was purchased from Nippon Gene Co., Ltd. (Tokyo, Japan). Tween 80 was purchased from MP Biomedicals LLC (Santa Ana, MO, USA).
Comparison of amino acid sequences of typical fungal Hog1 proteins
The amino acid sequences of Hog1 proteins in T. asahii, C. neoformans, C. albicans, and S. cerevisiae, and SakA protein in A. fumigatus were obtained from the Ensembl Fungi (http://fungi.ensembl.org/index.html). The Ensembl Fungi gene IDs of the proteins are shown in Supplementary Table 1. The functional domains of the proteins were estimated by the NCBI website (https://www.ncbi.nlm.nih.gov). The NCBI domain accession numbers of the proteins are shown in Supplementary Table 2. The phylogenic tree was constructed according to the maximum likelihood method using MEGA11 Software (https://www.megasoftware.net/). Amino acid sequence alignment was performed using MEGA11 Software.
Culture of T. asahii
The T. asahii strain (MPU129 ku70 gene-deficient mutant) used in this study was generated as previously reported30. The T. asahii MPU129 ku70 gene-deficient mutant was grown on SDA (1% hipolypeptone, 4% dextrose, and 1.5% agar) containing G418 (100 μg/mL) and incubated at 27 °C for 2 days.
Construction of the hog1 gene-deficient T. asahii mutant and the revertant
The plasmid for gene-deficient T. asahii strains was constructed according to a previous report34. To generate the hog1 gene-deficient strain, the 5′-UTR and 3′-UTR of the hog1 gene were introduced into a pAg1-NAT1 vector30 using the infusion method (In-Fusion HD Cloning Kit, Takara, Shiga, Japan). To generate the revertant, the hygromycin phosphotransferase gene (hph) cassette and the hog1 gene were introduced into pAg1-hog1(5′UTR)-NAT1-hog1(3′UTR). The primers used for PCR amplification of each DNA region are shown in Supplementary Table 3.
Gene transfer using electroporation was performed according to a previous report33. The 5′-UTR (hog1)-NAT1-3′-UTR (hog1) or 5′-UTR (hog1)-hog1-hph-3′-UTR (hog1) fragments were amplified by PCR with the primers shown in Supplementary Table 3. The T. asahii competent cells (40 µL) with the DNA fragments (90–180 ng) were added to a 0.2-cm gap cuvette (Bio-Rad Laboratories, Inc.) and electroporated (Time constant protocol: 1800 V, 5 ms) using a Gene Pulser Xcell (Bio-Rad Laboratories, Inc.). The cells were suspended by yeast peptone dextrose containing 0.6 M sorbitol and incubated at 27 °C for 3 h. After incubation, the cells were applied to SDA containing nourseothricin (300 µg/mL) or hygromycin B (300 µg/mL) and incubated at 27 °C for 3 days. The mutation into the genome of the candidates growing on the SDA containing drugs was confirmed by PCR with the primers shown in Supplementary Table 3. Information on the strains is provided in Table 3.
Temperature sensitivity test
The temperature sensitivity test was performed according to a previous report with slight modifications34. The T. asahii cells grown on SDA containing cefotaxime (100 μg/mL) at 27 °C for 1 day were suspended in physiologic saline solution with 0.01% Tween 80 and filtered through a 40-μm cell strainer (Corning Inc., Corning, NY, USA). The T. asahii cell suspension was adjusted to 1–1.2 on absorbance at 630 nm. Series of a tenfold dilution of the fungal suspension were prepared using saline. Five microliters of each cell suspension were spotted on the SDA. The agar plates were incubated at 27, 37, or 40 °C for 48 h, and photographs were obtained.
For growth on liquid medium, Sabouraud liquid medium (1% hipolypeptone, 4% dextrose) was used in this study. Cell suspensions of the T. asahii strains were prepared with Sabouraud liquid medium. The T. asahii suspensions (A630 = 0.005) were incubated at 27 °C, 37 °C, or 40 °C for 4 days and absorbance at 630 nm was measured using a microplate reader (iMark microplate reader; Bio-Rad Laboratories Inc., Hercules, CA, USA).
