Abstract
Oxidative stress (OS), which impacts lipid metabolic reprogramming, can affect the biological activities of cancer cells. How oxidative stress and phospholipid metabolism (OSPM) influence the prognosis of pancreatic cancer (PC) needs to be elucidated. The metabolic data of 35 pancreatic tumor samples, 34 para-carcinoma samples, and 31 normal pancreatic tissues were obtained from the previously published literature. Pan-cancer samples were obtained from The Cancer Genome Atlas (TCGA). And the Gene Expression Omnibus (GEO), International Cancer Genome Consortium (ICGC), ArrayExpress, and the Genotype-Tissue Expression (GTEx) databases were searched for more PC and normal pancreatic samples. The metabolites in PC were compared with normal and para-carcinoma tissues. The characteristics of the key OSPM genes were summarized in pan-cancer. The random survival forest analysis and multivariate Cox regression analysis were utilized to construct an OSPM-related signature. Based on this signature, PC samples were divided into high- and low-risk subgroups. The dysregulations of the tumor immune microenvironment were further investigated. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was conducted to investigate the expression of genes in the signature in PC and normal tissues. The protein levels of these genes were further demonstrated. In PC, metabolomic studies revealed the alteration of PM, while transcriptomic studies showed different expressions of OSPM-related genes. Then 930 PC samples were divided into three subtypes with different prognoses, and an OSPM-related signature including eight OSPM-related genes (i.e., SLC2A1, MMP14, TOP2A, MBOAT2, ANLN, ECT2, SLC22A3, and FGD6) was developed. High- and low-risk subgroups divided by the signature showed different prognoses, expression levels of immune checkpoint genes, immune cell infiltration, and tumor microenvironment. The risk score was negatively correlated with the proportion of TIL, pDC, Mast cell, and T cell co-stimulation. The expression levels of genes in the signature were verified in PC and normal samples. The protein levels of SLC2A1, MMP14, TOP2A, MBOAT2, ANLN, and SLC22A3 showed up-regulation in PC samples compared with normal tissues. After integrating metabolomics and transcriptomics data, the alterations in OSPM in PC were investigated, and an OSPM-related signature was developed, which was helpful for the prognostic assessment and individualized treatment for PC.
Similar content being viewed by others
Introduction
Pancreatic cancer (PC), a gastrointestinal malignancy, represents nearly 95% of pancreatic cancer cases1,2. It is widely recognized as a lethal malignancy. The 5-year survival rate of PC is about 10%3. Surgery is currently the main treatment for PC, but it is not suitable for patients with advanced disease. Although some patients with PC can benefit from radiotherapy and chemotherapy, the overall survival is still poor4. To date, few genetic mutations have been demonstrated to be connected with the occurrence of PC, and no clear mechanisms for the development of PC have been investigated. For a reduction in PC mortality, early diagnostic biomarkers and effective therapeutic targets are urgently needed5.
Oxidative stress (OS), with the overproduction of reactive oxygen species (ROS), is reported to be associated with oncogenesis6,7. ROS is produced in the process of mitochondrial respiration and is reported to be essential in signaling pathways8. The level of oxidative stress is linked to the oxidizing capacity and the antioxidation power. Excess ROS damages to cell components, including lipids9. Targeting OS is considered a novel treatment modality in various diseases, including cancer10. Current evidences support that the dysfunction of ROS is a risk factor and antioxidant supplementation is a preventive method for PC11,12.
Lipids, including triglycerides, cholesterol, and phospholipids, are essential for cancer metabolism and the activation of immune cells13,14. Currently, it is reported that lipid metabolic reprogramming including the increase of lipid uptake, storage and lipogenesis can result in rapid tumor growth and is viewed as a newly recognized hallmark of malignancy15. Lipid metabolic reprogramming can influence the biological activities of cancer cells and is reported to be of vital importance in solid tumors13,16. Phospholipids, the major component of cell membranes, are significant for cell structure and metabolism17. Additionally, phospholipids have been proposed as the inducers of cancer multidrug resistance18. Overall, oxidative stress, which compromises lipid function impairs and is related to lipid metabolic reprogramming, was found to function in the development and metastasis of cancer7,19,20. The disorders of phospholipid affect the development and drug sensitivity of PC21,22. The specific roles of OS and phospholipid metabolism (PM) in PC has not been investigated clearly.
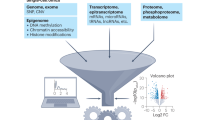
By integrating genomic, transcriptomic, proteomic, and metabolomic data, multi-omics studies have revealed the discovery of novel tumor prognostic markers23. Previous findings offer fresh insights into clinical prognostic assessment by providing valuable bioinformatics model24,25. Moreover, the utilization of machine learning algorithms on multi-omics data has demonstrated promising predictive capabilities in identifying tumor prognostic markers, which holds significant potential in delivering more accurate guidance for personalized treatment and survival prediction26,27. In this study, the altered metabolites in the occurrence of PC were comprehensively explored and the potential impacts of OSPM in the prognosis of PC were investigated. The significant genes in this process of OSPM were summed up from a pan-cancer perspective. Based on the function of OSPM, PC samples were classified into OSPM-active, OSPM-normal, and OSPM-inactive subtypes. Samples in the OSPM-active subtype had a poor prognosis. The specific alterations and discrepancies in the OSPM-active subtype were explored. Furthermore, an OSPM-related signature was constructed to identify the risk of PC patients. After detailed exploration and demonstration, the OSPM-related signature showed promising prognostic performance and diagnostic accuracy.
