Abstract
The rs2736100 (A > C) polymorphism of the second intron of Telomerase reverse transcriptase (TERT) has been confirmed to be closely associated with the risk of Lung cancer (LC), but there is still no unified conclusion on the results of its association with LC. This study included Genome-wide association studies (GWAS) and case–control studies reported so far on this association between TERT rs2736100 polymorphism and LC to clarify such a correlation with LC and the differences in it between different ethnicities and different types of LC. Relevant literatures published before May 7, 2022 on ‘TERT rs2736100 polymorphism and LC susceptibility’ in PubMed, EMbase, CENTRAL, MEDLINE databases were searched through the Internet, and data were extracted. Statistical analysis of data was performed in Revman5.3 software, including drawing forest diagrams, drawing funnel diagrams and so on. Sensitivity and publication bias analysis were performed in Stata 12.0 software. The C allele of TERT rs2736100 was associated with the risk of LC (Overall population: [OR] = 1.21, 95%CI [1.17, 1.25]; Caucasians: [OR] = 1.11, 95%CI [1.06, 1.17]; Asians: [OR] = 1.26, 95%CI [1.21, 1.30]), and Asians had a higher risk of LC than Caucasians (C vs. A: Caucasians: [OR] = 1.11 /Asians: [OR]) = 1.26). The other gene models also showed similar results. The results of stratified analysis of LC patients showed that the C allele was associated with the risk of Non-small-cell lung carcinoma (NSCLC) and Lung adenocarcinoma (LUAD), and the risk of NSCLC and LUAD in Asians was higher than that in Caucasians. The C allele was associated with the risk of Lung squamous cell carcinoma (LUSC) and Small cell lung carcinoma(SCLC) in Asians but not in Caucasians. NSCLC patients ([OR] = 1.27) had a stronger correlation than SCLC patients ([OR] = 1.03), and LUAD patients ([OR] = 1.32) had a stronger correlation than LUSC patients ([OR] = 1.09).In addition, the C allele of TERT rs2736100 was associated with the risk of LC, NSCLC and LUAD in both smoking groups and non-smoking groups, and the risk of LC in non-smokers of different ethnic groups was higher than that in smokers. In the Asians, non-smoking women were more at risk of developing LUAD. The C allele of TERT rs2736100 is a risk factor for LC, NSCLC, and LUAD in different ethnic groups, and the Asian population is at a more common risk. The C allele is a risk factor for LUSC and SCLC in Asians but not in Caucasians. And smoking is not the most critical factor that causes variation in TERT rs2736100 to increase the risk of most LC (NSCLC, LUAD). Therefore, LC is a multi-etiological disease caused by a combination of genetic, environmental and lifestyle factors.
Similar content being viewed by others
Introduction
Lung cancer (LC) is one of the cancers with a high mortality rate in the world, accounting for approximately one quarter of all cancer deaths1. And smoking is currently considered to be a major risk factor for it2. In addition, exposure to environmental factors such as radon, secondhand smoke and dust, asbestos, cooking fumes and air pollution are also the main causes of LC in non-smokers3,4,5,6. However, it’s not only the environmental factors but also genetic differences that contribute to LC susceptibility. Over the past two decades, multi-population Genome-wide association studies(GWAS) have identified dozens of risk loci for LC7,8, and most of these loci are concentrated in 5p15.33 (Telomerase reverse transcriptase—Cleft lip and cleft palate transmembrane protein 1)TERT-CLPTM1L region9,10,11,12. Several precise localization studies in the following years have also identified some new LC risk loci in this region13,14,15. Telomeres are consisted of repeated "TTAGGG" at the ends of chromosomes that gradually shorten in length with each round of cell division until cell cycle arrest is triggered, of which process is known as replicative senescence16,17,18,19. Telomeres can normally be elongated by the ribonucleoprotein telomerase to maintain the replication potential20,21. In human cancer cells, however, telomerase has been activated to escape the initial growth arrest and continue to divide22. Unlimited cell growth and proliferation following the activation of telomerase is one of the clinical cancer phenotypes23,24,25. It has been proved that long telomeres can promote the survival of cells with acquired oncogenic DNA alterations, thereby promoting tumorigenesis26,27,28. Telomerase is consisted of a catalytic protein component encoded by the TERT gene and an RNA template encoded by the Telomerase RNA component(TERC). Among them, TERT is located at the short arm 15.33 of chromosome 5 (5p15.33), which is responsible for encoding the catalytic subunit of telomerase29, regulating the expression level of telomerase, and maintaining telomere length, chromosomal stability and cell proliferation by adding "TTAGG" repeats at the end of the chromosomes30,31.
Variations of the TERT promoter are an important prerequisite for high telomerase expression to stabilize telomere length32, and this process has been observed in cancer cells23. Polymorphic genes in TERT and TERC have been reported to be associated with telomere length33,34,35, and longer telomere length contributes to increasing the risk of LC, especially for Lung adenocarcinoma (LUAD)36,37,38. In addition, the TERT gene is significantly overexpressed in LC tissues, which may also confirm the underlying mechanism of LC risk39. However, the association between LC risk and telomere length is inconclusive as telomere length varies with the histological type of LC40,41. Several single nucleotide polymorphisms (SNPs) in the TERT locus have been reported to be associated with cancer risk, and these SNPs are located in the exons or introns of TERT or its promoter42. The rs2736100 (A > C) polymorphism located in the second intron of TERT is the most common SNP in the TERT gene, and its association with cancer susceptibility, including LC, has been reported in various malignant tumors29. In TERT rs2736100, the C allele upregulates TERT expression in normal and LC tissues19 and is associated with longer telomere length35,43. Studies also have found that the increased telomere length of the C allele is associated with cancer44. Some studies have also shown an increased frequency of the C allele of TERT rs2736100 in LC patients9,45,46,47,48. These evidences imply that the C allele upregulates TERT expression, maintains and prolongs telomere length, and thus increases the risk of LC. In addition, some studies have conducted racial stratification analysis for different types of LC and proved that the influence of TERT variants in Asians is stronger than that in Caucasians45,49. These results in turn imply that the frequency of TERT rs2736100 variants varies across ethnic populations. However, there are some studies have not found the association between the C allele and LC50,51. The reasons for these different results may also be related to different ethnicities, countries, research methods, sample sizes, LC types, and linkage disequilibrium patterns. Therefore, there’s inconsistency in the results of the association of TERT rs2736100 with LC. While meta-analysis is an effective way to combine data to increase the sample size, obtain sufficient power to clarify inconsistent results in genetic association studies52.
Several meta-analyses have reported the association of the TERT rs2736100 polymorphism with LC, but these meta-analyses have some shortcomings: some meta-analyses have shown an increased frequency of the C allele of TERT rs2736100 in LC patients but they ignored the effect of different ethnic groups53,54; there are some meta-analyses of ethnic stratification of rs2736100, but most of them focused on different types of cancer, and they were not subjected to a stratified analysis of LC48; some studies have done racially stratified meta-analyses for different types of LC, however, they are outdated55. Therefore, there is still a lack of a unified conclusion on the association of TERT rs2736100 polymorphism with LC, especially the variability of this association in different ethnic populations and in different LC subtypes. This study included data from GWAS and case–control studies reporting the association of TERT rs2736100 (A > C) polymorphisms with LC up to date with the aim of clarifying its association with LC and the differences in this association between different ethnicities and different types of LC.