Drug sensitivity test using agar plate
The drug sensitivity test was performed according to a previous report with slight modifications34. Series of a tenfold dilution of the fungal suspension were prepared in the same manner as for the temperature sensitivity test described above. Five microliters of each cell suspension were spotted on the SDA containing NaCl (1.2 M), KCl (1.5 M), caffeine (0.65 mg/mL), Congo red (400 µg/mL), SDS (0.008%), H2O2 (3 mM), menadione (60 µM), TM (1 µg/mL), DTT (12 mM), MMS (0.05%), AMPH-B (0.4 µg/mL), FLCZ (18 µg/mL), or VRCZ (0.2 µg/mL). Each agar plate was incubated at 27 °C for 24–48 h, and photographs were obtained.
UV sensitivity test
Series of a tenfold dilution of the fungal suspension were prepared in the same manner as for the temperature sensitivity test described above. Five microliters of each cell suspension was spotted on the SDA. The SDA plate was exposed to UV light using a UV crosslinker (CX-2000; UVP LLC, Upland, USA). Each agar plate was incubated at 27 °C for 24 h, and photographs were obtained.
MIC determination
The MIC values were determined by a micro-liquid dilution method (CLSI, M27-A3) using antifungal susceptibility test plates (Yeast-like fungus DP ‘Eiken’; Eiken Chemical Co., Ltd., Tokyo, Japan)58. The T. asahii cells on SDA containing cefotaxime (100 μg/mL) was suspended in physiologic saline solution with 0.01% Tween 80. The T. asahii cell suspension was adjusted to McFarland 1. Each fungal suspension was diluted 20-fold in saline, and then diluted 100-fold in RPMI 1640 medium (Thermo Fisher Scientific Inc., MA, USA) containing 0.165 M MOPS (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) (RPMI MOPS) (pH 7.0). One hundred microliters of each cell suspension was applied to wells of the antifungal susceptibility test plates. The test plates were incubated at 37 °C for 48 h. The MIC values were determined from the results of two independent experiments.
Observation of T. asahii morphology
The morphology observation was performed according to a previous report34. The T. asahii cells were suspended in physiologic saline solution with 0.01% Tween 80 and filtered through a 40-μm cell strainer (Corning Inc.). The cell suspension (10 µL) adjusted to 1 on absorbance at 630 nm was added to Sabouraud dextrose liquid medium (90 µL) or harvested silkworm hemolymph (90 µL). The incubated solution at 27 °C for 48 h was placed on glass slides and covered by glass coverslips. Samples were examined with bright light under a microscope (CH-30; Olympus, Tokyo, Japan). The pictures were randomly obtained. The morphology of T. asahii was determined according to a previous report59. The numbers of the following 3 types of cells were counted: yeast (spherical cells including budding yeasts and may include blastoconidia); arthroconidia (≥ 3 round or rectangular cells connected with their interiors separated by septa); and hyphae (cells elongated to a length of at least 2 yeast cells possessing no septa).
Silkworm infection experiments
Silkworm infection experiments were performed according to a previous report with slight modifications30. Eggs of silkworms (Hu·Yo × Tukuba·Ne) were purchased from Ehime-Sanshu Co, Ltd. (Ehime, Japan). Silkworm fifth instar larvae were fed the artificial diet (Silkmate 2S; Ehime-Sanshu Co., Ltd.) overnight. The T. asahii grown on SDA plates incubated for 1 day at 27 °C was suspended in physiologic saline solution with 0.01% Tween 80 and filtered through a 40-μm cell strainer (Corning Inc.). A 50-µL suspension of T. asahii cells was administered to the silkworms and the silkworms injected with T. asahii cells were incubated at 37 °C.
LD50 measurement
The dose of T. asahii required to kill half of the silkworms (LD50) was determined according to a previous report with slight modifications30 T. asahii strains (1 × 102 to 5.5 × 104 cells/50 µL) were injected into the silkworm hemolymph and the silkworms were incubated at 37 °C. Survival of the silkworms (n = 4/group) at 72 h was recorded. The LD50 was determined from the combined data of 2 independent experiments by a simple logistic regression model using Prism 9.1.2 (GraphPad Software, LLC, San Diego, CA, USA, https://www.graphpad.com/scientific-software/prism/).
Cell viability of T. asahii in silkworm hemolymph
The T. asahii cells grown on SDA containing cefotaxime (100 μg/mL) were suspended in physiologic saline solution with 0.01% Tween 80 and filtered through a 40-μm cell strainer (Corning Inc.). Ten microliters of each cell suspension (A630 = 1–1.5) was added to 90 µL of filter-sterilized silkworm hemolymph. The cell suspensions were cultured at 37 °C for 4 days and collected every 24 h. The collected cell suspensions were diluted 2000-fold with physiologic saline and 100 μL was spread on SDA. The number of colonies on the SDA after 1 day incubation were counted.
