Abstract
Zoysia grass (Zoysia spp.) is the most widely used warm-season turf grass in Korea due to its durability and resistance to environmental stresses. To develop new longer-period greenness cultivars, it is essential to screen germplasm which maintains the greenness at a lower temperature. Conventional methods are time-consuming, laborious, and subjective. Therefore, in this study, we demonstrate an objective and efficient method to screen maintaining longer greenness germplasm using RGB and multispectral images. From August to December, time-series data were acquired and we calculated green cover percentage (GCP), Normalized Difference Vegetation Index (NDVI), Normalized Difference Red Edge Index (NDRE), Soil-adjusted Vegetation Index (SAVI), and Enhanced Vegetation Index (EVI) values of germplasm from RGB and multispectral images by applying vegetation indexs. The result showed significant differences in GCP, NDVI, NDRE, SAVI, and EVI among germplasm (p < 0.05). The GCP, which evaluated the quantity of greenness by counting pixels of the green area from RGB images, exhibited maintenance of greenness over 90% for August and September but, sharply decrease from October. The study found significant differences in GCP and NDVI among germplasm. san208 exhibiting over 90% GCP and high NDVI values during 153 days. In addition, we also conducted assessments using various vegetation indexes, namely NDRE, SAVI, and EVI. san208 exhibited NDRE levels exceeding 3% throughout this period. As for SAVI, it initially started at approximately 38% and gradually decreased to around 4% over the course of these days. Furthermore, for the month of August, it recorded approximately 6%, but experienced a decline from about 9% to 1% between September and October. The complementary use of both indicators could be an efficient method for objectively assessing the greenness of turf both quantitatively and qualitatively.
Similar content being viewed by others
Introduction
Zoysia grass, belonging to the Zoysia genus, is a popular turf grass used in Korea and consists of about 16 species. It is widely distributed along the Pacific from the northeast Asian region, such as Korea and Japan, showing a temperate climate, to the Southeast Asian region, such as the Philippines and Thailand, showing a tropical climate1. It grows low due to its strong lateral and underground spread and blooms in May to June and fruits in June to July but mostly relies on vegetative propagation because of its low germination rate. Zoysia grass in Korea maintains its green color for about six months from mid-April to mid-October, but during winter, it goes dormant and turns yellow or brown2. It has strong resistance to biological and non-biological stressors such as disease, insects, drought, and wear, making it easy to manage. However, it has slower growth, slower recovery rate after damage, and delayed green-up in the spring, as disadvantages3. The Zoysia grass species that grow natively in Korea include Zoysia japonica, Z. matrella, Z. sinica, Z. macrostachya, and hybrid Zoysia grass. Among these, Z. japonica, known as Korean lawn grass, is the most widely used species in Korea due to its strong adaptability to the environment, despite its slightly rough texture and low-density formation. Z. matrella is also commonly used in Korea, and it has received high evaluations in terms of turf quality due to its soft texture caused by fine leaves and excellent sod-forming ability. Z. sinica and Z. macrostachya are not commercially used yet, but their strong salt tolerance makes them valuable breeding materials. Hybrid Zoysia grass, showing intermediate characteristics between Z. japonica and Z. matrella or Z. sinica, has been confirmed to grow in the southern region of Korea. Z. tenuifloria is being used in Korea, although its native growing area has not been confirmed2,4.
The surrounding environment of grass has a significant impact on its growth, including factors such as moisture, temperature, light, and soil. When these environmental factors are not suitable for grass growth, such as in cases of drought, heat, cold, shade, or high salt, the grass becomes stressed, which can inhibit its growth and reduce its quality5,5,7. Therefore, many researchers have studied the physiological, morphological, and metabolic responses of grass to various environmental stresses to improve its internal adaptability8. Studies have been conducted to measure various parameters using the altered responses to stress as an indicator to identify grass species with excellent internal adaptability. The phenotypic diversity of perennial ryegrass (Lolium perenne L.) cultivars collected from six different regions have been compaired by exposing them to drought stress9. To evaluate the drought response, commonly used parameters for assessing drought tolerance such as growth reduction, photosynthesis reduction, water content reduction, and electrolyte leakage increase were measured. Leaf wilting and grass height were visually assessed and measured with tools9. Chlorophyll fluorescence was measured using a fluorometer to evaluate photosynthetic efficiency. To measure leaf water content and electrolyte leakage, a series of manual processes such as weighing after removing moisture or using a pressure chamber were performed. As a result, a new approach based on image analysis has been developed as an alternative to the existing methods and has begun to attract attention among researchers10.
The concept of imaging plants goes beyond simply taking photographs of plants. It involves quantitatively measuring a plant's phenotype through the interaction between the plant and light, including photon absorption, reflection, and emission11. To execute this imaging-based phenotyping technology, not only biology, but also interdisciplinary understanding and integration of sensor technology, computer vision, mathematics, and electronic engineering are essential. This approach allows for the objective acquisition of anatomical and physiological characteristics of plants through non-contact and non-destructive methods, surpassing human limitations in accuracy, sensitivity, and processing capacity11 Büchi et al.12 conducted experiments to visually compare the canopy cover area of plant cover obtained using two image analysis tools (Assess 2.0 and Canopeo) with that obtained through visual evaluation, finding a positive relationship between the two evaluations. However, they recognized the need to control environmental constraints such as light and shadow to obtain more objective results from the acquired images and emphasized that there should be an additional process to accurately measure canopy area, as there are limitations in distinguishing plant cover and weeds using only digital image analysis tools12.