Data and methods
Inclusion and exclusion criteria
Inclusion criteria
① They must be GWAS or case–control studies on TERT rs2736100 A/C gene polymorphism and LC susceptibility, the language should be English, and the detection methods and means should be accurately described; ② The gene frequency data can be used to calculate the Odds ratio(OR) and 95% Confidence interval(95% CI); ③ The distribution of genotype frequency of all controls conforms to Hardy–Weinberg(HWE)56; ④ The score of Newcastle Ottawa scale(NOS)57 should be no less than 7 (≥ 7).
Exclusion criteria
① Studies without allele-related data; ② Studies of the types of reviews, meta-analyses, conference reports and case reports; ③ Studies with pedigree as the reporting object; ④ same studies have published for multiple times, only the one with the most complete data will be included, and the others will be excluded.
Outcomes
The pre-specified primary outcomes were to investigate whether TERT rs2736100 A/C polymorphism increased the risk of LC in the overall population. The secondary outcomes were to determine whether there were differences in the intensity of the association between the TERT rs2736100 A/C polymorphism and LC (including various subtypes) between different ethnic groups.
Retrieval strategy
Relevant literatures on TERT rs2736100 polymorphism and LC susceptibility in PubMed, EMbase, CENTRAL, MEDLINE databases published before May 7, 2022 were searched by theme words and keywords. The language was limited to English.
Search terms in PubMed(Table 1/Table S1 in supplemental content): "Lung cancer" OR "LC"AND "rs2736100" OR "TERT" AND "polymorphism". Manual retrieval and literature tracing methods were used at the same time to expand the search scope.
Literature screening and data extraction
Two relatively independent researchers (X–ZW and WL) completed literature searching and screening according to the inclusion criteria. They cross checked and discussed them afterwards. For the literatures with different opinions, the third party (Y–ZC) made the decision. For some literatures with incomplete data, they tried to contact the author by e-mail to obtain the complete data. Finally, data extraction was carried out for the literatures being chosen after the final decision. These data include: author, year of publication, country, ethnicity, smoking status of subjects, type of LC, number of cases in case and control groups, frequency of each genotype in case and control groups, and the OR and 95% CI of each genotype.
Literature quality evaluation
The quality of the included literature was evaluated in the NOS57 (X–ZW and WL), and those with a score of no less than 7 were considered as literatures with high-quality.
Statistical methods
The HWE of the genotypes of the controls was detected by Pearson's chi-square test in SPSS 22.0 software. All results were statistically counted and analyzed in Revman 5.3 software, including drawing forest plots and funnel plots. When there was no heterogeneity among all studies or among all subgroups (P > 0.1 or I2< 50%), the fixed-effects model was used for statistical analysis; otherwise, the random-effects model was used for statistical analysis. The effect size and effect value of the statistical results were presented by OR value and 95% CI. Begg's Test and Egger's Test were performed in Stata 14.0 software to assess publication bias among studies, and sensitivity analysis was performed to assess the results of statistical analysis with greater heterogeneity. TSA 0.9.5.10 software was performed for the Trial sequential analysis(TSA) tests to evaluate the stability of the conclusion ((Type I error) probability = 5%, statistical test power = 80%, relative risk reduction = 20%).
Ethics and dissemination
This review does not require ethical approval because the included studies are published data and do not involve the patients’ privacy. The results of this review will be reported in accordance with the PRISMA extension statement and disseminated to a peer-reviewed journal.
Results
Characteristic of eligible studies
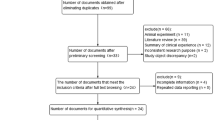
A total of 398 literatures were initially retrieved from the 4 databases and 43 studies in 40 literatures were finally included after the screening10,12,14,39,45,46,47,50,51,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88 , of which there were 25 GWAS in 22 literatures10,12,14,45,47,50,58,59,60,61,63,64,65,68,69,74,76,77,79,80,87,88. And a flow chart was made according to the PRISMA statement (Fig. 1). Among these studies, 12 in Caucasians and 31 in Asians were included. There were 99,941 LC patients (including 36,943 Caucasian patients and 62,998 Asian patients) and 131,856 controls (Tables 2, 3). All 43 studies had high NOS57 assessment scores (≥ 7), indicating that they are all at low risk of bias (Table 4).
Quantitative analysis
LC
The allelic model (C vs. A) was used to evaluate the association of TERT 2736100 with LC susceptibility. The random effects model was used for analysis as the test results showed that there was heterogeneity after the heterogeneity test (Overall population: P < 0.00001, I2= 83%; Caucasians: P < 0.0001, I2 = 73%; Asians: P < 0.00001, I2 = 74%) (Fig. 2a, Table 5). It was found that the C allele was associated with the risk of LC (Overall population: [OR] = 1.21, 95%CI [1.17, 1.25]; Caucasians: [OR] = 1.11, 95%CI [1.06, 1.17]; Asians: [OR] = 1.26, 95%CI [1.21, 1.30]), and Asians had a higher risk of LC than Caucasians (C vs. A: Caucasians: [OR] = 1.11 /Asians: [OR] = 1.26) (Fig. 2a, Table 5). The additive, heterozygous, dominant and recessive genetic models (CC vs. AA, CA vs. AA, CA + CC vs. AA and CC vs. AA + CA) were further used to evaluate the correlation between TERT 2736100 and LC since 29 of the 43 studies reported complete genotype frequencies. And the fixed-effects model (P > 0.1 or I2 < 50%) and random-effects model (P < 0.1 or I2 > 50%) were used to analyze each subgroup due to the different heterogeneity of each subgroup. Meta-analysis showed that people with "C" genotype had higher risks of LC than those with "A" genotype (P < 0.00001), and Asians had higher risks of LC than Caucasians (CC vs. AA: Caucasians: [OR] = 1.33/Asians: [OR] = 1.60; CA vs. AA: Caucasians: [OR] = 1.17/Asians: [OR] = 1.26; CA + CC vs. AA: Caucasians: [OR] = 1.22/Asians: [OR] = 1.34; CC vs. AA + CA: Caucasians: [OR] = 1.19/Asians: [OR] = 1.41) (Fig. 2b–e, Table 5). It’s also found that carriers of the CC genotype ([OR] = 1.56) were more likely to develop LC than carriers of the CA genotype ([OR] = 1.25) (Table 5).
Forest plots of LC. (a) Forest plot of the allele genetic model (C vs. A) (Random). (b) Forest plot of the additive genetic model (CC vs. AA) (Random). (c) Forest plot of the heterozygous genetic model (CA vs. AA) (Fixed). (d) Forest plot of the dominant genetic model (CA + CC vs. AA) (Random). (e) Forest plot of the recessive genetic model (CC vs. AA + CA) (Fixed).
