Abstract
Actinobacteria are well known as a rich source of diversity of bioactive secondary metabolites. Kutzneria, a rare actinobacteria belonging to the family Pseudonocardiaceae has abundance of secondary metabolite biosynthetic gene clusters (BGCs) and is one of important source of natural products and worthy of priority investigation. Currently, Kutzneria chonburiensis SMC256T has been the latest type-strain of the genus and its genome sequence has not been reported yet. Therefore, we present the first report of new complete genome sequence of SMC256T (genome size of 10.4 Mbp) with genome annotation and feature comparison between SMC256T and other publicly available Kutzneria species. The results from comparative and functional genomic analyses regarding the phylogenomic and the clusters of orthologous groups of proteins (COGs) analyses indicated that SMC256T is most closely related to Kutzneria sp. 744, Kutzneria kofuensis, Kutzneria sp. CA-103260 and Kutzneria buriramensis. Furthermore, a total of 322 BGCs were also detected and showed diversity among the Kutzneria genomes. Out of which, 38 clusters showing the best hit to the most known BGCs were predicted in the SMC256Tgenome. We observed that six clusters responsible for biosynthesis of antimicrobials/antitumor metabolites were strain-specific in Kutzneria chonburiensis. These putative metabolites include virginiamycin S1, lysolipin I, esmeraldin, rakicidin, aclacinomycin and streptoseomycin. Based on these findings, the genome of Kutzneria chonburiensis contains distinct and unidentified BGCs different from other members of the genus, and the use of integrative genomic-based approach would be a useful alternative effort to target, isolate and identify putative and undiscovered secondary metabolites suspected to have new and/or specific bioactivity in the Kutzneria.
Similar content being viewed by others
Introduction
Recent advances in high-throughput whole genome sequencing, genome mining and secondary metabolite biosynthetic gene clusters (BGCs) analyses have led to discovery of numerous bioactive metabolites, e.g. new and diverse antimicrobial agents, from microbial sources1,2,3. The most diverse and multiple secondary metabolite gene clusters have been investigated in a group of actinomycetes genomes, particularly in more difficultly isolated and cultured subgroups namely rare actinomycetes4,5,6. One of them includes a species belonging to the genus Kutzneria. Kutzneria is a genus of bacteria in Phylum Actinobacteria, Order Pseudonocardiales and Family Pseudonocardiaceae7. They are non-motile, aerobic, mesophilic, thermotolerant, Gram-positive and chemoorganotrophs. The majority of this species have been isolated from soils in different habitats, including tropical rainforest, mountain forest, deciduous forest, rhizosphere and sediment. Members of this genus are usually associated with soil, but little is known about their role within this surroundings. They are probably involved in the primary decomposition of plant material in soils7. To date, there is information of 15 Kutzneria strains on public databases (five strains with systematic classification: Kutzneria albida, Kutzneria viridogrisea, Kutzneria kofuensis, Kutzneria buriramensis and Kutzneria chonburiensis and ten strains without taxonomic description: Kutzneria species strains CA-103260, 744, A14, TSII, TM-S116, TM-B149, RMD-3Y-3-1, RDB-177, 306G04 and L1988), of which seven strains (Kutzneria albida, K. viridogrisea, K. kofuensis, K. buriramensis, K. chonburiensis, Kutzneria species strains CA-103260 and 744) have their genome sequences and the remainder eight strains (Kutzneria species strains A14, TSII, TM-S116, TM-B149, RMD-3Y-3-1, RDB-177, 306G04 and L1988) have 16S rRNA gene sequences deposited in public databases (see Supplementary Table S1)8. Because there are hardly any complete genome assemblies of members of the genus Kutzneria, only 3 genomes obtained from K. albida, K. chonburiensis and strain CA-103260 were fully sequenced and the remaining incomplete genome sequences were of K. buriramensis, K. viridogrisea, K. kofuensis and strain 7448. With the recent improvement of DNA sequencing technology, the quality and completeness of bacterial genomes assemblies have increased dramatically9. As a result, development of a suite of bioinformatics tools which can be applied to annotate and mine these genomes has become increasingly essential. A number of bioinformatics tools have been developed, including BAGEL10, ClustScan11, CLUSEAN12, NP searcher13, PRISM14, and antiSMASH15, to identify BGCs within the genome. Most of these tools count on algorithms that map the highly conserved sequences within the BGCs to their location. Genome mining approaches enable prediction of BGCs from genome data rapidly and effortlessly, and accelerate the process linking the product with their corresponding BGCs.