Methods
Data acquisition
The metabolic data of 35 pancreatic tumor samples, 34 para-carcinoma samples, and 31 normal pancreatic tissues were obtained from the previously published literature, completed by Liu et al.28. In total, 10,028 OS-related genes and 4,479 PM-related genes were searched in the Molecular Signatures Database (MSigDB)29 and GeneCard websites30. Specifically, the GeneCard platform provided 10,022 OS-related genes and 4477 PM-related genes. MSigDB platform provided two OS-related pathways (i.e. GOBP_RESPONSE_TO_OXIDATIVE_STRESS, WP_OXIDATIVE_STRESS_RESPONSE) and two PM-related pathways (i.e. GOBP_REGULATION_OF_PHOSPHOLIPID_METABOLIC_PROCESS, REACTOME_PHOSPHOLIPID_METABOLISM). “GOBP_RESPONSE_TO_OXIDATIVE_STRESS” pathway involves 437 OS-related genes, and “WP_OXIDATIVE_STRESS_RESPONSE” pathway involves 33 OS-related genes. “GOBP_REGULATION_OF_PHOSPHOLIPID_METABOLIC_PROCESS” pathway involves 37 PM-related genes, and “REACTOME_PHOSPHOLIPID_METABOLISM” pathway involves 211 PM-related genes. After removing duplicate genes, a final set of 10,028 OS-related genes and 4479 PM-related genes was obtained.
The gene expression data and clinical information were obtained from public databases. In total, PC datasets contained 930 PC samples made up of GSE28735, GSE57495, and GSE62452 from the Gene Expression Omnibus (GEO), MTAB-6134 from ArrayExpress, TCGA-PC samples from The Cancer Genome Atlas (TCGA), and ICGC-AU and ICGC-CA datasets from International Cancer Genome Consortium (ICGC). The Genotype-Tissue Expression (GTEx) database was searched to obtain normal pancreatic samples.
For pan-cancer summarization, mRNA expression, clinical information, single-nucleotide variation (SNV), copy number variation (CNV), and methylation data of common cancers were downloaded from TCGA database.
Identification of metabolic reprogramming in the occurrence of PC
Liu et al. detected and analyzed the metabolites in PC and presented the metabolic reprogramming in PC. We downloaded the published literature compiled by Liu et al., which contained the metabolomics data of normal and tumor tissues as supplementary materials28. This study involves the secondary mining of metabolomics data based on the supplementary materials provided in the article by Liu et al. After collecting and analyzing the metabolomics data, the alterations in metabolites caused by the occurrence of PC were further investigated. The detailed approaches are described as follows. The metabolic changes were investigated, and each fold change (FC) value was calculated after comparing tumor tissues with para-carcinoma tissues. Meanwhile, the differences between tumor tissues and normal tissues were also explored. On the basis of the p.adjust < 0.05 and FC > 2 or FC < 0.5, the metabolites with different contents in PC were obtained.
Identification of key OSPM-related genes in PC
Considering the prominent roles of OS and PM in the development of PC, we integrated the gene sets corresponding to OS and PM, resulting in the formation of OSPM genes. Then the OSPM-related differentially expressed genes (DEGs) were explored in three cohorts, namely GSE28735 (cohort1), GSE62452 (cohort2), and TCGA dataset combined with the GTEx dataset (cohort3). All the DEGs (criteria: FC > 2 or FC < 0.5) acquired from the three cohorts were taken into intersection. For further investigation of the prognostic values of the OSPM-related DEGs, univariate Cox regression analysis and Kaplan–Meier (KM) survival analysis were conducted. The prognostic OSPM-related DEGs were collected as candidate genes for the subsequent analyses.
The role of OSPM in pan-cancer
Given the potential function of OSPM in various cancers and the essential role of the OSPM-related genes in the process of OSPM, a comprehensive investigation of the OSPM-related genes was made in pan-cancer. First, the alteration in the expression level was assessed according to the FC, and the prognostic impact was distinguished by conducting univariate Cox regression analysis. Next, the CNV (amplified or deleted) was summed up. As for SNV, the mutation frequency (samples with SNV/all samples) was calculated and the detailed mutation types were exhibited. Then the status of promoter methylation in tumor samples was obtained after comparing with normal samples in pan-cancer. All the methods have been utilized and described before31,32,33.
OSPM-based cluster analysis
With the mRNA expression of the OSPM-related genes in PC, the function states of OSPM were estimated quantitatively with ssGSEA algorithm. Whether the discrepancies of OSPM could influence the survival of the sample was investigated after performing cluster analysis. During the process, “gplots” and “pheatmap” packages in R were employed to visualize the results of cluster analysis and relative expression levels of each OSPM-related genes34. Based on the ssGSEA scores obtained in the normal samples, PC samples were grouped into cluster1 with OSPM-active function, cluster2 with OSPM-inactive function, and cluster3 with OSPM-normal function.
Then the discrepancies in the different clusters were searched by performing the following analyses35,36: (1) Comparing the OSPM function scores by the Kruskal–Wallis test; (2) Comparing the survival probability by KM analysis; (3) Comparing expression levels of tumor suppressor and promoter genes through the Kruskal–Wallis test; and (4) Comparing the expression levels of immunological checkpoint genes (ICGs) by the Kruskal–Wallis test.
The estimation of the tumor immune microenvironment was conducted as follows 37: (1) ssGSEA was performed to quantify the immune-associated gene sets; (2) Spearman's correlation analysis was conducted determine the link between prognostic OSPM-related DEGs and immune cells; (3) The correlation between the OSPM function score and immune components was found by utilizing “ggstatsplot,” “data.table,” “dplyr,” “tidyr,” and “ggplot2” packages in R; and (4) MCPCOUNTER, XCELL, CIBERSORT, EPIC, CIBERSORT-ABS, and TIMER were performed to assess immune responses, and the statistically significant discrepancies among different clusters were exhibited.
Group division and OSPM-related signature construction
To develop a quantitative signature for guiding the clinical diagnosis and treatment of PC, an OSPM-related signature was constructed and validated. The training cohort (635 PC samples) included GSE57495, GSE28735, GSE62452, MTAB-6134, and TCGA-PC datasets, whereas the validation cohort (295 PC samples) included the ICGC-CA and ICGC-AU datasets. To make the signature concise and convenient, random survival forest analysis was conducted using the “randomForestSRC” package in R to select the most appropriate genes from all the prognostic OSPM-related DEGs to gain an optimal prognostic signature38. Multivariate Cox regression analysis was then conducted, and the coefficients helped to obtain the risk score of each sample39.