There are several vegetation indices used in remote sensing to monitor plant growth and assess vegetation health. Two widely used indices are the Normalized Difference Vegetation Index (NDVI) and the Soil Adjusted Vegetation Index (SAVI). The NDVI is a commonly used index for monitoring corn growth, managing farmland, and predicting yield. It is sensitive to variations in the growth of various crops. Osco et al.13, Santana et al.14, and Wan et al.15 have utilized NDVI in their studies related to corn growth, farmland management, and yield prediction. On the other hand, the SAVI is another popular vegetation index that provides more reliable results by minimizing the influence of soil effects. It takes into account the soil background reflectance, making it more appropriate than NDVI in certain situations. Braz et al.16 highlighted the advantages of SAVI, particularly its ability to mitigate the impact of soil variations. SAVI is often used in comparison with NDVI or as an alternative index for vegetation assessment8.
In recent years, there has been significant progress in optimizing ground-based high-throughput approaches for non-destructive estimation of spectral vegetation indices (SVIs). These SVIs, such as the normalized vegetation index (NDVI), simple ratio (SR), and chlorophyll indices (CI), serve as indicators of green biomass, leaf area index (LAI), chlorophyll levels, and photosynthesis rate. They are useful for assessing the senescence rate during crop maturation17,17,18,19,21. The advent of cost-effective, high-resolution multispectral sensors has led to the widespread use of unmanned aerial vehicles (UAVs) in crop physiology research. UAVs offer a practical approach for temporally quantifying spatial traits in large and diverse crop populations, thereby maximizing selection accuracy across various environmental factors. Multispectral sensors and RGB cameras mounted on UAVs have been employed to capture spectral imagery in different light reflectance bands (near-infrared, red, red-edge, green, and blue). These images have been used to detect biomass22, LAI23, plant density24, and photosynthetic activity25 in crops such as rapeseed, barley, and wheat. UAV-based multispectral imagery has also been utilized to estimate the rate of emergence and spring survival in wheat26.
In addition to UAVs, satellite-based vegetation data have been instrumental in crop growth monitoring11,27,28. More recently, UAV-based vegetation indices such as the enhanced vegetation index (EVI) and the normalized difference red edge (NDRE) have been employed to assess the physiological status of wheat and sorghum crops during maturation under drought conditions11. Since senescence plays a crucial role in the selection of stay-green genotypes, there is a need to develop accurate methods using cost-effective spectral data to predict senescence rate. Under extreme stress conditions, early senescence resulting from rapid breakdown of plant tissues and macromolecules, such as chlorophyll, can lead to significant yield losses. Borrell et al.10 demonstrated the negative impact of high senescence rates on grain yield under water-limited and heat conditions. Senescence is closely related to green leaf area, chlorophyll content, and canopy temperature. Selecting wheat genotypes with improved stay-green traits, characterized by delayed senescence, can enhance cultivar performance under stressed conditions29,30.
This study aims for estimating turfgrass green cover by utilizing high-resolution RGB and multispectral images obtained through UAV technology. The specific objectives of this research are to compare and introduce various methods for estimating percent green cover during turfgrass establishment using UAV-derived data, to demonstrate the accuracy and effectiveness of UAV-derived percent green cover estimation by comparing it with ground-level percent green cover data using different vegetative indexes and to find out the most tolerant grass to cold climate.
Materials and methods
Grass farming
To conduct this study, a turf cultivation site was created in a facility house located within Jeju National University in Aradong, Jeju City in Jeju Island. The grass used in the experiment was collected from the Forest Life Resources Conservation Field of Jeju National University and transplanted over a period of approximately nine days from July 29th to August 6th, 2020 (Table 1). Thirty-two pot trays (5 cm in height, and 12 cm in length per cell) were used, filled with agricultural biosolid and bedding biosolid. As shown in Fig. 1, one resource was planted in each of the 12 cells on both sides after leaving the middle 8 cells empty. The trays with the planted grass were randomly placed on a bed approximately 70 cm above the ground. Subsequently, watering was carried out using a sprinkler cooler and hose to promote normal grass growth. To maintain the grass, a ventilation fan was installed and the side windows of the facility house were kept open to maintain a temperature similar to the external environment (Fig. 2). In addition, to promote high-density growth of the grass, fertilization was carried out twice in May and June 2021, and seven weeding sessions were performed between April and September 2021 at the appropriate times (Fig. 3). Above all, we managed the grass genetic resources by ensuring that the grass in each designated area did not mix by checking and trimming any overlapping stems every day.
Image acquisition
For the one-year-old grass, monthly mid-month (August 13th, September 10th, October 15th, November 12th, and December 10th ) images were taken for five months from August to December 2021. A Phantom 4 Multispectral (WM336, DJI) unmanned aerial vehicle equipped with six 1–2.9″ CMOS sensors (Fig. 4) was used for the experiment, and the wavelength range information for each sensor is shown in Table 2. Prior to the shooting, weed removal and moss covering with sand were performed to prevent other plants from appearing in the images. The Shooting was performed using the DJI GO pro app, the ISO value was set to 400, and the white balance was automatically adjusted. The shooting started at 1 p.m. and lasted for about 2 h. To shoot the grass, a drone with a sensor attached was placed on a custom-made device and kept at a fixed distance vertically 2 m above the grass to ensure that data was acquired with a fixed interval (Fig. 5A). Additionally when moving the device to shoot the grass, a radiation correction target plate (Type 882 Woven polyester fabric, Group 8 Technology, United States) was also moved so that the target plate was visible in all images (Fig. 5B). Images were stored in different file formats depending on the sensor used, with RGB images stored in JPEG format and multi-spectral images stored in TIFF format. The size of all images was 1600 × 1300 pixels, which was the same for all sensors. A total of 285 temporal data points (57 per session) were obtained over five sessions.