LC subtypes
A further stratified analysis of these LC studies was performed since there were four different disease types in LC studies: Non-small-cell lung carcinoma(NSCLC, N = 21), Small cell lung carcinoma (SCLC, N = 7), Lung adenocarcinoma(LUAD, N = 17) and Lung squamous cell carcinoma(LUSC, N = 13). Meta-analysis of the allele model (C vs. A) found that the C allele was associated with the risk of NSCLC (Overall population: [OR] = 1.27, 95%CI [1.22, 1.33]; Caucasians: [OR] = 1.19, 95%CI [1.09, 1.31]; Asians: [OR] = 1.28, 95%CI [1.22, 1.34]), and Asians had a higher risk of NSCLC than Caucasians (C vs. A: Caucasians: [OR] = 1.19/Asians: [OR] = 1.28) (Fig. S1 in supplemental content, Table 6). In SCLC patients, the C allele was associated with the risk of SCLC only in Asians (Overall population: [OR] = 1.03, 95%CI [0.98, 1.09]; Caucasians: [OR] = 1.00, 95%CI [0.94, 1.06]; Asians: [OR] = 1.11, 95%CI [1.01, 1.22]) (Fig. S1 in supplemental content, Table 6). In LUAD patients, the C allele was associated with the risk of developing LUAD (Overall population: [OR] = 1.32, 95%CI [1.26, 1.38]; Caucasians: [OR] = 1.22, 95%CI [1.16, 1.28]; Asians: [OR] = 1.34, 95%CI [1.27, 1.41]), and Asians had a higher risk of developing LUAD than Caucasians (C vs. A: Caucasians: [OR] = 1.22/Asians: [OR] = 1.34) (Fig. S2 in supplemental content, Table 6). In LUSC patients, the C allele was associated with LUSC risk in Asians but not in Caucasians (Overall population: [OR] = 1.09, 95%CI [1.06, 1.13]; Caucasians: [OR] = 1.04, 95%CI [0.99, 1.10]; Asians: [OR] = 1.13, 95%CI [1.08, 1.18]) (Fig. S2 in supplemental content, Table 6). It’s also found that NSCLC patients ( [OR] = 1.27) had a stronger disease association than SCLC patients ( [OR] = 1.03) when the OR values were compared (Fig. S1 in supplemental content, Table 6), and LUAD patients ([OR] = 1.32) had a stronger disease association than LUSC patients ([OR] = 1.09) (Fig. S2 in supplemental content, Table 6).
Analysis of smoking status in LC patients
Among the included studies, 25 reported smoking or non-smoking in LC patients, of which 9 reported smoking history in LC patients and 16 reported no smoking history in LC patients. Therefore, a stratified analysis of smoking in LC patients in these 25 studies was conducted to clarify whether smoking caused variation in TERT rs2736100 and increased the risk of LC. Meta-analysis of the allele model (C vs. A) found that the C allele was associated with the risk of LC in both the smoking group and the non-smoking group (Smoking: [OR] = 1.16, 95%CI [1.09, 1.23]; Non-smoking: [OR] = 1.34, 95%CI [1.26, 1.41]), and the risk of LC in the non-smoking group was higher than that in the smoking group (C vs. A: Smoking: [OR] = 1.16/Non-smoking: [OR] = 1.34), and it was also found that non-smokers had the highest risk of LC in Asians ([OR] = 1.36, 95%CI [1.27, 1.46]) (Fig. S3 in supplemental content, Table 7).
A further stratified analysis of the smoking status of patients with different types of LC was performed due to the presence of different types of LC in the included studies. For NSCLC, the TERT polymorphism (C vs. A) was associated with the risk of NSCLC in both smoking group and non-smoking group (Smoking: [OR] = 1.20, 95%CI [1.05, 1.36]; Non-smoking: [OR] = 1.33, 95%CI [1.18, 1.50]), and the non-smoking group had a higher risk of NSCLC than the smoking group (C vs. A: Smoking: [OR] = 1.20/Non-smoking: [OR] = 1.33), and it’s also found that non-smokers had the highest risk of NSCLC in Asians ( [OR] = 1.35, 95%CI [1.17, 1.55]) (Fig. S4 in supplemental content, Table 7). For LUAD, the TERT polymorphism (C vs. A) was associated with the risk of LUAD in both the smoking group and the non-smoking group (Smoking: [OR] = 1.26, 95%CI [1.16, 1.37]; Non-smoking: [OR] = 1.37, 95%CI [1.20, 1.56]), and the risk of developing LUAD in the non-smoking group was higher than that in the smoking group (C vs. A: Smoking: [OR] = 1.26/Non-smoking: [OR] = 1.37). In addition, the risk of LUAD was found to be the highest among non-smokers in Caucasians ( [OR] = 1.40, 95%CI [1.17, 1.68]) (Fig. S5 in supplemental content, Table 7). For LUSC and SCLC, TERT polymorphisms (C vs. A) were not associated with the risk of LUSC and SCLC in both smoking group and non-smoking group in all populations (P > 0.05) (Table 7).
Sensitivity analysis
For LC, the sensitivity analysis results of the allele, additive, heterozygous, dominant and recessive genetic models (C vs. A, CC vs. AA, CA vs. AA, CA + CC vs. AA and CC vs. AA + CA) showed that none of the studies had significant sensitivity, indicating that there’s no significant difference in the result of the meta-analysis after removing any study (Fig. S6, Tables S2–S6 in supplemental content). For NSCLC, SCLC, LUAD and LUSC, the sensitivity analysis of the allele model (C vs. A) also showed no significant sensitivity (Fig. S7, Tables S7, S8 in supplemental content).
Heterogeneity analysis
For LC, there was some heterogeneity in the overall population analysis results for the allele, additive, heterozygous, dominant and recessive genetic models (C vs. A, CC vs. AA, CA vs. AA, CA + CC vs. AA and CC vs. AA + CA) (P < 0.1 or I2> 50%), and this heterogeneity mainly exists in Asians (Table 5). In the stratified analysis, the allele model (C vs. A) of NSCLC and LUAD analysis results in the overall population also showed a certain degree of heterogeneity (P < 0.1 or I2> 50%), and this heterogeneity mainly existed in Asians (Table 6).
Publication bias
For LC, the funnel plots of the allele, additive, heterozygous, dominant and recessive genetic models (C vs. A, CC vs. AA, CA vs. AA, CA + CC vs. AA and CC vs. AA + CA) were all roughly symmetrical, suggesting there’s no apparent bias (Fig. S8 in supplemental content). In terms of NSCLC, SCLC, LUAD and LUSC, the funnel plots of the allele model (C vs. A) were all roughly symmetrical (Fig. S9 in supplemental content). Additionally, the results of publication bias for all genetic models suggested that there were no obvious biases (PBegg > 0.05, PEgger > 0.05) (Tables 5, 6/Figs. S10–S12 in supplemental content).
Trial sequential analysis (TSA)
For LC, TSA analysis of the allele, additive, heterozygous, dominant and recessive genetic models (C vs. A, CC vs. AA, CA vs. AA, CA + CC vs. AA and CC vs. AA + CA) showed Z-curve (blue line) crossed both the traditional boundary (green dashed line) and the TSA boundary (red line) (Figs. S13–S17 in supplemental content). In terms of NSCLC, SCLC, LUAD and LUSC, TSA analysis of the allele model (C vs. A) in the overall and Asian populations also showed the same results (Figs. S18–S21 in supplemental content). Similar results were found in TSA analysis of the allele model (C vs. A) for patients with LC, NSCLC, and LUAD in terms of smoking status (Figs. S22–S24 in supplemental content). These results showed the overall stability and credibility of the results of this meta-analysis. The TSA results of NSCLC, SCLC, LUAD and LUSC in Caucasians cannot be comprehensively analyzed due to the reasons such as small sample size or the absence of complete gene frequencies in some of the original data reported in the literature. In addition, TSA results for smoking status in SCLC and LUSC couldn’t be comprehensively analyzed because of these reasons as well.