The risk score corresponding to each sample was determined, and the median score helped to distinguish high- and low-risk subgroups in the training and test cohorts, respectively. Then the discrepancies of the high- and low-risk subgroups were investigated in the two cohort as follows: (1) Sample division was visualized through principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE); (2) Survival probability was compared through KM analysis40,41; (3) The expression levels of the genes in the signature were compared between high- and low-risk subgroups and exhibited in heatmap; (4) The diagnostic accuracy was demonstrated by developing time-dependent receiver operating characteristic (ROC) curves and obtaining the AUC values42; (5) The expression levels of ICGs were compared, and the immune response was estimated through the methods mentioned above43,44. In addition, the GEPIA website was utilized to compare the expression levels of the model genes in PC samples with different clinical stages45.
Cell culture and qRT-PCR
Human pancreatic cell line (HPDE6-C7) and PC cell lines (BxPC-3, CFPAC-1, Panc-1, and Mia-Paca-2) were obtained for PCR experiments of model genes. HPDE6-C7, Panc-1, and Mia-Paca-2 were cultured in DMEM medium. BxPC-3 was maintained in 1640 medium. CFPAC-1 was maintained in IMDM medium. All these mediums were supplemented with 10% fetal bovine serum at 37 °C and 5% –CO2. Total RNA was extracted utilizing TRIzol reagent and synthesized into cDNA utilizing M-MLV reverse transcriptase under the instruction of the manufacturer. qRT-PCR was performed using SYBR Green assay and β-actin (Forward: CCTGGGCATGGAGTCCTGTG, Reverse: TCTTCATTGTGCTGGGTGCC) was selected as an endogenous reference. The specific PCR primer sequences are as follows: SLC2A1 (FORWARD: ATGAAGGAAGAGAGTCGGCAGATG, REVERSE: AGCACCACAGCGATGAGGATG), MMP14 (FORWARD: TGCGTCCATCAACACTGCCTAC, REVERSE: CGCCTCATCAAACACCCAATGC), TOP2A (FORWARD: GCACCAGCACATCAAAGGAAGC, REVERSE: ATAGCAGCATCATCTTCAGGACCAG), MBOAT2 (FORWARD: CCATCTCCAAATACTGCGGTTGTTC, REVERSE: GCTCATCAATGTTGTACTCCACAGG), ANLN (FORWARD: GTTCTCCAAGTCCTGTGTCTCCTC, REVERSE: TGCAGTTGCTTCCAATCTTGAGTTC), ECT2 (FORWARD: AGCTCCACTCCAGTTCCTTCAAAG, REVERSE: GGTCCACCACGTTGTCCTTCC), SLC22A3 (FORWARD: CGTGTGGCTAGAACTACCTCTGATC, REVERSE: ACTGTCTCTGGCAAGGCAATACC), FGD6 (FORWARD: CAACTCGGAGACACCACCACAG, REVERSE: CTACCAACAAGCCTTGCCAATCAC).
Single-cell expression level, immunohistochemistry, and immunofluorescence of model genes in PC
Tumor samples are composed of a variety of cells. Tumor Immune Single-cell Hub 2 (TISCH2), a scRNA-seq database focusing on tumor microenvironment (TME), was searched for identifying of major cell types and the expression levels of model genes in different cells in PC46. The Human Protein Atlas (HPA: https://www.proteinatlas.org/) was utilized for further exploration about the protein levels of model genes47,48. Additionally, the HPA was also utilized to exhibit the cellular localization on the basis of immunofluorescence.
Results
Identification of metabolic alterations and OSPM-related genes in PC
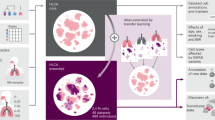
The workflow is depicted in Fig. 1. The alteration of metabolites plays a vital role in tumor initiation and progression. The discrepancies of the content of metabolite between pancreatic tumor samples and para-carcinoma samples were investigated, and 99 metabolites with significant changes were exhibited in the heatmap (Fig. 2A). Similarly, 60 metabolites altered significantly between tumor samples and normal samples, as shown in another heatmap (Fig. 2B). After taking an intersection, 35 shared metabolites were obtained (Fig. 2C). Of note, the shared metabolites primarily belonged to diacylglycerol, glycerophosphocholine, and glycerophosphoethanolamine. It is well-known that PM mainly includes the metabolism of glycerophospholipids and sphingophospholipids. And diacylglycerol is the major substrates for the synthesis of phosphatidylglycerol in glycerophospholipid metabolism. Conversely, glycerophospholipids can be converted into glycerophosphocholine and glycerophosphoethanolamine by the action of phospholipase B, after removing the fatty acids. As a result, PM showed a proper role in the development of PC. Due to the influence of OS on PM, the following investigations were mainly focused on the two biological processes.
Metabolomics analyses for exploring the altered metabolites. (A) The altered metabolites (Cancer vs. Normal); (B) The altered metabolites (Cancer vs. Para-cancer); (C) The common differential metabolites. The heatmaps were created with the help of TBtools (version 1.120, https://bio.tools/tbtools).
After taking an intersection of the 10,028 OS-related genes and 4,479 PM-related, 3,197 OSPM-related genes were used in the following analyses (Fig. 3A). Then DEGs between tumor samples and normal samples were explored in GSE28735 (cohort1), GSE62452 (cohort2), and TCGA dataset combined with GTEx dataset (cohort3; Supplementary Figs. 1–3). In total, 72 intersected OSPM-related DEGs were obtained (Supplementary Table 1; Fig. 3B). Univariate Cox regression analysis was conducted, and 22 prognostic OSPM-related DEGs were selected (Fig. 3C). KM survival analysis was performed for further demonstration of the prognostic value of these DEGs. In total, 21 prognostic OSPM-related DEGs were screened as candidate genes (Fig. 3D).
Identification of key OSPM-related genes in PC. (A) The intersection of OS-related genes and PM-related genes; (B) The common OSPM-related differentially expressed genes (DEGs); (C,D) Univariate Cox regression analysis and Kaplan–Meier survival analysis for demonstrating the prognostic value of OSPM-related DEGs.