Image preprocessing
There are some preprocessing steps performed for image analysis. First, distortion correction was performed to flatten out the radial distortion caused by camera lens characteristics. Second, the radiometric correlation was necessary to equalize the light conditions for all acquired images, as uneven lighting conditions can affect the accurate characterization of plant features31,32. Third, histogram equalization was executed to improve the contrast of the RGB image, which had become dark after radiometric correction, making it difficult to discern the color of the grass33. Fourth, image alignment was performed on the NIR and Red images used to derive NDVI. This was necessary because of the slight physical differences in the camera positions used in this study, which caused the two images to be captured with slightly different positions. Therefore, the Red image had to be shifted by the number of pixels that differed from the NIR image to align the two images.
Distortion correction
The first is distortion correction, where the acquired image shows a radial distortion in the form of a convex bulge in the center (Fig. 6A). To solve this problem, initial processing was performed using the software Pix4D Mapper (Pix4D SA, Switzerland) to correct the camera lens distortion (Fig. 6B).
Radiometric calibration and histogram equalization
The light conditions vary from day to day, even for data acquired on the same day, depending on the weather (Fig. 7). The process required to equalize the light conditions is radiometric calibration. It uses a digital numerical value for the image and a fixed reflectance value. It converts every digital number in the image to the reflectance of an object’s surface. This is calculated using the best-fit equation for each band of a homogeneous object. In this study, a professional radiometric calibration reference tarp with uniform reflectance was used, and the surface reflectance value of the radiometric calibration tarp was provided by Group 8 Technology manufacturing company. The camera responses of the RGB and multispectral bands, which are necessary information for radiometric calibration, were obtained from the work of Burggraaff et al.31 and Lu et al.34, respectively. Finally, the exponential model for radiometric calibration of RGB and the linear model for that of multispectral images were developed by using the empirical line calibration equation35. With this information, we obtained digital numerical values for each image and proceeded with the radiometric correction.
In addition, it was applied for RGB images with radiometric calibration to perform histogram equalization which straightens the histogram of pixels whose brightness values are clustered towards the dark side of the image (Fig. 8) using MATLAB (R2015b, MathWorks).
Image alignment
For the multispectral image calibrated, image alignment was performed to match the NIR band image by shifting the RED band image based on the NIR (Near Infrared) band image (Fig. 9).
Their metadata was used to determine the difference in pixels between the RED and NIR band images in rows and columns. The metadata includes information on the Relative Optical Center X and Y pixel positions, which indicate the physical distance from the NIR lens. In this case, the values for both are zero. It was exported from the raw data and used ‘pyexiv2’ library to convert a TIFF file to an XMP file in Python. The ‘pyexiv2’ library is a Python binding for the Exiv2 library, which is a C + + library for manipulating image metadata.
The image was processed using in-house software written in Python 3.7.10. The software was used to extract the digital numerical values of the images, apply the radiometric calibration equation, and align the positions of the multispectral images.
Image analysis
GCP (green cover percentage) computation using RGB images
To calculate the greenness rate of the grass for each round, it is necessary to know the total canopy area and green cover area of each grass and calculate it using the formula below (Eq. 1).
First, the total canopy area of the grass was obtained using NIR images. NIR images show a better contrast between plants and soil than other images, making it easier to identify the boundaries of plant leaves36. The green area of the plant was extracted using a color-based index, ExG (Excess Green), on a RGB images that had undergone histogram equalization, in MATLAB. Only the green areas were extracted. Finally, to separate the grass and soil, Otus threshold method was applied to the NIR images and the RGB images with ExG applied, in MATLAB. The method divided the pixels in the images into two categories, black and white, and in this experiment, the black pixels represented the background and the white pixels represented the grass or the green area of the grass (Fig. 10). Subsequently, four rectangular regions of interest (ROI) with a size of 48.5 × 132 pixels were repeatedly specified in the grass area of the image, and the number of white pixels inside each ROI was counted to determine the total area and green area of the grass as shown in Fig. 11. This process was performed using self-developed software based on Python for this experiment51.
Various vegetation indexes computation using multispectral images
In this study, to monitor the degree of stress in the grass over time, we selected the NDVI (Normalized Difference Vegetation Index), which is the most widely used vegetation index. This index is based on the basic principle that healthy vegetation absorbs a lot of light in the visible red region and reflects a lot of light in the near-infrared region, which can be used to determine leaf area index, chlorophyll content, photosynthetic absorbed radiation, growth status, and vitality of vegetation.To calculate the NDVI of the grass, the radiometrically corrected RED and NIR images were used in the equation below (Eq. 2).