Statistical analysis
All experiments were performed at least twice and representative results are shown. The significance of differences between groups in the silkworm infection experiments in Fig. 7a was calculated by the log-rank test based on curves determined using the Kaplan–Meier method with Prism 9.1.2. P < 0.05 was considered significant. Statistically significant differences between 0 and 4 days in Fig. 8a were evaluated using Student’s t-test. Statistically significant differences between groups in Fig. 8b were evaluated using Tukey’s test.
Data availability
The datasets generated in the present study are available from the corresponding author upon reasonable request.
References
Sugita, T., Nishikawa, A., Ichikawa, T., Ikeda, R. & Shinoda, T. Isolation of Trichosporon asahii from environmental materials. Med. Mycol. 38, 27–30 (2000).
Sugita, T. et al. Genetic diversity and biochemical characteristics of Trichosporon asahii isolated from clinical specimens, houses of patients with summer-type-hypersensitivity pneumonitis, and environmental materials. J. Clin. Microbiol. 39, 2405–2411 (2001).
Zhang, E., Sugita, T., Tsuboi, R., Yamazaki, T. & Makimura, K. The opportunistic yeast pathogen Trichosporon asahii colonizes the skin of healthy individuals: Analysis of 380 healthy individuals by age and gender using a nested polymerase chain reaction assay. Microbiol. Immunol. 55, 483–488 (2011).
Colombo, A. L., Padovan, A. C. B. & Chaves, G. M. Current knowledge of Trichosporon spp. and Trichosporonosis. Clin. Microbiol. Rev. 24, 682–700 (2011).
Gouba, N., Raoult, D. & Drancourt, M. Eukaryote culturomics of the gut reveals new species. PLoS One 9, e106994 (2014).
Cho, O., Matsukura, M. & Sugita, T. Molecular evidence that the opportunistic fungal pathogen Trichosporon asahii is part of the normal fungal microbiota of the human gut based on rRNA genotyping. Int. J. Infect. Dis. 39, 87–88 (2015).
Walsh, T. J., Melcher, G. P., Lee, J. W. & Pizzo, P. A. Infections due to Trichosporon species: New concepts in mycology, pathogenesis, diagnosis and treatment. Curr. Top. Med. Mycol. 5, 79–113 (1993).
Duarte-Oliveira, C. et al. The cell biology of the Trichosporon–host interaction. Front. Cell Infect. Microbiol. 7, 118 (2017).
Krcmery, V. et al. Hematogenous trichosporonosis in cancer patients: Report of 12 cases including 5 during prophylaxis with itraconazol. Support Care Cancer 7, 39–43 (1999).
Kimura, M. et al. Micafungin breakthrough fungemia in patients with hematological disorders. Antimicrob. Agents Chemother. 62, 10–128 (2018).
Kimura, M. et al. Factors associated with breakthrough fungemia caused by Candida, Trichosporon, or fusarium species in patients with hematological disorders. Antimicrob. Agents Chemother. 66, e0208121 (2022).
Toriumi, Y., Sugita, T., Nakajima, M., Matsushima, T. & Shinoda, T. Antifungal pharmacodynamic characteristics of amphotericin B against Trichosporon asahii, using time-kill methodology. Microbiol. Immunol. 46, 89–93 (2002).
Iturrieta-González, I. A., Padovan, A. C. B., Bizerra, F. C., Hahn, R. C. & Colombo, A. L. Multiple species of Trichosporon produce biofilms highly resistant to triazoles and amphotericin B. PLoS One 9, e109553 (2014).
Winkler, A. et al. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell 1, 163–173 (2002).
Day, A. M. & Quinn, J. Stress-activated protein kinases in human fungal pathogens. Front. Cell Infect. Microbiol. 9, 261 (2019).
Blomberg, A. Yeast osmoregulation—Glycerol still in pole position. FEMS Yeast Res. 22, 1–20 (2022).
Bahn, Y.-S., Kojima, K., Cox, G. M. & Heitman, J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16, 2285–2300 (2005).
Bahn, Y.-S., Kojima, K., Cox, G. M. & Heitman, J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17, 3122–3135 (2006).