The role of OSPM in pan-cancer
Due to the significant role of the 21 prognostic OSPM-related DEGs, they were viewed as the functional genes in the OSPM progress. From a pan-cancer perspective, the general summarization of the 21 OSPM function genes was made. First, the mRNA levels of these genes in tumor samples were compared with those in normal samples. MELK, ANLN, and TOP2A were markedly up-regulated in almost all types of tumors (Fig. 4A). Then the prognostic performances of the 21 genes were explored and exhibited in the heatmap. Genes with hazard ratio (HR) > 1 and p < 0.05 were risky genes, while those with HR < 1 and p < 0.05 were protective genes (Fig. 4B). The CNV and SNV characteristics of 21 genes in pan-cancer were displayed in Fig. 4C–E. Among all types of tumor samples, LUAD, LUSC, SKCM, STAD, and UCEC patients exhibited the most prominent SNV phenomena (Fig. 4D). Additionally, among the 21 genes, CTNND2 showed the highest mutation rate in these cancers, and missense mutation was the most common mutation type (Fig. 4E). Promoter methylation levels revealed that CTNND2 exhibited hypermethylation in CESE, COAD, and HNSC. Conversely, SLPI showed hypomethylation in nearly all cancers, with the exception of PRAD (Fig. 4F).
OSPM-related genes in pan-cancer. The mRNA alteration (A), the prognostic value (B), the CNV frequency (C), the SNV frequency (D), the mutation types (E), and status of promoter methylation (F) of OSPM-related genes. The heatmaps were created with the help of R language (version 4.0.3, https://www.r-project.org/).
OSPM-based cluster analysis
To further illustrate the role of OSPM in PC, the ssGSEA was performed using a score derived from the expression of 21 OSPM function genes. Based on the enrichment scores in normal samples, 930 PC samples were divided into three clusters (C1, C2, and C3). Subsequently, the enrichment scores of the OSPM function were compared. C1, C2, and C3 were the OSPM-active, OSPM-inactive, and OSPM-normal cluster (scores: C1 > C3 > C2; Fig. 5A). The heatmap presented herein showcases the expression levels of the 21 OSPM function genes (Fig. 5B). For example, SLCO1B3 exhibited a clear upregulation in C1 and downregulation in C2 and C3 (Fig. 5B). The survival probabilities in the three clusters demonstrated statistically significant differences, with C2 showing the highest survival probability, followed by C3, and then C1 (Fig. 5C). Regarding the expression of tumor suppressor genes and oncogenes, COL24A1, FAT2, CENPJ, ECT2, PXDN, CCND1, and ADAMTS12 exhibited elevated expression in C1. Conversely, KMT2C displayed increased expression specifically in C2 (Fig. 5D).The differential expressions of ICGs might also contribute to the prognosis of pancreatic cancer (PC). HAVCR2, TNFSF4, TNFRSF9, PDCD1LG2, CD80, TNFSF9, ICOS, CD70, IDO1, and PDCD1 exhibited significantly higher expression in C1 compared to C2. Conversely, HHLA2 displayed reduced expression in C1 (Fig. 6A). Furthermore, the immune response showed distinct variations among the three clusters. Following the application of various algorithms, the infiltration patterns of immune cells, which exhibited statistically significant differences, are illustrated in the heatmap presented herein(Supplementary Fig. 4). The relationship between the expression of the 21 OSPM function genes and the infiltration of immune cells was illustrated in Fig. 6B. The findings revealed that MMP9, MMP14, and COL5A2 exhibited positive correlations with nearly all types of immune cells, while TCEA3, SLPI, and MBOAT2 displayed negative correlations with these immune cells. To provide a concise overview of the immune response, Spearman's correlation analysis was conducted. The results indicated a negative correlation between the OSPM function score and the proportion of tumor-infiltrating lymphocytes (TILs) (r = − 0.30, p = 2.09e−21), T cell co-stimulation (r = − 0.26, p = 3.08e−16), pDC (r = − 0.25, p = 9.64e−15), and Mast cell (r = − 0.23, p = 7.20e−13; Fig. 6C–G).
OSPM-based cluster analysis. The discrepancies of OSPM function scores (A), OSPM function genes’ expressions (B), survival probabilities (C), and the expression of tumor suppressor genes and oncogenes in three clusters (D). The heatmaps were created with the help of R language (version 4.0.3, https://www.r-project.org/).
The OSPM-related alteration of immune status in PC. (A) Comparing the expression of ICGs in the three PC subtypes; (B) The link between OSPM-related genes and immune cells; (C) The correlation between the OSPM function score and immune components; The correlation between OSPM function score and the proportion of TIL (D), T cell co-stimulation (E), pDC (F), and Mast cell (G).
Identification of the OSPM-related signature
For correct establishment and complete validation of the OSPM-related signature, the training and test cohorts were grouped. In the training cohort, random survival forest analysis helped identify the most appropriate genes (i.e., SLC2A1, MMP14, TOP2A, MBOAT2, ANLN, ECT2, SLC22A3, and FGD6) related to survival of PC patients, and an OSPM-related signature was identified (Fig. 7A). The following multivariate Cox analysis helped identify the risk score of each sample. The computational formula of risk score is as follows. The risk score = 0.229994697234363*TOP2A + 0.124787795719623*SLC2A1 + 0.220093179394101*MMP14 + 0.0671974510904209*MBOAT2 + 0.0484008588233876*ANLN − 0.17194413420108*ECT2 + 0.0362333570976207*SLC22A3 + 0.0192255505103104*FGD6. The median risk score of training cohort is 0.002583997. Based on the median risk score, samples were grouped into high- and low-risk subgroups (Fig. 7B and C). The PCA and t-SNE demonstrated the distinction between the two subgroups (Fig. 7D and E). The survival analysis indicated that samples with high-risk scores had poor prognoses (Fig. 7F). The eight genes in the OSPM-related signature showed different expression in the two subgroups. All these genes had increased expression in the high-risk samples (Fig. 7G). Interestingly, the expression of ECT2, FGD6, MMP14, and SLC2A1 were statistically different in the patients with different tumor stage, which implied that ECT2, FGD6, MMP14, and SLC2A1 might be associated with the development of PC (Supplementary Fig. 5). The AUC value of the ROC curve was 0.682 for 5-year survival in the training cohort, which implied its accurate diagnosis (Fig. 7H). The immune responses in high- and low-risk subgroups also exhibited discrepancies. As is depicted in the heatmap, the infiltration of various immune cells in the two subgroups differed from each other, which is one of the root causes of the prognosis (Supplementary Fig. 6A). Furthermore, the expression level of the ICG might be another root cause of the prognosis of PC. As depicted in Supplementary Fig. 6B, SIGLEC15, HAVCR2, PDCD1LG2, YTHDF1, CD274, CD80, TNFRSF14, TNFSF4, TNFRSF9, and CD70 showed increased expression in the high-risk subgroup.