Also, we use the Normalized Difference Red Edge Index (NDRE), Soil-adjusted Vegetation Index (SAVI), and Enhanced Vegetation Index (EVI) to monitor the grass over time. The equations below were used to calculate each VIs (Vegetation Indexes).
[L is the soil brightness correction factor defined as 0.5 to accommodate most land cover types.]
[G is the gain factor, which is typically set to 2.5; C1 and C2 are coefficients used to reduce atmospheric influences, which commonly used values are C1 = 6 and C2 = 7.5; L is the canopy background adjustment factor, which is commonly used value is L = 1.]
After that, as in the case of selecting the ROI for the greening rate in the grass area in the VI images, we selected four ROIs of the same size as before and excluded the background to extract the VIs values of the pixels corresponding to the grass (Fig. 12). The VIs for each grass was then represented by the average value of the extracted VIs values This process was also conducted using software based on Python, which was independently developed for this experiment.
Statistics analysis
We used Excel (version 2205, Microsoft) to uniformly process values greater than 100% in the greening rate to 100% prior to statistical analysis. This was done to address an issue where during the process of removing moss, some areas were not fully removed, leading to a slight difference in the location of the green area within the grass, resulting in experimental errors in the calculation of the greening rate.
We performed statistical analysis on the temporal data of the greening rate, NDVI, NDRE, SAVI, and EVI values using the R programming language. Since the data did not meet the assumption of normality, nonparametric Kruskal–Wallis tests were used to determine statistical significance. Subsequently, post hoc analysis was conducted using the rank-based Dunn test, and p-values were adjusted using the Benjamini-Hochberg (BH) method. The correlation between the greening rate and NDVI, NDRE, SAVI, and EVI for each session was analyzed using the Pearson correlation coefficient.
Results
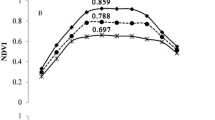
We conducted a study between August and December, where we measured GCP, NDRE, NDVI, SAVI, and EVIfor five months. Our analysis using the Kruskal–Wallis test showed that there were statistically significant differences in GCP and VIs data for each genetic resource at every measurement point, with a p-value of less than 0.05 (Table 3, Figs. 13, 14, 15, 16, 17). We presented the changes in GCP and NDVI for each genetic resource in Tables 4 and 5, respectively. These tables illustrate how the rankings of genetic resources differed and how browning rates varied among resources for each measurement period. We also observed differences in the size of the variance in GCP and NDVI between repetitions for each genetic resource by month. Until October, the GCP was relatively consistent across all genetic resources. However, starting in October, we began to see differences among genetic resources, with the differences becoming most pronounced in November. In contrast, the NDVI showed a certain degree of consistency among genetic resources, as shown in Fig. 14.
We were able to confirm the changes and trends of each germplasm over a five-month period that can be found by comparing Table 4 and Fig. 13. We observed that the differences in GCP rates between each germplasm were generally similar, with a few exceptions. In particular, the differences were maintained at 95% or higher in August and September. However, in contrast to the previous months, when san180 and san568 germplasms showed a GCP of 100%, they showed GCP of 67.22% and 60.97%, respectively, in October, a sharp decrease compared to September. On the other hand, san303 germplasm showed a GCP of 96.90% in August, ranking low, but it increased to 98.37% in October. Similarly, although the GCP of san187 germplasm decreased from 95.99% in August to 91.11% in September, it increased again to 94.87% in October. Based on the fact that the GCP of san180 and san568 germplasms decreased while those of san303 and san187 germplasms increased during the same period, we were able to observe that the changes in GCP vary depending on the germplasm over time. Looking at the GCP in November, the germplasm with the highest GCP was san208, which showed a GCP of 94%. San208 continuously showed a high GCP of 97.48% in August, 97.81% in September, and 96.86% in October, and although it slightly decreased in November, the rate of decrease was less than that of other germplasms.
Upon examining the RGB and GCP images for Suncheon, san218, and san187 in December we noticed that the soil part (sand) was identified as green. However, we believe that during the process of lightly washing away the sand dust on the grass that covers the moss before taking pictures, the moss was exposed again on the sand surface. To solve this distortion, we applied various VIs such as NDRE, SAVI, and EVI (Figs. 15, 16, 17, Tables 6, 7, 8). The results of NDRE show that san135, san180, san187, and san218 represent high values. On the other hand, the values of SAVI represent that san128 and san398 are higher values than others. Lastly, EVI applied results suggested that san128 got a high value. The average result of NDRE for san208 from August to December was found to be about 3% 5% respectively (Table 6). In constrast, SAVI decreased from August (38%) to December (4%) (Table 7). Furthermore, EVI remained slightly decreased during this time period (Table 8).
The differences and patterns of changes in NDVI for each genetic resource, provided a comprehensive understanding of the changes in NDVI over the 5-month period (Table 5 and Fig. 14). In August, the san568 genetic resource showed the highest NDVI value of 0.85, while the san9dangugdae genetic resource showed the lowest NDVI value of 0.45. Checking the NDVI values in October, it can be observed that the changes in NDVI values vary greatly depending on the genetic resource, similar to the GCP. The NDVI value of san568 decreased to 0.45, while the san208 genetic resource showed the best value of 0.65. The san9dangugdae genetic resource consistently showed the lowest NDVI value, which continued until November. In December, the highest NDVI value was 0.32 for the san218 genetic resource, while the lowest NDVI value was 0.08 for the san128 genetic resource.