Summary of all the results
Due to the large amount of data in this study, a summative forest plot of all the results was created to show the statistical results more visually and more clearly, see Fig. 3.
Discussion
Current studies have reported that gene polymorphisms in TERT and TERC are associated with telomere length33,34,35, and longer telomeres length contributes to an increased risk of LC36,37,38. The increased telomere length of the C allele of the rs2736100 (A > C) polymorphism in the second intron of TERT is related to cancer44. A number of research reports have also reported that the frequency of the C allele of TERT rs2736100 increases in patients with LC9,45,46,47,48. It’s showed that the C allele can upregulate the expression of TERT, maintain and prolong telomere length, thereby increasing the risk of LC. However, due to the existence of factors such as ethnic differences, different types of LC, environmental pollution and smoking, the association between TERT rs2736100 polymorphism and LC still lacks a unified conclusion. This study included the data of GWAS and case–control studies on the association of rs2736100 polymorphism with LC that have been reported so far to clarify the association between this polymorphism and LC and the differences in the association between different ethnic groups and different types of LC.
43 studies (including 99,941 LC patients and 131,856 healthy controls) were included in this meta-analysis. The association of TERT polymorphisms with LC susceptibility was first evaluated by using the allele, additive, heterozygous, dominant and recessive genetic models (C vs. A, CC vs. AA, CA vs. AA, CA + CC vs. AA and CC vs. AA + CA). And the results showed that the C allele and "C" genotype were associated with the risk of LC comparing with the A allele and "A" genotype in the overall population. These results are consistent with those of previous GWAS studies10,12,14,45,47,50,59,61,64,65,68,69,74,76,77,79,80,87,88. It indicates that people with C allele are more likely to suffer from LC, and C allele and "C" genotype are the risk factors for LC, and the C allele increases the risk of LC by extending telomere length. However, there are some GWAS that haven’t found the association between the C allele and LC58,59,60,63. The reasons for these different results may also be related to different ethnicities, countries, research methods, sample sizes, LC types, and linkage disequilibrium patterns. Previous studies also reported that the impact of TERT variation in Asians was stronger than that in Caucasians45,55. Another study showed that rs2735947 was the most significant SNP in the Caucasians rather than rs273610049. Our findings also confirmed that the C allele and "C" genotype frequencies were indeed higher in Asians than in Caucasians, suggesting that Asians may have longer telomeres that leads to an increased risk of LC.
Since telomere length can vary with the histological type of LC40,41, different types of LC may have different degrees of association with TERT gene polymorphism due to their different pathological types. Therefore, a stratified analysis of the included LC studies was performed. Previous studies have found that longer telomere length contributes to increase the risk of LC, especially for NSCLC and LUAD36,37,38, and the C allele can increase the risk of NSCLC65,83. The results of our study also suggested that the C allele was associated with the risk of NSCLC. It indicates that the population carrying the C allele are more susceptible to NSCLC due to telomere lengthening. And it’s also found in our study that Asians had a higher risk of NSCLC than Caucasians, proving that Asians may have longer telomeres, which contribute to an increased risk of NSCLC. Some studies47,69 found that TERT rs2736100 wasn’t associated with the risk of SCLC in Caucasians, but Hu et al10 found that TERT rs2736100 could increase the risk of developing SCLC in Asians. Our results showed that the C allele was only associated with the risk of SCLC in Asians. It suggests that the C allele is a risk factor for SCLC in Asians but not in Caucasians, and the reason may be strongly related to the fact that Asian populations may have longer telomeres. When the OR values of NSCLC and SCLC were compared, it was found that NSCLC patients had a stronger disease association than SCLC patients. A previous study92 identified a locus on chromosome 5p15.33 that was significantly associated with the risk of LUAD in NSCLC, but not with other major histological types. Another study found that TERT s2736098 was significantly associated with an increased risk of SCLC in the Chinese population instead of rs273610089. These findings, combined with our results, suggested that Asian populations and NSCLC patients may have longer telomeres, which triggered the risk of cancer, and TERT rs2736100 is of a higher value as a genetic marker for diagnosing the pathogenesis of NSCLC than SCLC.
NSCLC is the most common type of LC, and LUAD is the most prevalent subtype of NSCLC73. Previous studies90 have found rs2736100 to be a risk factor associating with increased susceptibility to LC, especially for LUAD. results of this study also showed that the C allele was associated with the risk of LUAD, confirming that the risk of developing LUAD is also strongly associated with telomere lengthening36,37,38. The results of this study also showed that Asians had a higher risk of LUAD than Caucasians, suggesting that Asians may possess longer telomeres, which contribute to an increased risk of LUAD. Some studies have found that there’s no such a risk association among LUSC patients49. Several other studies47,69 also showed that TERT rs2736100 wasn’t associated with the risk of developing LUSC in Caucasians. However, in some studies on Asians10, the C allele of TERT rs2736100 was found to increase the risk of developing LUSC. Results of this study also showed that the C allele was associated with LUSC risk in Asians but not Caucasians. It proves that the C allele is a risk factor for LUSC in Asians but not in Caucasians and the reason has a lot to do with the fact that Asian populations may has longer telomeres. It’s found that patients with LUAD had a stronger disease association than patients with LUSC. Previous studies have confirmed that rs2736100 was more associated with LUAD than with LUSC69,91, which is consistent with our findings. Similarly, there are studies92 have identified a locus on chromosome 5p15.33 that is clearly associated with the risk of LUAD but not with other major histological types. These evidences demonstrate that Asian populations and patients with LUAD may have longer telomeres, thereby triggering the risk of cancer, and TERT rs2736100 has a higher value as a genetic marker for diagnosing the pathogenesis of LUAD than LUSC.