Identification of the OSPM-related signature in the training cohort. (A) Random survival forest analysis for the selection of the most appropriate genes combination; (B) Group division of PC samples; (C) The link between risk score and survival; (D) PCA analysis; (E) t-SNE analysis; (F) The survival analysis in the high- and low-risk subpopulations; (G) Heatmap exhibiting the expression of the signature genes; (H) ROC curves for demonstrating the diagnostic accuracy of the signature. The heatmaps were created with the help of R language (version 4.0.3, https://www.r-project.org/).
To validate the performance of the OSPM-related signature, samples in the test cohort were grouped into high- and low-risk subgroups according to the median risk score of 0.002583997 (Fig. 8A and B). The analyses described above were performed again in the test cohort. Samples in high- and low-risk subgroups also showed distinct separation through PCA and t-SNE (Fig. 8C and D). High-risk subpopulations also had a poor prognosis (Fig. 8E). All the genes in the signature showed increased expression in the high-risk subgroup (Fig. 8F). The AUC value of the ROC curve in the test cohort were 0.753 for 5-year survival (Fig. 8G). Also, the immune responses in the test cohort are depicted in Supplementary Fig. 6C. The expression of ICGs showed a similar trend in the test cohort. TIGIT, HAVCR2, PDCD1LG2, TNFRSF4, TNFSF4, TNFRSF9, and CD70 were up-regulated in the high-risk subgroup in the test cohort (Supplementary Fig. 6D). All these results in the test cohort were similar to those in the training cohort, which indicated the satisfactory performance of the OSPM-related signature.
Validation of the OSPM-related signature in the test cohort. (A) Group division; (B) The link between risk score and survival; (C) PCA analysis; (D) t-SNE analysis; (E) The survival analysis in the high- and low-risk subpopulations; (F) Heatmap exhibiting the expression of the signature genes; (G) ROC curves for demonstrating the diagnostic accuracy of the signature. The heatmaps were created with the help of R language (version 4.0.3, https://www.r-project.org/).
The expression of the eight genes in the OSPM-related signature
For further investigation of the eight genes (i.e., SLC2A1, MMP14, TOP2A, MBOAT2, ANLN, ECT2, SLC22A3, and FGD6) in the OSPM-related signature, the qRT-PCR was conducted. And the results indicated that the expression levels of these genes in PC were different with that in normal samples. For ANLN, its expression was higher in CFPAC-1 while lower in BxPC-3 and Mia-Paca-2. For ECT2, its expression was lower in CFPAC-1, BxPC-3 and Mia-Paca-2. For FDG6 and MBOAT2, the expression levels were higher in CFPAC-1 while lower in Panc-1 and Mia-Paca-2. For MMP14 and SLC2A1, the expression levels were higher in BxPC-3 and CFPAC-1 while lower in Panc-1 and Mia-Paca-2. For SLC22A3, its expression was higher in BxPC-3 and CFPAC-1 while lower in Mia-Paca-2. For TOP2A, its expression was higher in CFPAC-1 while lower in BxPC-3, Panc-1 and Mia-Paca-2 (Fig. 9).
Single-cell expression level, immunohistochemistry, and immunofluorescence of model genes in PC
To investigate the tumor microenvironment, single-cell expression levels of the eight genes in the signature were explored. The expression of MBOAT2 primarily existed in the epithelium and the expression of MMP14 mainly existed in fibroblasts, while the expression of FGD6 primarily existed in the progenitor (Supplementary Fig. 7A). Regarding the protein levels of these genes, SLC2A1, MMP14, TOP2A, ANLN, and SLC22A3 exhibited higher expression in tumor samples compared to normal samples. Additionally, immunofluorescence helped determine the cell location of these proteins (Supplementary Fig. 7B).
Discussion
PC, as a pancreatic malignancy, is projected to be the second leading cause of cancer-related deaths by 203049. Owing to the lack of a reliable diagnostic method, most PC patients are diagnosed at an advanced stage50. The incidence of pancreatic cancer was reported to be around 57,600 with 47,050 deaths in the United States in 202051. Surgery is now the only curative treatment method for PC; therefore, the underlying cause and precise mechanisms of PC needed to be explored. Searching for more effective biomarkers for the diagnosis and treatment of PC patients is urgent. OS is the imbalance of the production and clearance of ROS. High levels of ROS lead to severe oxidative damage in proteins, lipids, and DNA52,53,54. Phospholipids, are especially susceptible to ROS attack55,56. It has been discovered that OS promotes lipid peroxidation and can cause cell damage57. Regulating the dysfunction of OS which is related to the dysfunction of phospholipids is supposed to be a novel approach for cancer treatment20,58,59.
Given the occurrence of the metabolism disorder in tumor cells, the metabolite alteration in PC samples was explored. After taking an intersection of the different metabolites between tumor samples and normal samples and different metabolites between tumor samples and para-carcinoma samples, the key metabolites differentially generated in PC were identified. Among all the metabolites, diacylglycerol, glycerophosphocholine, and glycerophosphoethanolamine were the main differentially-produced metabolites in PC samples. As a result, the disorder of PM was regarded as essential in the development of PC.