Kim, S.-Y. et al. Hrk1 plays both Hog1-dependent and -independent roles in controlling stress response and antifungal drug resistance in Cryptococcus neoformans. PLoS One 6, e18769 (2011).
Day, A. M., Herrero-de-Dios, C. M., MacCallum, D. M., Brown, A. J. P. & Quinn, J. Stress-induced nuclear accumulation is dispensable for Hog1-dependent gene expression and virulence in a fungal pathogen. Sci. Rep. 7, 14340–14411 (2017).
Day, A. M. et al. Blocking two-component signalling enhances Candida albicans virulence and reveals adaptive mechanisms that counteract sustained SAPK activation. PLoS Pathog. 13, e1006131 (2017).
Wang, L. et al. SPT20 regulates the Hog1-MAPK pathway and is involved in Candida albicans response to hyperosmotic stress. Front. Microbiol. 11, 213 (2020).
Correia, I., Wilson, D., Hube, B. & Pla, J. Characterization of a Candida albicans mutant defective in All MAPKs highlights the major role of Hog1 in the MAPK signaling network. J. Fungi (Basel) 6, 230 (2020).
Husain, F., Pathak, P., Román, E., Pla, J. & Panwar, S. L. Adaptation to endoplasmic reticulum stress in Candida albicans relies on the activity of the Hog1 mitogen-activated protein kinase. Front. Microbiol. 12, 794855 (2021).
Eisman, B. et al. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell 5, 347–358 (2006).
Su, C., Lu, Y. & Liu, H. Reduced TOR signaling sustains hyphal development in Candida albicans by lowering Hog1 basal activity. Mol. Biol. Cell 24, 385–397 (2013).
Unoje, O., Yang, M., Lu, Y., Su, C. & Liu, H. Linking Sfl1 regulation of hyphal development to stress response kinases in Candida albicans. mSphere 5, e00672-19 (2020).
Chen, H., Zhou, X., Ren, B. & Cheng, L. The regulation of hyphae growth in Candida albicans. Virulence 11, 337–348 (2020).
Matsumoto, Y. et al. A novel silkworm infection model with fluorescence imaging using transgenic Trichosporon asahii expressing eGFP. Sci. Rep. 10, 10991–11011 (2020).
Matsumoto, Y. et al. Development of an efficient gene-targeting system for elucidating infection mechanisms of the fungal pathogen Trichosporon asahii. Sci. Rep. 11, 18270–18310 (2021).
Matsumoto, Y. & Sekimizu, K. Silkworm as an experimental animal to research for fungal infections. Microbiol. Immunol. 63, 41–50 (2019).
Kaito, C., Murakami, K., Imai, L. & Furuta, K. Animal infection models using non-mammals. Microbiol. Immunol. 64, 585–592 (2020).
Matsumoto, Y., Nagamachi, T., Yoshikawa, A., Yamada, T. & Sugita, T. A joint PCR-based gene-targeting method using electroporation in the pathogenic fungus Trichosporon asahii. AMB Express 12, 91 (2022).
Matsumoto, Y. et al. A critical role of calcineurin in stress responses, hyphal formation, and virulence of the pathogenic fungus Trichosporon asahii. Sci. Rep. 12, 16126–16212 (2022).
Lee, K.-T. et al. Distinct and redundant roles of protein tyrosine phosphatases Ptp1 and Ptp2 in governing the differentiation and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 13, 796–812 (2014).
Romanov, N. et al. Identifying protein kinase-specific effectors of the osmostress response in yeast. Sci. Signal. 10, eaag2435 (2017).
Kirkland, M. E. et al. Host lung environment limits Aspergillus fumigatus germination through an SskA-dependent signaling response. mSphere 6, e0092221 (2021).
Maeng, S. et al. Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans. Eukaryot. Cell 9, 360–378 (2010).
Nascimento, A. C. M. O. B. et al. Mitogen activated protein kinases SakA (HOG1) and MpkC collaborate for Aspergillus fumigatus virulence. Mol. Microbiol. 100, 841–859 (2016).
Bloom, A. L. M., Goich, D., Knowles, C. M. & Panepinto, J. C. Glucan unmasking identifies regulators of temperature-induced translatome reprogramming in C. neoformans. Sphere 6, e01281-20 (2021).
Singh, K. K. The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic. Biol. Med. 29, 1043–1050 (2000).