First report in discovery of important secondary metabolites based on genome mining was investigated in K. albida DSM43870T. The antiSMASH, bioinformatics analysis tool for the rapid genome-wide identification, annotation and analysis of secondary metabolite BGCs, is able to identify BGCs responsible for the biosynthesis of aculeximycin that shows antimicrobial activity against Gram-positive bacteria, fungi and mosquito larvae16. Recently, the genome mining technique has also been applied to characterize BGCs in order to discover new 30-membered macrolides (epemicins A and B) in the culture of Kutzneria sp. CA-103260, which genome contains BGC similarly to the macrolide aculeximycin BGC found in K. albida6. These compounds showed antimicrobial activity against methicillin resistant Staphylococcus aureus (MRSA) and showed chemical structure related to aculeximycin core with some structural diversity. As a consequence, these findings present the potential of rare actinobacteria to produce new and structurally diverse metabolites. In this work, we therefore present the first report of complete genome sequence of Kutzneria chonburiensis SMC256T by performing the genome sequencing using Oxford Nanopore MinION long read and Illumina short read sequencing technologies. Subsequently, the complete genome is subjected to annotation, functional and genomic features analyses. Furthermore, we carried out the comparative genome analysis of K. chonburiensis with other related Kutzneria species to obtain an in-depth comparison of metabolic potential of the representatives of this genus, and would allow to identify diversity and unique of the secondary metabolite biosynthetic gene clusters among them.
Results
Complete genome sequence characteristics of Kutzneria chonburiensis and its comparison to other rare Kutzneria species
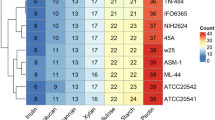
The repeat regions obtained by short-read sequencing frequently make the step of assembly complicated and resulted in underestimation of the repetitive contents. To overcome this limitation of short reads, a hybrid strategy, utilizing sequence data generated by the long- and short-read sequencing methods, has been used in an attempt to complete genome assembly in the Kutzneria chonburiensis strain SMC256T. The complete sequenced genome of K. chonburiensis reveals a circular chromosome of 10,411,187 bp (10.4 Mbp) and GC content of 69.86% as depicted on a circular map (Fig. 1). The whole complete genome sequence is subject to be analyzed using the RAST server system to predict and assign functions to the genes and the results illustrated the total genes in the genome: 9,564 protein-coding sequences (CDS), 74 tRNA-coding genes, 9 rRNA coding genes, and 1 tmRNA genes (Table 1). At the present moment, only 7 sequenced genomes of Kutzneria species are available in public databases, NCBI. The 3 complete genome sequences (1 contig) are merely from Kutzneria albida DSM43870T, Kutzneria chonburiensis SMC256T and Kutzneria sp. CA-103260. The remaining are incomplete genome sequences (draft genome) of Kutzneria buriramensis DSM45791T (65 contigs), Kutzneria viridogrisea NBRC15561T (11 contigs), Kutzneria kofuensis DSM43851T (5 contigs) and Kutzneria sp. 744 (534 contigs). The completeness assessment of the assemblies with BUSCO shows genome assemblies of very good quality excepted for Kutzneria sp. 744. In fact, they all have a BUSCO score higher than 98% without any missing genes and between 0 and 4 fragmented genes excepted for Kutzneria sp. 744 that has a BUSCO score of 87.3% with many missing genes and many fragmented genes. Furthermore, the BUSCO results at the annotation level show little difference with what was observed at the assembly level (> 98% for all annotation excepted for Kutzneria sp. 744 with 86.2% (Table 2). For genome comparison, all available genome sequences were retrieved and used for genome sequence comparison with that of K. chonburiensis through a circular map created by BLAST method (Fig. 2). This circular map shows gaps and low sequence similarity regions (pale color shades) indicating wide variable regions among the compared genomes. The results of this annotation exhibit the genome lengths between 9.87 and 11.96 Mbp, GC% of 69.86–70.6%, and CDS of 8894–11,066 genes. There are repeat regions within the genome sequences of Kutzneria sp. 744, Kutzneria buriramensis, Kutzneria albida and Kutzneria viridogrisea but not in K. chonburiensis, K. kofuensis and Kutzneria sp. CA-103260. To determine genome similarity among the Kutzneria species, the pairwise genome average nucleotide identity (ANI) values calculated by JSpecies were used as an index for genome comparisons (Table 1). This ANI comparison between the genome of K. chonburiensis with those of other species showed