Due to the influence of OS on PM, the potential mechanisms of OSPM-related genes in PC were investigated. After differential expression analysis, univariate Cox regression analysis, and KM survival analysis, 21 prognostic OSPM-related DEGs of 3,197 OSPM-related genes emerged as OSPM function genes. Subsequently, ssGSEA was performed to assess the OSPM function of all samples, and then PC samples were divided into three clusters based on the ssGSEA function score of normal samples. Samples in cluster 1 (OSPM-active cluster), with relatively high ssGSEA function score, had a poor prognosis. In the in-depth exploration, the discrepancies of the expression of tumor suppressor genes and oncogenes (i.e., COL24A1, FAT2, CENPJ, ECT2, PXDN, CCND1, ADAMTS12, and KMT2C) and ICGs (HAVCR2, TNFSF4, TNFRSF9, PDCD1LG2, CD80, TNFSF9, ICOS, CD70, IDO1, PDCD1, and HHLA2) may result in different prognoses.
In addition, the 21 genes were summarized in pan-cancer. Compared with their corresponding normal samples, these genes were differentially expressed in tumor samples and were related with their prognosis. The high frequencies of CNV and SNV and the methylation level of promoters might be related to the alteration of the gene expression levels. The above findings shed new light on the in-depth research concerning OSPM in cancer.
In view of the impact of 21 prognostic OSPM-related DEGs on PC patients, an OSPM-related prognostic signature was developed utilizing the most appropriate variables selected from the 21 genes. Eight genes (i.e., SLC2A1, MMP14, TOP2A, MBOAT2, ANLN, ECT2, SLC22A3, and FGD6) were included in the signature. Interestingly, all the eight genes had increased expression levels in the high-risk samples, and the protein levels of SLC2A1, MMP14, TOP2A, ANLN, and SLC22A3 also increased in tumor samples compared with normal samples. Additionally, the expressions of ECT2, FGD6, MMP14, and SLC2A1 were related to the staging of the tumor. The potential roles of these eight genes in PC have been identified in other studies, and all these genes were demonstrated to combine with unfavorable functions in pancreatic cancer. Solute carrier family 2 member 1 (SLC2A1), known as glucose transporter 1 (GLUT1), is reported to express highly in many cancers and promote cancer growth60,61,62. It is reported to be highly enriched in active glucose metabolism subtype, which is associated with lower survival rate63. Matrix metalloproteinase 14 (MMP14) was also identified as a prognostic gene in PC64. Certainly, knock-down of MMP14 was proven to prevent the invasion of pancreatic cancer65. DNA topoisomerase 2-alpha (TOP2A), as the downstream molecule of DGCR5, could influence the sensitivity of pancreatic cancer cells to gemcitabine66. Membrane-bound O-acyltransferase domain-containing 2 (MBOAT2), correlated with decreasing infiltration of CD8 + T cells and KRAS activation, has been demonstrated as an unfavorable biomarker in pancreatic cancer67. Anillin (ANLN) promoted the progression of pancreatic cancer through inducing EZH2 up-regulation by mediating the miR-218-5p/LASP1 signaling68. Epithelial cell transforming 2 (ECT2) was regulated by Yes-associated protein 1 and mediates pancreatic cancer progression and metastasis69. Ephrin-2 was found to promote the migration and invasion of PC through targeting of miR-55770. Solute carrier family 22 member 3 (SLC22A3) showed up-regulation in pancreatic cancer when compared with non-neoplastic pancreatic tissues, and patients with high expression of SLC22A3 may benefit from nucleoside analogs71. Faciogenital dysplasia 6 (FGD6) was found to promote the proliferation, macropinocytosis, and tumor growth both in vitro and in vivo in pancreatic cancer72. Our results suggested that the expression of MBOAT2 mainly existed in the epithelium, and the expression of MMP14 primarily existed in fibroblasts, while the expression of FGD6 mainly existed in progenitor.
Based on the OSPM-related signature, PC samples were divided into high- and low-risk subgroups. Further investigation of the discrepancies between these subgroups indicated that the high-risk subgroup showed statistically different immune response compared with the low-risk subgroup. Immune system disorders in PC may result in the poor prognosis. Also, the expressions of ICGs (i.e., SLC2A1, FGD6, MMP14, MBOAT2, TOP2A, ANLN, SLC22A3, and CD70) were up-regulated in the high-risk subgroup in both the training cohort and the test cohort. All these discrepancies related the risk of PC samples were potential targets for the treatment of PC.
This study has a few limitations that need to be addressed. Our signature was developed according to the analyses of retrospective data. Although the expression levels of genes in the signature were explored at both transcriptional level and translational level according to the online databases, additional basic exploration is required for further demonstration. Additionally, large clinical cohorts are needed to confirm the accuracy of the signature.
Conclusion
In this study, metabolomics analyses uncovered alterations in PM, while transcriptomic studies revealed differential expression of OSPM-related genes in PC. Then three OSPM-based PC subtypes were identified, and an OSPM-related signature was developed, which helped to unmask the OSPM-based discrepancies of prognosis, tumor microenvironment, immune response, and the expression of immune checkpoint genes in PC. Finally, the expression levels of genes in the signature were further demonstrated. All these findings are valuable for estimating prognosis and facilitating individualized treatment in pancreatic cancer.
Data availability
The metabonomic data used in the current study can be downloaded from the supplementary materials of Liu’s study (https://doi.org/10.3389/fonc.2022.991051). The genomics and transcriptomics data used in the current study can be downloaded from the TCGA (https://portal.gdc.cancer.gov/), GEO (https://www.ncbi.nlm.nih.gov/geo/), ICGC (https://dcc.icgc.org/), ArrayExpress (https://www.ebi.ac.uk/biostudies/arrayexpress) platforms.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70(1), 7–30 (2020).
Chi, H. et al. Cuprotosis programmed-cell-death-related lncRNA signature predicts prognosis and immune landscape in PAAD patients. Cells 11(21), 3436 (2022).
Rawla, P., Sunkara, T. & Gaduputi, V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J. Oncol. 10(1), 10–27 (2019).
Conroy, T. et al. Current standards and new innovative approaches for treatment of pancreatic cancer. Eur. J. Cancer 57, 10–22 (2016).
Le, N., Sund, M. & Vinci, A. Prognostic and predictive markers in pancreatic adenocarcinoma. Dig. Liver Dis. 48(3), 223–230 (2016).
Moloney, J. N. & Cotter, T. G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 80, 50–64 (2018).
Sosa, V. et al. Oxidative stress and cancer: An overview. Ageing Res. Rev. 12(1), 376–390 (2013).