Epidemiological surveys showed that although smoking was identified as a major environmental risk factor for LC worldwide, only a small proportion of smokers develop LC during their lifetime. In contrast, a large proportion of LC cases have no history of smoking93,94. LC in never-smokers differs from LC in smokers, and a large proportion of LC patients in never-smokers carry genetic variants in oncogenes95. Recent studies have shown that the genetic susceptibility of never-smokers to LC is associated with genetic variants with pan-cancer risk effects, and that gene-environment interactions are important in LC etiology96. Tumor suppressor genes are normally expressed in healthy cells due to key regulators of cell division, such as cyclin and cyclin-dependent kinases, as well as other cell cycle checkpoints that limit this process97. However, when oncogenes triggered by environmental factors are activated and tumor suppressor genes are turned off, the control of cell division is altered, and cancer starts from a single cell98,99. Studies have shown that multiple environmental risk factors such as smoking, heavy alcohol consumption, high intake of red meat and fat, low fiber intake , indoor and outdoor air pollution, and exposure to chemicals and radiation can contribute to genomic instability100,101,102,103,104. Genomic instability leads to nucleotide dysfunction, such as base substitution, base loss, nucleotide deletion, insertion or amplification of base pairs, which further induce DNA breaks, chromosomal remodeling or translocation. And if the damage is not fixed, it can lead to irreversible cell mutation and continuous growth105,106. In LC studies, CT and TT genotype carriers of miR-26a-1 rs7372209 and miR-16-1 rs1022960 who have been exposed to cooking fumes have a higher risk of LC than those who have not been exposed107. Another study evaluating the association between gene-radon interactions among uranium miners and LC indicated that the OR interaction effect of SNP rs6891344 and rs11747272 with chromosomes 5q23.2 was estimated to be 3.9 and 3.4, suggesting that uranium miners exposed to the radioactive gas radon are more susceptible to LC108. These evidences suggest that a variety of environmental factors other than smoking can also cause genetic variants that lead to LC. Therefore, a stratified analysis on the smoking status of LC patients included in the study was conducted to clarify whether smoking or non-smoking caused variation in TERT rs2736100 and increased the risk of LC. The results showed that the C allele was associated with the risk of LC in both smokers and non-smokers, and the risk of LC in non-smokers was higher than that in smokers. It’s been reported that rs2736100 is the most significant variation among non-smokers, while rs2736100 is less significant than rs3601944649 among smokers, which confirms that TERT variation has a stronger impact on non-smokers than on smokers45,109.A study also showed that TERT SNP was a risk factor for LC in never smokers110. Similarly, a case–control study also showed that the C allele increased the risk of LC in never smokers111. Therefore, smoking is not the most critical factor to cause variation in TERT rs2736100 and increase the risk of LC.
To further clarify this genetic difference between smokers and non-smokers, we performed a stratified analysis of different types of LC in different ethnic groups as the telomere length and the frequency of TERT gene variants were different in different ethnic groups and different histological types of LC40,41. The results of this study also showed that TERT polymorphism (C vs. A) was associated with the risk of NSCLC in both smokers and non-smokers, and the risk of NSCLC in non-smokers was higher than that in smokers. For LUAD, the same result existed: TERT polymorphism (C vs. A) was associated with the risk of LUAD in both smokers and non-smokers, and the risk of LUAD in non-smokers was higher than that in smokers. Previous studies have also found that there are non-tobacco related risk factors in the pathogenesis of NSCLC. These possible risk factors include: the exposure to cooking fume, hormones and viral infection112. Subramanian113 mentioned before that LUAD was the most common type among never smokers. Therefore, non-smokers are more likely to be at the risk of NSCLC and LUAD due to variation in TERT rs2736100 leading to telomere lengthening. It’s confirmed that smoking does cause variation in TERT rs2736100, which increases the risk of most LC (NSCLC, LUAD), however, it’s not the most critical factor. Evidence shows that82 education level, BMI, prior diagnosis of COPD, occupational exposure to pesticides, duration of smoking, exposure to a large number of cooking emissions, dietary factors (including less fish and shrimp, vegetables, soy products and nuts) and the excessive intake of meat in LC patients are all related to the development of LC. When combined with many environmental and lifestyle factors, TERT rs2736100 is still significantly associated with LC82. Therefore, LC (NSCLC, LUAD) is a multi-etiological disease caused by a combination of genetic and lifestyle factors. Comparing with different ethnic groups, it’s found that the risk of LC and NSCLC in the non-smokers was the highest in Asians. Combined with the results above, it’s proved that the Asian non-smoking populations may be more likely at the risk of LC and NSCLC due to the elevated frequency of TERT rs2736100 C allele combined with environmental factors that cause telomere lengthening. But for LUAD, non-smokers were found to have the highest risk of developing LUAD in Caucasians rather than Asians. The reason for this is still related to the small sample size of non-smokers in Caucasians, and the fact that there’s not only one pathological type of LUAD in NSCLC but also many other types such as LUSC and large cell lung cancer (LCLC), which can lead to inconsistent results in the analysis of NSCLC and LUAD. In addition, the majority of non-smoking LUAD patients included in this study are Asian females (Asian females: N = 9618/Overall: N = 12,327), indicating that non-smoking females in the Asians are more likely to have the risk of LUAD. Previous studies have also confirmed that LUAD is more common in females114,115. Patel et al. showed that among the never-smoking LC patients, the number of females exceeded that of males116. There was evidence confirmed that the common genetic variation of TERT-CLPTM1L was associated with the risk of LUAD in non-smoking Asian females45. This can be explained by the following assumptions: females are more likely to be exposed to second-hand smoking, and exposed to coal for cooking at home and hormone replacement therapy. All these reasons can lengthen telomere to avoid apoptosis and ultimately lead to cancer117.
For LUSC and SCLC, TERT polymorphisms (C vs. A) were not associated with the risk of them in all populations, both in smokers and in non-smokers. Therefore, smoking may not cause variation in TERT rs2736100 that increase the risk of LUSC and SCLC. The cause of variation in TERT rs2736100 leading to LUSC and SCLC remains to be further clarified.
Limitations of this study: ① This meta-analysis is based on the research reports of different ethnic groups and different types of LC, which will inevitably produce some heterogeneity; ② The methods of gene detection and genotyping used in all studies were different, and there will be some differences in data results; ③ In terms of sample size, this study is sufficient in general. However, after subgroup analysis according to different LC types and ethnicity, the results signify that the sample size of SCLC and LUSC is still small. This will inevitably produce some false negative results for SCLC and LUSC; ④ Although this study discussed the effects of smoking, environment, lifestyle and other factors on LC in details, from the perspective of smoking status, the sample size of smoking patients reported in these studies is still relatively small, especially those of SCLC and LUSC studies. Therefore, to some extent, the reliability of the results of the correlation between smoking and the risk of SCLC and LUSC will be affected; ⑤ All the literatures included in this study are in English, not in the other languages.
Conclusion
In conclusion, the C allele of TERT rs2736100 is a risk factor for LC, NSCLC, and LUAD in different ethnic groups, and the risk is more common in Asians. Moreover, the C allele is a risk factor for LUSC and SCLC in Asians but not in Caucasians. Among the different types of LC, NSCLC patients have stronger risk correlation than SCLC patients, and LUAD patients have a stronger disease risk correlation than LUSC patients. Asians have a more common risk of various types of LC because they may have longer telomeres than Caucasians. The C allele is correlated with the risk of LC, NSCLC and LUAD in smokers and non-smokers, and the risk of LC in non-smokers of different ethnic groups is more common than that in smokers. In the Asians, non-smoking females are more at the risk of developing LUAD. Therefore, smoking does cause variation in TERT rs2736100 and increases the risk of most LC (NSCLC, LUAD), but it’s not the most critical factor.
LC (NSCLC, LUAD) is a multi-etiological disease caused by a combination of genetic, environmental and lifestyle factors. Of course, it’s necessary to integrate and analyze the data of studies with a larger sample size to draw more reliable conclusions in the future.
Data availability
Data supporting our findings are contained within the manuscript.