the results of < 95%, implying that K. chonburiensis does not belong to the same species with Kutzneria sp. 744 (89% ANI), K. kofuensis (88.6% ANI), Kutzneria sp. CA-103260 (87.6% ANI), K. buriramensis (87.42% ANI), K. albida (84.65% ANI) and K. viridogrisea (84.65% ANI). Based on phylogenomic analysis using the whole-genome multi-locus sequence typing (MLST), Kutzneria species formed a unique group in the family Pseudonocardiaceae in which K. chonburiensis is closely related to Kutzneria sp. 744, K. kofuensis, Kutzneria sp. CA-103260 and K. buriramensis, respectively and rather far from K. viridogrisea and K. albida (Fig. 3). In addition, functional roles of genes present in these genomes were annotated, assigned and categorized using RAST and tools in PATRIC (Pathosystems Resource Integration Center) server platform as seen in Table 2. It is noted that among the total ~ 10,000 annotated genes, nearly 60% of the genes encoding proteins with known functions have been assigned. The remaining (40%) have been predicted to code for hypothetical proteins. According to the RAST database, the majority of genes have been functionally classified into metabolism, protein processing, stress response, energy, DNA processing, RNA processing, cellular processing, membrane transport, cell envelops, regulation and cell signaling, and miscellaneous. By this analysis, the most abundant genes were distributed in the category of metabolism (Table 2). Further studying the clusters of orthologous groups of proteins (COGs: a family of orthologous protein-coding genes) within genomes of the Kutzneria was represented in a clustering heat map (Fig. 4). The orthologous protein-coding genes were assigned and mapped into 23 functional COG classes with their gene abundance (represented by colors) over the members of this group. It is noteworthy that K. albida and K. viridogrisea clearly displayed the most similar heat map patterns of COGs in the same clusters. These gene clusters belonged to (L) lipid transport and metabolism, (Q) secondary metabolites biosynthesis, transport and catabolism, (E) amino acid transport and metabolism, (F) nucleotide transport and metabolism, (H) coenzyme transport and metabolism, (J) translation, ribosomal structure and biogenesis, (B) chormatin structure and dynamics, (O) post-translational modification, protein turnover, chaperones, (L) replication, recombination and repair, (V) defense mechanisms, and (P) inorganic ion transport and metabolism. On the other hand, although K. chonburiensis was closest to K. kofuensis, Kutzneria sp. CA-103260, K. buriramensis and Kutzneria sp. 744, it showed the distinct pattern of COGs among them in which the genes were located mostly in the categories of (U) intracellular trafficking, secretion and vesicular transport, and (G) carbohydrate transport and metabolism, and some were identified to be (S) unknown function (Fig. 4).
A circular map displays distribution of the genome annotations of Kutzneria chonburiensis strain SMC256T. From outer to inner rings: contig, CDS on the forward strand, CDS on the reverse strand, RNA genes, antimicrobial resistance genes, virulence factors, GC content and GC skew. The color codes of the CDS on the forward and reverse strand indicate the subsystem that these genes belong to.
Genomic comparison of Kutzneria chonburiensis strain SMC256T to other Kutzneria sp. genomes. The gaps and color shades in the circular map exhibit regions of no and low similarity, respectively. From outside: Kutzneria viridogrisea, K. albida, K. buriramensis, Kutzneria sp. CA103260, K. kofuensis, Kutzneria sp.744 and K. chonburiensis.
Secondary metabolite biosynthetic gene clusters (BGCs) of Kutzneria chonburiensis and the other species of the genus Kutzneria
To elucidate high potential genes involved in the secondary metabolite biosynthesis in bacterial genomes, antiSMASH version 6.0 has been used to rapid genome-wide identification, annotation and analysis of the secondary metabolite biosynthesis gene clusters (BGCs) in the Kutzneria spceis15. The results revealed that 322 gene clusters were identified and distributed among the Kutzneria genomes and grouped into 30 BGC types as seen in Fig. 5 and Supplementary Table S2. Among them, the hybrid clusters, terpene, Type I PKS (T1PKS), non-ribosomal peptide synthetase cluster (NRPS), other unspecified ribosomally synthesised and post-translationally modified peptide product cluster (RiPP-like), NRPS-like fragment (NRPS-like), Class I lanthipeptide clusters like nisin (Lantipeptide I), Class II lanthipeptide clusters like mutacin II (Lantipeptide II) and Redox-cofactors such as PQQ (Redox-cofactor) were widespread in all Kutzneria species, especially hybrid clusters, terpene, T1PKS and NRPS were the predominant BGCs of the Kutzneria