Kim, J. H., Chan, K. L., Faria, N. C. G., Martins, M. L. & Campbell, B. C. Targeting the oxidative stress response system of fungi with redox-potent chemosensitizing agents. Front. Microbiol. 3, 88 (2012).
Hernández-Elvira, M. et al. Tunicamycin sensitivity-suppression by high gene dosage reveals new functions of the yeast Hog1 MAP kinase. Cells 8, 710 (2019).
Huang, S., Zhang, D., Weng, F. & Wang, Y. Activation of a mitogen-activated protein kinase Hog1 by DNA damaging agent methyl methanesulfonate in yeast. Front. Mol. Biosci. 7, 581095 (2020).
Malacrida, A. M. et al. Hospital Trichosporon asahii isolates with simple architecture biofilms and high resistance to antifungals routinely used in clinical practice. J. Méd. Mycol. 33, 101356 (2023).
Mulè, A. et al. Trichosporon asahii infective endocarditis of prosthetic valve: A case report and literature review. Antibiotics 12, 1181 (2023).
Jung, K.-W., Strain, A. K., Nielsen, K., Jung, K.-H. & Bahn, Y.-S. Two cation transporters Ena1 and Nha1 cooperatively modulate ion homeostasis, antifungal drug resistance, and virulence of Cryptococcus neoformans via the HOG pathway. Fungal Genet. Biol. 49, 332–345 (2012).
Cheetham, J. et al. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol. Biol. Cell 18, 4603–4614 (2007).
Yaakoub, H., Mina, S., Calenda, A., Bouchara, J.-P. & Papon, N. Oxidative stress response pathways in fungi. Cell. Mol. Life Sci. 79, 333–427 (2022).
Dunayevich, P. et al. Heat-stress triggers MAPK crosstalk to turn on the hyperosmotic response pathway. Sci. Rep. 8, 15168 (2018).
Ibe, C. & Munro, C. A. Fungal cell wall proteins and signaling pathways form a cytoprotective network to combat stresses. J. Fungi 7, 739 (2021).
Probst, C. et al. A fungal lytic polysaccharide monooxygenase is required for cell wall integrity, thermotolerance, and virulence of the fungal human pathogen Cryptococcus neoformans. PLoS Pathog. 19, e1010946 (2023).
Xu, X. et al. Glucosamine stimulates pheromone-independent dimorphic transition in Cryptococcus neoformans by promoting Crz1 nuclear translocation. PLoS Genet. 13, e1006982 (2017).
Ko, Y.-J. et al. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot. Cell 8, 1197–1217 (2009).
Aoki, K. et al. Hyphal growth in Trichosporon asahii is accelerated by the addition of magnesium. Microbiol. Spectr. 11, e04242-22 (2023).
Polvi, E. J. et al. Metal chelation as a powerful strategy to probe cellular circuitry governing fungal drug resistance and morphogenesis. PLoS Genet. 12, e1006350 (2016).
Juvvadi, P. R. et al. Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents. Nat. Commun. 10, 4275–4318 (2019).
Clinical and Laboratory Standards Institute (CLSI). M27-A3 Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Standard 3rd edn. (Clinical and Laboratory Standards Institute, 2008).
Kurakado, S. et al. Role of arthroconidia in biofilm formation by Trichosporon asahii. Mycoses 64, 42–47 (2021).
Acknowledgements
We thank Eri Sato, Hiromi Kanai, and Sachi Koganesawa (Meiji Pharmaceutical University) for their technical assistance in rearing the silkworms. This study was supported by JSPS KAKENHI grant number JP20K07022 and JP23K06141 (Scientific Research (C) to Y.M.) and in part by the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development, AMED (Grant number JP20fk0108135h0201 and 23fk0108679h0401 to T.S.).
Author information
Authors and Affiliations
Contributions
Study conception and design: Y.M., Y.S. Acquisition of data: Y.M., Y.S. Analysis and interpretation of data: Y.M., Y.S., T.N., A.Y. Drafting of manuscript: Y.M., Y.S. Critical revision: Y.M., T.N., A.Y., T.S. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Matsumoto, Y., Sugiyama, Y., Nagamachi, T. et al. Hog1-mediated stress tolerance in the pathogenic fungus Trichosporon asahii. Sci Rep 13, 13539 (2023). https://doi.org/10.1038/s41598-023-40825-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-40825-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.