Abbreviations
- LC:
-
Lung cancer
- GWAS:
-
Genome-wide association studies
- TERT:
-
Telomerase reverse transcriptase
- CLPTM1L:
-
Cleft lip and cleft palate transmembrane protein 1
- TERC:
-
Telomerase RNA component
- SNP:
-
Single nucleotide polymorphism
- HWE:
-
Hardy–Weinberg equilibrium
- NOS:
-
Newcastle Ottawa scale
- OR:
-
Odds ratio
- 95% CI:
-
95% Confidence interval
- TSA:
-
Trial sequential analysis
- SCLC:
-
Small cell lung carcinoma
- NSCLC:
-
Non-small-cell lung carcinoma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- LCLC:
-
Large cell lung cancer
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer Statistics, 2019. CA Cancer J Clin. 69, 7–34 (2019).
Moolgavkar, S. H. et al. Impact of reduced tobacco smoking on lung cancer mortality in the United States during 1975–2000. J. Natl. Cancer Inst. 104, 541–548 (2012).
Torres-Durán, M. et al. Residential radon and lung cancer characteristics in never smokers. Int. J. Radiat. Biol. 91(8), 605–610 (2015).
Gorlova, O. Y. et al. Never smokers and lung cancer risk: A case-control study of epidemiological factors. Int. J. Cancer. 118(7), 1798–1804 (2006).
Markowitz, S. B., Levin, S. M., Miller, A. & Morabia, A. Asbestos, asbestosis, smoking, and lung cancer. New findings from the North American insulator cohort. Am. J. Respir. Crit. Care Med. 188(1), 90–96 (2013).
Liao, Y., Xu, L., Lin, X. & Hao, Y. T. Temporal trend in lung cancer burden attributed to ambient fine particulate matter in Guangzhou, China. Biomed. Environ. Sci. 30(10), 708–717 (2017).
Bosse, Y. & Amos, C. I. A decade of GWAS results in lung cancer. Cancer Epidemiol. Biomark. Prevent. 27(4), 363–379 (2017).
Wang, J. et al. Genetic predisposition to lung cancer: comprehensive literature integration, meta-analysis, and multiple evidence assessment of candidate-gene association studies. Sci Rep. 21, 8371 (2017).
McKay, J. D. et al. Lung cancer susceptibility locus at 5p15.33. Nature Genet. 40(12), 1404–1406 (2008).
Hu, Z. et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nature Genet. 43(8), 792–796 (2011).
Walsh, K. M. et al. Fine-mapping of the 5p15.33, 6p22.1-p21.31, and 15q25.1 regions identifies functional and histology-specific lung cancer susceptibility loci in African-Americans. Cancer Epidemiol. Biomark. Prevent. 22(2), 251–260 (2013).
Hosgood, H. D. 3rd. et al. Interactions between household air pollution and GWAS-identified lung cancer susceptibility markers in the Female Lung Cancer Consortium in Asia (FLCCA). Hum Genet. 134(3), 333–341 (2015).
Wang, Z. et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum. Mol. Genet. 23(24), 6616–6633 (2014).
Dong, J. et al. Fine mapping of chromosome 5p15.33 identifies novel lung cancer susceptibility loci in Han Chinese. Int. J. Cancer 141(3), 447–456 (2017).
Kachuri, L. et al. Fine mapping of chromosome 5p15.33 based on a targeted deep sequencing and high density genotyping identifies novel lung cancer susceptibility loci. Carcinogenesis 37(1), 96–105 (2016).
Autexier, C. & Lue, N. F. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75, 493–517 (2006).
de Lange, T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes. Dev. 19, 2100–2110 (2005).
Diotti, R. & Loayza, D. Shelterin complex and associated factors at human telomeres. Nucleus 2, 119–135 (2011).
Mocellin, S. et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: A feld synopsis and metaanalysis. J. Natl. Cancer Inst. 104(11), 840–854 (2012).
Liu, L., Lai, S., Andrews, L. G. & Tollefsbol, T. O. Genetic and epigenetic modulation of telomerase activity in development and disease. Gene 340, 1–10 (2004).
Wyatt, H. D. M., West, S. C. & Beattie, T. L. InTERTpreting telomerase structure and function. Nucl. Acids Res. 38, 5609–5622 (2010).
Bell, R. J. et al. Understanding TERT promoter mutations: A common path to immortality. Mol. Cancer Res. 14(4), 315–323 (2016).
Kim, N. W. et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994).
Ding, Z. et al. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 148, 896–907 (2012).
Stewart, S. A. & Weinberg, R. A. Telomerase and human tumorigenesis. Semin. Cancer Biol. 10(6), 399–406 (2000).
Shen, M. et al. A prospective study of telomere length measured by monochrome multiplex quantitative PCR and risk of lung cancer. Lung Cancer 73, 133–137 (2011).
Bull, C. F. et al. Folate deficiency induces dysfunctional long and short telomeres; both states are associated with hypomethylation and DNA damage in human WIL2-NS cells. Cancer Prev. Res. 7, 128–138 (2014).
Halaschek-Wiener, J. et al. Reduced telomere length variation in healthy oldest old. Mech. Ageing Dev. 129, 638–641 (2008).
Rafnar, T. et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 41(2), 221–227 (2009).
Liu, Y. et al. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. CB 10(22), 1459–1462 (2000).
Oh, H. et al. Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. Proc. Natl. Acad. Sci. U. S. A. 98(18), 10308–10313 (2001).
Boukamp, P., Popp, S. & Krunic, D. Telomere-dependent chromosomal instability. J. Investig. Dermatol. Symp. Proc. 10(2), 89–94 (2005).
Codd, V. et al. Common variants near TERC are associated with mean telomere length. Nat. Genet. 42, 197–199 (2010).
Soerensen, M. Genetic variation and human longevity. Dan. Med. J. 2012, 59 (2012).
Codd, V. et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 45, 422–427 (2013).
Seow, W. J. et al. Telomere length in white blood cell DNA and lung cancer: A pooled analysis of three prospective cohorts. Can. Res. 74(15), 4090–4098 (2014).
Yuan, J. M. et al. Leukocyte telomere length in relation to risk of lung adenocarcinoma incidence: Findings from the Singapore Chinese Health Study. Int. J. Cancer 142(11), 2234–2243 (2018).
Telomeres, C. et al. Association between telomere length and risk of cancer and non-neoplastic diseases: A mendelian randomization study. JAMA Oncol 3(5), 636–651 (2017).
Wei, R. et al. TERT polymorphism rs2736100-C Is associated with EGFR mutation-positive non-small cell lung cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 21(22), 5173–5180 (2015).
Jang, J. S. et al. Telomere length and the risk of lung cancer. Cancer Sci. 99, 1385–1389 (2008).
Sanchez-Espiridion, B. et al. Telomere length in peripheral blood leukocytes and lung cancer risk: A large case-control study in Caucasians. Cancer Res. 74, 2476–2486 (2014).
Gaspar, T. B. et al. Telomere maintenance mechanisms in cancer. Genes 9, 241 (2018).
Iles, M. M. et al. The effect on melanoma risk of genes previously associated with telomere length. J. Natl. Cancer Inst. 106, dju267 (2014).
Haycock, P. C. et al. Association between telomere length and risk of cancer and non-neoplastic diseases: A Mendelian Randomization Study. JAMA Oncol. 3, 636–651 (2017).
Hsiung, C. A. et al. The 5p1533 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 6, e1001051 (2010).