Based on the analysis of the correlation between the growth rate and NDVI for each month (Fig. 18), November and December showed a significant correlation with a p-value less than 0.05 and an r square-values of 0.61 and 0.31.
Discussion
Turfgrass is a valuable ground cover globally, providing numerous environmental, economic, and social benefits. To assess overall plant health and establishment rate over time, green cover percent, which measures the percentage of green vegetation per unit area, is a crucial indicator. Turfgrass producers and consumers prefer species and cultivars with faster establishment from plugs or sprigs and higher GCP. The conventional method of visually assessing turfgrass plots to estimate the amount of green cover, either on a percent or 1–9 scale, has become a standard and is regularly employed by turfgrass researchers. Low temperatures can have a detrimental effect on grass, but certain grasses exhibit higher resistance due to increased enzyme activity, unsaturated fatty acid content, and anthocyanin production18.
In this study, we aim to objectively evaluate the quality of grass by proposing criteria for its assessment using an RGB camera and a Multi-spectral sensor. The amount and quality of green in grass per unit area and the biomass of the plant have been shown to be correlated with its cold tolerance37 Furthermore, color is a component of visual evaluation for grass quality38. Through analysis of RGB images, it is possible to perform evaluations similar to those made by the human eye, and in particular, to determine the proportion of green area by counting the number of pixels, allowing us to assess the amount of green in the grass. The Normalized Difference Vegetation Index (NDVI) is a widely used index for the quantitative estimation of vegetation, which uses the reflectance of near-infrared and red wavelengths to quantify plant growth39. Moreover, NDVI is the most commonly used index for greenness and can estimate the amount of chlorophyll that makes plants appear green by using reflectance, which allows us to assess the quality of grass greenness using NDVI in this study40. All genetic resources showed a GCP of over 95% in August. However, looking at the changes until October, it was found that some genetic resources (san303 and san187) showed a slight increase in greenness rate, indicating that these two genetic resources continued to grow relatively longer compared to other genetic resources, given that the growing period of Korean grass is from April to mid-October. However, most of the genetic resources showed a decrease in the greenness rate in October. This was more pronounced in the final result in December, where the san208 genetic resource maintained a very excellent greenness rate of over 90%, while the san125 genetic resource showed a greenness rate of 0.4%, showing a very clear difference from san208. These results indicate that traits related to changes in greenness rate are expressed differently in each genetic resource. During the research conducted from August to December 2021 at a grass cultivation site, the graph showing the highest temperature, lowest temperature, and average temperature obtained from the nearby weather observatory (Fig. 2) shows a tendency for the temperature to decrease as time passes from August to December, especially showing a rapid decrease in temperature in mid-October. By looking at the temperature graph (Fig. 2) and Fig. 19 together, it can be seen that in August and September, all grass genetic resources showed good quality green color at appropriate temperatures for growth, but in mid-October, the temperature decreased rapidly and the color of the grass changed from green to yellow or yellow–brown in most genetic resources. Low-temperature stress is one of the most harmful environmental stress factors for grass, leading to decreased photosynthesis due to photoinhibition, accumulation of carbohydrates within the cell, dehydration due to cell membrane damage caused by ice crystals formed inside and outside the cell at temperatures below freezing, changes in cell membrane composition, and protein denaturation, which can cause the grass to wither and die in severe cases41. Grass that is resistant to low temperatures has higher levels of the enzyme activity associated with carbon metabolism, the unsaturated fatty acid content in the lipids that make up the cell membrane, and accumulation of proline, which maintains membrane stability and scavenges reactive oxygen species, than susceptible grasses5. In addition, grasses produce and accumulate anthocyanin, an antioxidant, as a secondary metabolite in response to low-temperature stress, which reduces oxidative damage and gives winter grass a red color visible to the naked eye18 In our study, we found that In November and December, the grass color changed noticeably to not only yellow but also red and reddish-blue. The change in grass color to red is due to the synthesis, transportation, and accumulation of anthocyanin, a secondary metabolite, and antioxidant known to prevent oxidative damage to grass caused by the large number of reactive oxygen species generated inside the plant under low temperatures. A study shows that the accumulation of anthocyanin is related to the high cold tolerance of grass18.
Bremer et al.'s42 studied, the quality of grass was evaluated by calculating the NDVI from canopy images of grass obtained through a hyperspectral sensor, which showed a high correlation (R2 = 0.88, p < 0.0001) with the visually evaluated green cover area of the grass. They also mentioned that the RED image, which is part of the NDVI, affects the density difference and chlorophyll content of grass, and that additional research is needed, particularly using the NIR image, to predict the water stress status of plants before stress symptoms appear. In this study, considering the growth period of Korean grass, during the growing season in August and September, all grasses were observed to have high NDVI values as they appeared green. However, from October to December, during the non-growth season when the grass entered dormancy and lost its green color, the NDVI values gradually decreased. Among the genetic resources studied, san218 showed the highest NDVI value in December. In September, it showed a very low NDVI value of 0.42, but in December, it showed a higher NDVI value of 0.32 compared to other genetic resources. In contrast, san9dangugdae showed a higher NDVI value of 0.45 in August compared to san218, but a relatively low NDVI value of 0.12 in December among the genetic resources. Like the rate of greening, the rate of change in NDVI values also showed a significant difference in the change patterns of genetic resources. However, while the NDVI values of other genetic resources decreased as the greening rate decreased, only san208 showed a high greening rate of over 90% from August to December, but the NDVI value gradually decreased over time. This seems to be because san208 showed an increase in the number of other pigments produced under low-temperature stress, reducing the relative proportion of chlorophyll and decreasing the NDVI value. Therefore, additional analysis of leaf pigment composition for san208 is needed to explain why it still showed high greening compared to other genetic resources.