Jabs, T. Reactive oxygen intermediates as mediators of programmed cell death in plants and animals. Biochem. Pharmacol. 57(3), 231–245 (1999).
Łuczaj, W., Gęgotek, A. & Skrzydlewska, E. Antioxidants and HNE in redox homeostasis. Free Radic Biol Med 111, 87–101 (2017).
Forman, H. J. & Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 20(9), 689–709 (2021).
McGrath, J., Kane, L. E. & Maher, S. G. The influence of MicroRNA-31 on oxidative stress and radiosensitivity in pancreatic ductal adenocarcinoma. Cells 11(15), 2294 (2022).
Yu, J. H. & Kim, H. Oxidative stress and cytokines in the pathogenesis of pancreatic cancer. J. Cancer Prev. 19(2), 97–102 (2014).
Beloribi-Djefaflia, S., Vasseur, S. & Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 5(1), e189 (2016).
Hubler, M. J. & Kennedy, A. J. Role of lipids in the metabolism and activation of immune cells. J. Nutr. Biochem. 34, 1–7 (2016).
Cheng, C. et al. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. (Lond) 38(1), 27 (2018).
Iwamoto, H. et al. Cancer lipid metabolism confers antiangiogenic drug resistance. Cell Metab. 28(1), 104-117.e5 (2018).
Suetsugu, S., Kurisu, S. & Takenawa, T. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol. Rev. 94(4), 1219–1248 (2014).
Kopecka, J. et al. Phospholipids and cholesterol: Inducers of cancer multidrug resistance and therapeutic targets. Drug Resist. Update 49, 100670 (2020).
Klaunig, J. E. Oxidative stress and cancer. Curr. Pharm. Des. 24(40), 4771–4778 (2018).
Gorrini, C., Harris, I. S. & Mak, T. W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 12(12), 931–947 (2013).
Logan, K. A. et al. Synthesis of a gemcitabine-modified phospholipid and its subsequent incorporation into a single microbubble formulation loaded with paclitaxel for the treatment of pancreatic cancer using ultrasound-targeted microbubble destruction. Eur. J. Pharm. Biopharm. 165, 374–382 (2021).
Entrialgo-Cadierno, R. et al. The phospholipid transporter PITPNC1 links KRAS to MYC to prevent autophagy in lung and pancreatic cancer. Mol. Cancer 22(1), 86 (2023).
Yuan, Q. et al. Integration of transcriptomics, proteomics, and metabolomics data to reveal HER2-associated metabolic heterogeneity in gastric cancer with response to immunotherapy and neoadjuvant chemotherapy. Front. Immunol. 13, 951137 (2022).
Chi, H. et al. Machine learning to construct sphingolipid metabolism genes signature to characterize the immune landscape and prognosis of patients with uveal melanoma. Front. Endocrinol. (Lausanne) 13, 1056310 (2022).
Chi, H. et al. Circadian rhythm-related genes index: A predictor for HNSCC prognosis, immunotherapy efficacy, and chemosensitivity. Front. Immunol. 14, 1091218 (2023).
Jin, W. et al. Ensemble deep learning enhanced with self-attention for predicting immunotherapeutic responses to cancers. Front Immunol 13, 1025330 (2022).
Peng, G. et al. Identification and validation of neurotrophic factor-related genes signature in HNSCC to predict survival and immune landscapes. Front. Genet. 13, 1010044 (2022).
Liu, C. et al. Tissue metabolomics identified new biomarkers for the diagnosis and prognosis prediction of pancreatic cancer. Front. Oncol. 12, 991051 (2022).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1(6), 417–425 (2015).
Stelzer, G. et al. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 54, 1.30.1-1.30.33 (2016).
Liu, Z. et al. Systematic analysis of the aberrances and functional implications of ferroptosis in cancer. iScience 23(7), 101302 (2020).
Che, X. et al. Analysis of genomes and transcriptomes of clear cell renal cell carcinomas identifies mutations and gene expression changes in the TGF-beta pathway. Front. Genet. 13, 953322 (2022).
Miao, Y. et al. Machine learning identification of cuproptosis and necroptosis-associated molecular subtypes to aid in prognosis assessment and immunotherapy response prediction in low-grade glioma. Front. Genet. 13, 951239 (2022).
Qi, X. et al. Potential application of pyroptosis in kidney renal clear cell carcinoma immunotherapy and targeted therapy. Front. Pharmacol. 13, 918647 (2022).
Cui, Y. et al. Determination and characterization of molecular heterogeneity and precision medicine strategies of patients with pancreatic cancer and pancreatic neuroendocrine tumor based on oxidative stress and mitochondrial dysfunction-related genes. Front. Endocrinol. (Lausanne) 14, 1127441 (2023).
Dong, Y. et al. Identification and characterization of a novel molecular classification incorporating oxidative stress and metabolism-related genes for stomach adenocarcinoma in the framework of predictive, preventive, and personalized medicine. Front. Endocrinol. (Lausanne) 14, 1090906 (2023).
Chen, X. et al. Comprehensive characterization of extracellular matrix-related genes in PAAD identified a novel prognostic panel related to clinical outcomes and immune microenvironment: A silico analysis with in vivo and vitro validation. Front. Immunol. 13, 985911 (2022).
Wang, J. et al. Identification and verification of the ferroptosis- and pyroptosis-associated prognostic signature for low-grade glioma. Bosn. J. Basic Med. Sci. 22(5), 728–750 (2022).
Zhang, B. et al. Characterization of neuroendocrine regulation- and metabolism-associated molecular features and prognostic indicators with aid to clinical chemotherapy and immunotherapy of patients with pancreatic cancer. Front. Endocrinol. (Lausanne) 13, 1078424 (2022).
Huang, X., et al., An Aggrephagy-Related LncRNA Signature for the Prognosis of Pancreatic Adenocarcinoma. Genes (Basel), 2023. 14(1).
Pei, S. et al. Integrating single-cell RNA-seq and bulk RNA-seq to construct prognostic signatures to explore the role of glutamine metabolism in breast cancer. Front. Endocrinol. (Lausanne) 14, 1135297 (2023).