Myneni, A. A. et al. Genetic polymorphisms of TERT and CLPTM1L and risk of lung cancer–a case-control study in a Chinese population. Lung Cancer 80, 131–137 (2013).
Wang, Y., Broderick, P., Matakidou, A., Eisen, T. & Houlston, R. S. Role of 5p133 (TERT-CLPTM1L), 6p2133 and 15q251(CHRNA5-CHRNA3) variation and lung cancer risk in never-smokers. Carcinogenesis 31, 234–238 (2010).
Zhang, X., Chen, Y., Yan, D., Han, J. & Zhu, L. TERT gene rs2736100 and rs2736098 polymorphisms are associated with increased cancer risk: A meta-analysis. Biochem. Genet. 60(1), 241–266 (2022).
Li, Z. et al. Fine mapping in TERT-CLPTM1L region identified three independent lung cancer susceptibility signals: A large-scale multi-ethnic population study. Mol. Carcinog. 57(10), 1289–1299 (2018).
Chen, X. F. et al. Multiple variants of TERT and CLPTM1L constitute risk factors for lung adenocarcinoma. Genet. Mol. Res. 11(1), 370–378 (2012).
Li, X. et al. Rs2853677 modulates Snail1 binding to the TERT enhancer and affects lung adenocarcinoma susceptibility. Oncotarget 7, 37825–37838 (2016).
Pabalan, N. A. Meta-analysis in cancer genetics. Asian Pac. J. Cancer Prev. 11(1), 33–38 (2010).
Li, H. et al. The TERT rs2736100 polymorphism increases cancer risk: A meta-analysis. Oncotarget 8(24), 38693–38705 (2017).
Snetselaar, R., van Oosterhout, M. F. M., Grutters, J. C. & van Moorsel, C. H. M. Telomerase reverse transcriptase polymorphism rs2736100: a balancing act between cancer and non-cancer disease, a meta-analysis. Front. Med. 5, 41 (2018).
Yang, J. & Jiao, S. Increased lung cancer risk associated with the TERT rs2736100 polymorphism: an updated meta-analysis. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 35(6), 5763–5769 (2014).
Wang, J. & Shete, S. Testing departure from Hardy-Weinberg proportions. Methods Mol. Biol. 1666, 83–115 (2017).
Higgins, J. P. & Green, S. Cochrane Handbook for Systematic Reviews of Interventions (John Wiley & Sons, Hoboken, 2011).
Bae, E. Y. et al. Replication of results of genome-wide association studies on lung cancer susceptibility loci in a Korean population. Respirology 17(4), 699–706 (2012).
Brenner, D. R. et al. Hierarchical modeling identifies novel lung cancer susceptibility variants in inflammation pathways among 10,140 cases and 11,012 controls. Hum Genet. 132(5), 579–589 (2013).
Broderick, P. et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 69(16), 6633–6641 (2009).
Cheng, Y. et al. Risk assessment models for genetic risk predictors of lung cancer using two-stage replication for Asian and European populations. Oncotarget 8(33), 53959–53967 (2016).
Furuie, H., Arimura-Omori, M., Hamada, N., Yanagihara, T. & Kiyohara, C. The association of aging-related polymorphisms with susceptibility to lung cancer: A case-control study in a Japanese population. Asian Pac. J. Cancer Prev. 22(4), 1279–1285 (2021).
Ito, H. et al. Association between a genome-wide association study-identified locus and the risk of lung cancer in Japanese population. J. Thorac. Oncol. 7(5), 790–798 (2012).
Jaworowska, E. et al. Smoking related cancers and loci at chromosomes 15q25, 5p15, 6p22.1 and 6p21.33 in the Polish population. PLoS ONE 6(9), e25057 (2011).
Jin, G. et al. Common genetic variants on 5p15.33 contribute to risk of lung adenocarcinoma in a Chinese population. Carcinogenesis 30(6), 987–990 (2009).
Kohno, T. et al. Contribution of the TP53, OGG1, CHRNA3, and HLA-DQA1 genes to the risk for lung squamous cell carcinoma. J. Thorac. Oncol. 6(4), 813–817 (2011).
Lan, Q. et al. Longer telomere length in peripheral white blood cells is associated with risk of lung cancer and the rs2736100 (CLPTM1L-TERT) polymorphism in a prospective cohort study among women in China. PLoS ONE 8(3), e59230 (2013).
Lan, Q. et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat. Genet. 44(12), 1330–1335 (2012).
Landi, M. T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma [published correction appears in Am J Hum Genet. 2011 Jun 10;88(6):861]. Am. J. Hum. Genet. 85(5), 679–691 (2009).
Li, H. et al. Prediction of lung cancer risk in a Chinese population using a multifactorial genetic model. BMC Med. Genet. 13, 118 (2012).
Liu, S. G. et al. Association of genetic polymorphisms in TERT-CLPTM1L with lung cancer in a Chinese population. Genet. Mol. Res. 14(2), 4469–4476 (2015).
Machiela, M. J. et al. Genetic variants associated with longer telomere length are associated with increased lung cancer risk among never-smoking women in Asia: A report from the female lung cancer consortium in Asia. Int. J. Cancer. 137(2), 311–319 (2015).
Mandour, I., Hussein, S. A. M., Essam, R. & El-Hossainy, M. A. Study of genetic variants in chromosome 5p15.33 region in non-smoker lung cancer patients. Adv. Respir. Med. 88(6), 485–494 (2020).
McKay, J. D. et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 40(12), 1404–1406 (2008).
Miki, D. et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat. Genet. 42(10), 893–896 (2010).
Pande, M. et al. Novel genetic variants in the chromosome 5p15.33 region associate with lung cancer risk. Carcinogenesis 32(10), 1493–1499 (2011).
Seow, W. J. et al. Association between GWAS-identified lung adenocarcinoma susceptibility loci and EGFR mutations in never-smoking Asian women, and comparison with findings from Western populations. Hum. Mol. Genet. 26(2), 454–465 (2017).
Shiraishi, K. et al. Association of variations in HLA class II and other loci with susceptibility to EGFR-mutated lung adenocarcinoma. Nat. Commun. 7, 12451 (2016).
Shiraishi, K. et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat. Genet. 44(8), 900–903 (2012).
Truong, T. et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J. Natl. Cancer Inst. 102(13), 959–971 (2010).
Wang, F. et al. TERT rs2736100T/G polymorphism upregulates interleukin 6 expression in non-small cell lung cancer especially in adenocarcinoma. Tumour Biol. 35(5), 4667–4672 (2014).
Wang, X. et al. Combining telomerase reverse transcriptase genetic variant rs2736100 with epidemiologic factors in the prediction of lung cancer susceptibility. J. Cancer. 7(7), 846–853 (2016).
Xing, Y. L. et al. Case-control study on impact of the telomerase reverse transcriptase gene polymorphism and additional single nucleotide polymorphism (SNP)- SNP interaction on non-small cell lung cancers risk in Chinese Han population. J. Clin. Lab. Anal. 30(6), 1071–1077 (2016).
Yang, P. et al. A rigorous and comprehensive validation: Common genetic variations and lung cancer. Cancer Epidemiol. Biomark. Prev. 19(1), 240–244 (2010).