Significant results were only obtained for November when examining the correlation between the greenness rate and NDVI. This indicates that there is almost no correlation between the trait of maintaining quality greenness by occupying a certain area with grass (greenness rate data confirmed by RGB camera) and the trait of maintaining quality greenness by the amount of chlorophyll in the leaves themselves (NDVI based on multi-spectral imagery). The correlation that appeared in November, when the growth status of the grass was extremely poor, is believed to have occurred because both the qualitative state (NDVI) and quantitative state (greenness rate) of the grass showed a correlation. Previously, even if the qualitative state of the NDVI value was low, there appeared to be many grass cultivars with high quantitative states that could not show a correlation. In December, even superior cultivars showed a decrease in both the qualitative and quantitative states, which is presumed to be the reason why the correlation did not appear again. Therefore, it is necessary to select the exact observation time to confirm the correlation between the traits obtained from the two images.
In a study conducted by Barboza et al.43, researchers used multiple linear regression analysis to determine the most accurate estimates for fresh biomass. They considered three input parameters: plant width, height, and vegetation index (either NDVI or NDRE). While NDRE showed saturation during the reproductive stages, the results of the analysis revealed that NDVI performed better than NDRE in estimating fresh biomass. This means that NDVI provided more reliable and accurate predictions of biomass compared to NDRE. Despite its limitations during the reproductive stages, NDVI was found to be a more suitable vegetation index for estimating fresh biomass in this particular study. In our study, we examined various vegetation indices, including NDRE, SAVI, and EVI. The results of NDRE indicated that san135, san180, san187, and san218 had high values. Conversely, the values of SAVI showed that san128 and san398 had higher values compared to others. Lastly, when EVI was applied, san128 had a high value. On average, the NDRE results for san208 from August to December ranged from approximately 3% to 5%. In contrast, SAVI decreased from August (38%) to December (4%). Furthermore, EVI showed a slight decrease during this time period. The variation may be due to the fact that the NDRE index is better suited for evaluating chlorophyll content in larger trees, while the NDVI index is more reliable for assessing biomass and health in smaller trees. The choice of index depends on the specific characteristics of the trees under study, such as their size and the desired parameter to be measured.
Boiarskii and Hasegawa conducted a study in 2019 where they compared two layers with different indices, specifically NDVI and NDRE, and observed variations in vegetation activity. The study found that NDVI proved to be particularly effective in analyzing large land areas to assess vegetation density and the overall greenness of crops. It provided insights into crop health, cultivation effectiveness, and seeding rates. On the other hand, NDRE allowed for the visualization of chlorophyll content in leaves. This suggests that different indices may be more suitable for different crops, plant densities, seeding rates, and growth stages44.
The response of plants to low-temperature stress can serve as indicators for research on cold tolerance. One method is to analyze digital images using RGB sensors to observe grass color and growth, while multispectral sensors can be used to evaluate grass quality in terms of color, density, and vegetation vitality. Multispectral imaging can provide information on chlorophyll content, canopy structure, and leaf area index at different wavelengths, and vegetation indices derived from them can be combined to assess plant moisture and nutrient status under environmental stress45,42,47. Fluorescence sensors can also be used to measure leaf chlorophyll fluorescence to evaluate the photosynthetic apparatus' state under low-temperature stress. A laser-induced chlorophyll fluorescence measurement method has also been proposed for outdoor measurement where ambient light and temperature are not constant48,49. Moreover, LIDAR/laser sensors can generate 3D images that can be used to examine plant structure, biomass, and other factors, providing a way for real-time research on the morphological changes in grass that are exposed to low-temperature stress50,51.
Conclusion
This study suggests that the choice between using RGB-based greenness and NDVI-based greenness for evaluating grass greenness may vary depending on the purpose. When evaluating greenness from an economic standpoint, it may be more practical to use RGB-based greenness, which provides an intuitive estimate of the amount of greenness similar to visual observation. However, to select grass that maintains a high level of greenness, using NDVI-based greenness, which evaluates the grass from a biological perspective, can complement the RGB-based greenness. Therefore, it is recommended to use both greenness measures in a complementary manner to select grass that can maintain good greenness over time. Additionally, considering the time when the two measures show a correlation can be advantageous in selecting grass using both measures. Finally, by using the greenness measures of this study, it is expected that time and energy spent on grass quality evaluation can be reduced while selecting excellent quality grass through an objective and reproducible method.
References
Cai, H. et al. Isolation, characterization and mapping of simple sequence repeat markers in zoysiagrass (Zoysia spp.). Theor. Appl. Genet. 112, 158–166 (2005).
Bae, E.-J., Lee, K.-S., Park, Y.-B., Kim, D. S. & Yang, G.-M. Characteristics of seed variation of zoysiagrasses (Zoysia spp.) native to South Korea. Weed Turfgrass Sci. 2, 267–273 (2013).