Chi, H. et al. T-cell exhaustion signatures characterize the immune landscape and predict HCC prognosis via integrating single-cell RNA-seq and bulk RNA-sequencing. Front. Immunol. 14, 1137025 (2023).
Yuan, Q., Zhang, W. & Shang, W. Identification and validation of a prognostic risk-scoring model based on sphingolipid metabolism-associated cluster in colon adenocarcinoma. Front. Endocrinol. (Lausanne) 13, 1045167 (2022).
Zhang, P. et al. Integrating multiple machine learning methods to construct glutamine metabolism-related signatures in lung adenocarcinoma. Front. Endocrinol. (Lausanne) 14, 1196372 (2023).
Tang, Z. et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45(W1), W98-w102 (2017).
Han, Y. et al. TISCH2: Expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment. Nucleic Acids Res. 51, D1425–D1431 (2022).
Uhlén, M. et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteom. 4(12), 1920–1932 (2005).
Uhlén, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347(6220), 1260419 (2015).
Rahib, L. et al. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74(11), 2913–2921 (2014).
Tesfaye, A. A. et al. The evolution into personalized therapies in pancreatic ductal adenocarcinoma: Challenges and opportunities. Expert Rev. Anticancer Ther. 18(2), 131–148 (2018).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424 (2018).
Cao, S. S. & Kaufman, R. J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal 21(3), 396–413 (2014).
Juan, C. A. et al. The chemistry of reactive oxygen species (ROS) revisited: Outlining their role in biological macromolecules (DNA, Lipids and Proteins) and induced pathologies. Int. J. Mol. Sci. 22(9), 4642 (2021).
Cervantes-Cervantes, M. P. et al. Copper increases the damage to DNA and proteins caused by reactive oxygen species. Biol. Trace Elem. Res. 103(3), 229–248 (2005).
Fang, J., Seki, T. & Maeda, H. Therapeutic strategies by modulating oxygen stress in cancer and inflammation. Adv Drug Deliv Rev 61(4), 290–302 (2009).
Paradies, G. et al. Oxidative stress, cardiolipin and mitochondrial dysfunction in nonalcoholic fatty liver disease. World J. Gastroenterol. 20(39), 14205–14218 (2014).
Cruz-Gregorio, A. et al. Lipid metabolism and oxidative stress in HPV-related cancers. Free Radic. Biol. Med. 172, 226–236 (2021).
Postovit, L. et al. Harnessing oxidative stress as an innovative target for cancer therapy. Oxid. Med. Cell. Longev. 2018, 6135739 (2018).
Avolio, R. et al. Modulation of mitochondrial metabolic reprogramming and oxidative stress to overcome chemoresistance in cancer. Biomolecules 10(1), 135 (2020).
Sun, H. W. et al. GLUT1 and ASCT2 as predictors for prognosis of hepatocellular carcinoma. PLoS ONE 11(12), e0168907 (2016).
Berlth, F. et al. Both GLUT-1 and GLUT-14 are independent prognostic factors in gastric adenocarcinoma. Ann. Surg. Oncol. 22(Suppl 3), S822–S831 (2015).
Goldman, N. A. et al. GLUT1 and GLUT8 in endometrium and endometrial adenocarcinoma. Mod. Pathol. 19(11), 1429–1436 (2006).
Lee, S. H. et al. MCT4 as a potential therapeutic target to augment gemcitabine chemosensitivity in resected pancreatic cancer. Cell Oncol. (Dordr) 44(6), 1363–1371 (2021).
Wang, H. et al. Identification of prognostic genes in the pancreatic adenocarcinoma immune microenvironment by integrated bioinformatics analysis. Cancer Immunol. Immunother. 71(7), 1757–1769 (2022).
Liang, Z. W. et al. M2-phenotype tumour-associated macrophages upregulate the expression of prognostic predictors MMP14 and INHBA in pancreatic cancer. J. Cell Mol. Med. 26(5), 1540–1555 (2022).
Liu, S. L. et al. DGCR5 is activated by PAX5 and promotes pancreatic cancer via targeting miR-3163/TOP2A and activating Wnt/β-catenin pathway. Int. J. Biol. Sci. 17(2), 498–513 (2021).
Li, Z. et al. Identification of MBOAT2 as an unfavorable biomarker correlated with KRAS activation and reduced CD8(+) T-cell infiltration in pancreatic cancer. J. Oncol. 2022, 4269733 (2022).
Wang, A. et al. ANLN-induced EZH2 upregulation promotes pancreatic cancer progression by mediating miR-218-5p/LASP1 signaling axis. J. Exp. Clin. Cancer Res. 38(1), 347 (2019).
Li, C. et al. Epithelial cell transforming 2 is regulated by Yes-associated protein 1 and mediates pancreatic cancer progression and metastasis. Am. J. Physiol. Gastrointest. Liver Physiol. 320(3), G380-g395 (2021).
Zhang, Y. et al. EFNB2 acts as the target of miR-557 to facilitate cell proliferation, migration and invasion in pancreatic ductal adenocarcinoma by bioinformatics analysis and verification. Am. J. Transl. Res. 10(11), 3514–3528 (2018).
Mohelnikova-Duchonova, B. et al. The association between the expression of solute carrier transporters and the prognosis of pancreatic cancer. Cancer Chemother. Pharmacol. 72(3), 669–682 (2013).
Zhang, Y. F. et al. A low amino acid environment promotes cell macropinocytosis through the YY1-FGD6 axis in Ras-mutant pancreatic ductal adenocarcinoma. Oncogene 41(8), 1203–1215 (2022).
Author information
Authors and Affiliations
Contributions
All authors are solely responsible for the content and writing of the manuscript. All authors made significant contributions to the design, data collection and interpretation, and manuscript preparation and revision of this study. All the authors reviewed the final manuscript and agreed to publish it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, H., Guo, H., Sun, J. et al. Multi-omics analyses based on genes associated with oxidative stress and phospholipid metabolism revealed the intrinsic molecular characteristics of pancreatic cancer. Sci Rep 13, 13564 (2023). https://doi.org/10.1038/s41598-023-40560-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-40560-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.