Yin, Z. et al. Genetic polymorphisms of TERT and CLPTM1L, cooking oil fume exposure, and risk of lung cancer: A case-control study in a Chinese non-smoking female population. Med. Oncol. 31(8), 114 (2014).
Yoo, S. S. et al. The effect of susceptibility variants, identified in never-smoking female lung cancer cases, on male smokers. Korean J. Intern. Med. 35(4), 929–935 (2020).
Yoon, K. A. et al. A genome-wide association study reveals susceptibility variants for non-small cell lung cancer in the Korean population. Hum. Mol. Genet. 19(24), 4948–4954 (2010).
Zhao, Z. et al. Significant association of 5p15.33 (TERT-CLPTM1L genes) with lung cancer in Chinese Han population. Exp. Lung Res. 39(2), 91–98 (2013).
Zhao, M. M. et al. Genetic variations in TERT-CLPTM1L genes and risk of lung cancer in a Chinese population. Asian Pac. J. Cancer Prev. 15, 2809–2813 (2014).
Yuan, Y. et al. Association between TERT rs2736100 polymorphism and lung cancer susceptibility: Evidence from 22 case-control studies. Tumour Biol. 35(5), 4435–4442 (2014).
Zanetti, K. A. et al. Genome-wide association study confirms lung cancer susceptibility loci on chromosomes 5p15 and 15q25 in an African-American population. Lung Cancer 98, 33–42 (2016).
Landi, M. T. et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 16(2), 430–441 (2010).
Lam, W. K., White, N. W. & Chan-Yeung, M. M. Lung cancer epidemiology and risk factors in Asia and Africa State of the Art. Int. J. Tuberc. Lung. Dis. 8, 1045–1057 (2004).
Spitz, M. R., Wei, Q., Dong, Q., Amos, C. I. & Wu, X. Genetic susceptibility to lung cancer the role of DNA damage and repair. Cancer Epidemiol. Biomark. Prev. 12, 689–698 (2003).
Lee, Y. J. et al. Lung cancer in never smokers: change of a mindset in the molecular era. Lung Cancer 72(1), 9–15 (2011).
Hung, R. J. et al. Lung cancer risk in never-smokers of european descent is associated with genetic variation in the 515.33 tert-clptm1ll region. J. Thorac. Oncol. 14(8), 1360–1369 (2019).
Mader, S. S., Windelspecht, M. & Cox, D. Essentials of Biology 15th edn. (McGraw-Hill Higher Education, New York, 2015).
Massagué, J. G1 cell-cycle control and cancer. Nature 432(7015), 298–306 (2004).
Negrini, S., Gorgoulis, V. G. & Halazonetis, T. D. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11(3), 220–228 (2010).
Kopp, T. I., Vogel, U. & Andersen, V. Associations between common polymorphisms in CYP2R1 and GC, Vitamin D intake and risk of colorectal cancer in a prospective case-cohort study in Danes. PLoS ONE 15(2), e0228635 (2020).
Bagot, R. C. & Meaney, M. J. Epigenetics and the biological basis of gene x environment interactions. J. Am. Acad. Child Adolesc. Psychiatry. 49(8), 752–771 (2010).
Dos Reis Filho, A. P., Silveira, M. A. D., Demarco, N. R. & D’Arce, L. P. G. Increased DNA damage, instability and cytokinesis defects in occupationally exposed car painters. In Vivo 33(6), 1807–1811 (2019).
Parsa, N. Environmental factors inducing human cancers. Iran J. Public Health. 41(11), 1–9 (2012).
Sankpal, U. T. et al. Environmental factors in causing human cancers: Emphasis on tumorigenesis. Tumour Biol. 33(5), 1265–1274 (2012).
Ferguson, L. R. et al. Genomic instability in human cancer: Molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Semin. Cancer Biol. 35(Suppl), S5–S24 (2015).
Langie, S. A. et al. Causes of genome instability: The effect of low dose chemical exposures in modern society. Carcinogenesis 36(Suppl 1), S61–S88 (2015).
Yin, Z. et al. Interaction between polymorphisms in Pre-MiRNA genes and cooking oil fume exposure on the risk of lung cancer in chinese non-smoking female population. PLoS ONE 10(6), e0128572 (2015).
Rosenberger, A. et al. Genetic modifiers of radon-induced lung cancer risk: A genome-wide interaction study in former uranium miners. Int. Arch. Occup. Environ. Health. 91(8), 937–950 (2018).
Timofeeva, M. N. et al. Influence of common genetic variation on lung cancer risk: Meta-analysis of 14 900 cases and 29 485 controls. Hum. Mol. Genet. 21(22), 4980–4995 (2012).
Wang, Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 40(12), 1407–1409 (2008).
Liao, Y. et al. VSIG4 expression on macrophages facilitates lung cancer development. Lab. Invest. 94(7), 706–715 (2014).
Gealy, R. et al. Comparison of mutations in the p53 and K-ras genes in lung carcinomas from smoking and nonsmoking women. Cancer Epidemiol. Biomark. Prev. 8(4 Pt 1), 297–302 (1999).
Subramanian, J. & Govindan, R. Lung cancer in never smokers: A review. J. Clin. Oncol. 25(5), 561–570 (2007).
Henschke, C. I. et al. Early lung cancer action project: Annual screening using single-slice helical CT. Ann. N. Y. Acad. Sci. 952, 124–134 (2001).
Samet, J. M. et al. Lung cancer in never smokers: Clinical epidemiology and environmental risk factors. Clin. Cancer Res. 15(18), 5626–5645 (2009).
Patel, J. D., Bach, P. B. & Kris, M. G. Lung cancer in US women: a contemporary epidemic. JAMA 291(14), 1763–1768 (2004).
Subramanian, J. et al. Review of ongoing clinical trials in non-small-cell lung cancer: a status report for 2012 from the ClinicalTrials.gov Web site. J. Thorac. Oncol. 8(7), 860–865 (2013).
Funding
This work is supported by National Natural Science Foundation of China (82160861), Guizhou Provincial Basic Research Program(Natural Science) (Qiankehe Foundation-ZK[2023]General 411), Academic New Seedling Project of Guizhou University of Traditional Chinese Medicine (Guike Cooperative Academic New Seedling [2023]-22), Innovation and Entrepreneurship Training Program for College Students of Guizhou University of Traditional Chinese Medicine, China (Gui Zhong Yi Da Chuang He Zi (2022) No. 90), and Project of Education Department of Guizhou Province, China (Guizhou Education Technology 2022-023).
Author information
Authors and Affiliations
Contributions
This study is initiated by X.W.; X.W. will develop the search strategies, conduct data collection, and analyze independently. G.H., W.L. and Y.C. will revise it. All authors have approved the fifinal manuscript. Conceptualization: X.W.; Methodology: X.W., W.L.; Software: X.W.; Supervision: Y.C.; Writing – original draft: X.W.; Writing – review and editing: X.W., G.H., W.L., Y.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, X., Huang, G., Li, W. et al. Ethnicity-specific association between TERT rs2736100 (A > C) polymorphism and lung cancer risk: a comprehensive meta-analysis. Sci Rep 13, 13271 (2023). https://doi.org/10.1038/s41598-023-40504-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-40504-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.