Brian, M. et al. Heritability Estimates for Turfgrass Performance and Stress Response in Zoysia spp. Crop Science 49(6), 2113–2118. https://doi.org/10.2135/cropsci2008.12.0705 (2009).
Patton, A. J., Schwartz, B. M. & Kenworthy, K. E. Zoysiagrass (Zoysia spp.) history, utilization, and improvement in the United States: a review. Crop Sci. 57, S37 (2017).
Huang, B., DaCosta, M. & Jiang, Y. Research advances in mechanisms of turfgrass tolerance to abiotic stresses: From physiology to molecular biology. Crit. Rev. Plant Sci. 33, 141–189 (2014).
Jiang, Y., Duncan, R. R. & Carrow, R. N. Assessment of low light tolerance of seashore paspalum and bermudagrass. Crop Sci. 44, 587–594 (2004).
Lee, G., Carrow, R. N. & Duncan, R. R. Growth and water relation responses to salinity stress in halophytic seashore paspalum ecotypes. Sci. Hortic. 104, 221–236 (2005).
Fan, J. et al. Mechanisms of environmental stress tolerance in turfgrass. Agronomy 10, 522 (2020).
Cui, Y., Wang, J., Wang, X. & Jiang, Y. Phenotypic and genotypic diversity for drought tolerance among and within perennial ryegrass accessions. HortScience 50, 1148–1154 (2015).
Kimball, J. A., Tuong, T. D., Arellano, C., Livingston Iii, D. P. & Milla-Lewis, S. R. Assessing freeze-tolerance in St. Augustinegrass: Temperature response and evaluation methods. Euphytica 213, 110 (2017).
Li, L., Zhang, Q. & Huang, D. A review of imaging techniques for plant phenotyping. Sensors 14, 20078–20111 (2014).
Büchi, L., Wendling, M., Mouly, P. & Charles, R. Comparison of visual assessment and digital image analysis for canopy cover estimation. Agron. J. 110, 1289–1295 (2018).
Osco, L. P. et al. Leaf nitrogen concentration and plant height prediction for maize using UAV-based multispectral imagery and machine learning techniques. Remote Sens. 12, 3237 (2020).
Santana, D. C. et al. UAV-based multispectral sensor to measure variations in corn as a function of nitrogen topdressing. Remote Sens. Appl. Soc. Environ. 23, 100534 (2021).
Wan, W. et al. Spatiotemporal patterns of maize drought stress and their effects on biomass in the Northeast and North China Plain from 2000 to 2019. Agric. For. Meteorol. 315, 108821 (2022).
Braz, A. M., Águas, T. D. A. & Mirandola Garcia, P. H. Análise de índices de vegetação NDVI e SAVI e índice de área foliar (IAF) para a comparação da cobertura vegetal na bacia hidrográfica do Córrego Ribeirãozinho, município de Selvíria–MS. Rev. Percurso 7, 5 (2015).
Deery, D., Jimenez-Berni, J., Jones, H., Sirault, X. & Furbank, R. Proximal remote sensing buggies and potential applications for field-based phenotyping. Agronomy 4, 349–379 (2014).
Jin, H.-X. et al. A survey of enhanced cold tolerance and low-temperature-induced anthocyanin accumulation in a novel Zoysia japonica biotype. Plants 11, 429 (2022).
Jin, X., Li, Z., Feng, H., Xu, X. & Yang, G. Newly combined spectral indices to improve estimation of total leaf chlorophyll content in cotton. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 7, 4589–4600 (2014).
Magney, T. S., Eitel, J. U. H., Huggins, D. R. & Vierling, L. A. Proximal NDVI derived phenology improves in-season predictions of wheat quantity and quality. Agric. For. Meteorol. 217, 46–60 (2016).
Tattaris, M., Reynolds, M. P. & Chapman, S. C. A direct comparison of remote sensing approaches for high-throughput phenotyping in plant breeding. Front. Plant Sci. 7, (2016).
Bendig, J. et al. Estimating Biomass of barley using crop surface models (CSMs) derived from UAV-based RGB imaging. Remote Sens. 6, 10395–10412 (2014).
Verger, A. et al. Green area index from an unmanned aerial system over wheat and rapeseed crops. Remote Sens. Environ. 152, 654–664 (2014).
Jin, X., Liu, S., Baret, F., Hemerlé, M. & Comar, A. Estimates of plant density of wheat crops at emergence from very low altitude UAV imagery. Remote Sens. Environ. 198, 105–114 (2017).
Guillen-Climent, M. L., Zarco-Tejada, P. J., Berni, J. A. J., North, P. R. J. & Villalobos, F. J. Mapping radiation interception in row-structured orchards using 3D simulation and high-resolution airborne imagery acquired from a UAV. Precis. Agric. 13, 473–500 (2012).
Sankaran, S., Khot, L. R. & Carter, A. H. Field-based crop phenotyping: Multispectral aerial imaging for evaluation of winter wheat emergence and spring stand. Comput. Electron. Agric. 118, 372–379 (2015).
Liu, C., Shang, J., Vachon, P. W. & McNairn, H. Multiyear crop monitoring using polarimetric RADARSAT-2 data. IEEE Trans. Geosci. Remote Sens. 51, 2227–2240 (2013).
Zhang, J. et al. Applications of unmanned aerial vehicle based imagery in turfgrass field trials. Front. Plant Sci. 10, 279 (2019).
Borrell, A. K. et al. Stay-green alleles individually enhance grain yield in sorghum under drought by modifying canopy development and water uptake patterns. New Phytol. 203, 817–830 (2014).
Hassan, M. et al. Time-series multispectral indices from unmanned aerial vehicle imagery reveal senescence rate in bread wheat. Remote Sens. 10, 809 (2018).
Burggraaff, O. et al. Standardized spectral and radiometric calibration of consumer cameras. Opt. Express 27, 19075 (2019).
Guo, Y. et al. Radiometric calibration for multispectral camera of different imaging conditions mounted on a UAV platform. Sustainability 11, 978 (2019).
Stark, J. A. Adaptive image contrast enhancement using generalizations of histogram equalization. IEEE Trans. Image Process. 9, 889–896 (2000).
Lu, H., Fan, T., Ghimire, P. & Deng, L. Experimental evaluation and consistency comparison of UAV multispectral minisensors. Remote Sens. 12, 2542 (2020).
Wang, C. & Myint, S. W. A simplified empirical line method of radiometric calibration for small unmanned aircraft systems-based remote sensing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 8, 1876–1885 (2015).
Agostini, A. et al. A cognitive architecture for auto-matic gardening. Comput. Electron. Agr. 138, 69–79. https://doi.org/10.1016/j.compag.2017.04.015 (2017).
Schafleitner, R. et al. Field Screening for variation of drought tolerance in Solanum tuberosum L. by agronomical. Physiol. Genet. Anal. Potato Res. 50, 71–85 (2007).
Kim, K. N., Park, W. K. & Nam, S. Y. Comparison of establishment vigor, uniformity, rooting potential and turf quality of sods of kentucky bluegrass, perennial ryegrass, tall fescue and cool-season grass mixtures grown in sand soil. Kor. J. Turfgrass Sci. 17(4), 129–146 (2003).
Rouse, J. W, Haas, R. H., Scheel, J. A. & Deering, D. W. Monitoring vegetation systems in the great plains with ERTS. In proceedings. 3rd earth resource technology satellite (ERTS) symposium 48–62 (1974).
Walter, A., Liebisch, F. & Hund, A. Plant phenotyping: From bean weighing to image analysis. Plant Methods 11, 14 (2015).
Borawska-Jarmułowicz, B., Mastalerczuk, G., Pietkiewicz, S. & Kalaji, M. H. Low temperature and hardening effects on photosynthetic apparatus efficiency and survival of forage grass varieties. Plant Soil Environ. 60, 177–183 (2014).
Bremer, D. J., Lee, H., Su, K. & Keeley, S. J. Relationships between normalized difference vegetation index and visual quality in cool-season turfgrass: II. Factors affecting NDVI and its component reflectances. Crop Sci. 51, 2219–2227 (2011).
Barboza, T. O. C. et al. Performance of vegetation indices to estimate green biomass accumulation in common bean. AgriEngineering 5, 840–854 (2023).
Boiarskii, B. Comparison of NDVI and NDRE indices to detect differences in vegetation and chlorophyll content. J. Mech. Contin. Math. Sci. spl1, (2019).
Narmilan, A. et al. Predicting canopy chlorophyll content in sugarcane crops using machine learning algorithms and spectral vegetation indices derived from UAV multispectral imagery. Remote Sens. 14, 1140 (2022).
Adam, E., Mutanga, O. & Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: a review. Wetl. Ecol. Manag. 18, 281–296 (2010).
Xue, J. & Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 1–17 (2017).
Fracheboud, Y. & Leipner, J. The application of chlorophyll fluorescence to study light, temperature, and drought stress. in Practical Applications of Chlorophyll Fluorescence in Plant Biology (eds. DeEll, J. R. & Toivonen, P. M. A.) 125–150 (Springer US, 2003). https://doi.org/10.1007/978-1-4615-0415-3_4.
Wu, L., Wang, L., Shi, C. & Yin, D. Detecting mangrove photosynthesis with solar-induced chlorophyll fluorescence. Int. J. Remote Sens. 43, 1037–1053 (2022).
Rivera, G., Porras, R., Florencia, R. & Sánchez-Solís, J. P. LiDAR applications in precision agriculture for cultivating crops: A review of recent advances. Comput. Electron. Agric. 207, 107737 (2023).
Al-Tamimi, N. et al. Capturing crop adaptation to abiotic stress using image-based technologies. Open Biol. 12, 210353 (2022).
Funding
Basic Science Research Program supported this research through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2019R1A6A1A11052070).
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.S.C.; Methodology, K.U. and T.T.T.; Investigation, S.M. and Y.S.C.; Data curation, K.K., S.M. and T.T.T.; Writing—original draft preparation, K.K. and S.M.; Writing—review and editing, K.K., S.M., G.D.H. and Y.S.C.; Supervision, Y.S.C.; Project Administration, Y.S.C..; All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ku, KB., Mansoor, S., Han, G.D. et al. Identification of new cold tolerant Zoysia grass species using high-resolution RGB and multi-spectral imaging. Sci Rep 13, 13209 (2023). https://doi.org/10.1038/s41598-023-40128-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-40128-2
This article is cited by
-
Enhancing citrus fruit yield investigations through flight height optimization with UAV imaging
Scientific Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.




 : no germplasm, it is empty.
: no germplasm, it is empty.
