genus (Fig. 5b). In fact, the largest number of BGCs in all analyzed species was the hybrid clusters (~ 25% in dark blue) and the second most abundant BGCs was the terpene compounds (~ 14% in orange) (Fig. 5a). Furthermore, out of 30 BGC types, 15 types were identified in the genome of K. chonburiensis, which include tRNA-dependent cyclodipeptide synthases (CDPS), pheganomycin-style protein ligase-containing cluster (guanidinotides), lantipeptide class I, lantipeptide class II, NRPS, NRPS-like, oligosaccharide cluster (oligosaccharide), redox-cofactor, RiPP-like, T1PKS, type II PKS (T2PKS), type III PKS (T3PKS), terpene, thioamide-containing non-ribosomal peptide (thioamide-NRP) and hybrid clusters (Fig. 5a). A small number of putative bacteriocins, mainly belonging to the class II lanthipeptide were also determined in the genome of K. chonburiensis, as shown in Fig. 6 and Supplementary Table S3. The heat map in Fig. 7 representing abundance and clustering of BGCs among the Kutzneria genomes revealed strain/species-specific biosynthetic gene clusters of each Kutzneria species (red boxes) and slightly separated members of the Kutzneria into 2 subgroups: K. albida and K. viridogrisea in the same group whereas K. buriramensis, K. kofuensis, Kutzneria sp. 744, Kutzneria sp. CA-103260 and K. chonburiensis in another group. Moreover, the principal component analysis (PCA) derived on BGCs data as shown in Fig. 8 also clearly supported that these Kutzneria species formed separate clusters (cluster 1: K. viridogrisea and K. albida; cluster 2: K.chonburiensis, Kutzneria sp. 744, K. kofuensis, K. buriramensis and Kutzneria sp. CA-103260) similarly to patterns of cladogram (Fig. 3) and clustering heat map (Fig. 7). Among all BGCs detected, 235 gene clusters were predicted directly through the antiSMASH to be responsible for biosynthesis of known metabolites (compounds). Geosmin, lankacidin C and hopene are common secondary metabolites found in all analyzed Kutzneria (see Supplementary Table S4). Interestingly, the Kutzneria chonburiensis genome possesses several specific putative BGCs among the genus that matched the known biosynthetic gene clusters in MIBiG at various % sequence similarity. These matched known BGCs are reported to be responsible for biosynthesis of virginiamycin S1 (77% similarity), lysolipin I (32% and 73% similarities), esmeraldin (44% similarity), rakicidins A and B (22% similarity), aclacinomycin (20% similarity) and streptoseomycin (4% similarity) (Fig. 9). Additionally, according to protein-blasting of these gene clusters against the NCBI database, we found the similar gene clusters located in other microorganisms, including Micromonospora sp. NBS 11–29 (40% similarity to gene cluster coding for putative virginiamycin), Streptomyces auratus AGR0001(58% similarity to genes coding for putative lysolipin), Burkholderia ubonensis MSMB2035(48% similarity to genes coding for putative esmeraldin), Streptomyces guanduensis CGMCC4.2022 (19% similarity to genes coding for putative lanthipeptide class II), Streptomyces sp. BK387 (25% similarity to genes coding for putative lanthipeptide class I) and Saccharothrix tamanrassetensis (21% similarity to genes coding for unidentified compound), respectively.
The 30 different types of secondary metabolites biosynthetic gene clusters (BGCs) in genomes of Kutzneria species. The proportion of different BGC types (a) and of same BGC type (b) among Kutzneria chonburiensis, K. kofuensis, K. buriramensis, K. albida, K. viridogrisea, Kutzneria sp.744 and Kutzneria sp. CA-103260.
The 3 putative bacteriocin gene clusters of Kutzneria chonburiensis strain SMC256T predicted by BAGEL 4.0 (AOI is the area of interest where the bacteriocin gene clusters had been identified by the program. AOI 1 and AOI 2 were best hit to lanthipeptide class II BGCs (LanM, LanC as indicated by red arrows) and AOI 3 was hit to a putative bacteriocin-family protein, as pointed by red arrow).
Putative species-specific antimicrobial/antitumor biosynthetic gene clusters in Kutzneria chonburiensis obtained by antiSMASH. The identified gene clusters were compared with known clusters in the MIBiG database to identify the putative compounds produced by these clusters. Gene clusters for (a) virginiamycin S1, (b) lysolipin I, (c) esmeraldin, (d) rakicidins A/B, (e) aclacinomycin, and (f) streptoseomycin.
Discussion
During the period of exploration of actinomycetes biodiversity and searching for new antimicrobials in Thailand, Kutzneria chonburiensis strain SMC256T was isolated and taxonomically identified from mountain soil. Because it is the latest type-strain and the potential producer of novel secondary metabolites in the rare group of Kutzneria, the insights into this genome will be noteworthy to uncover the diversity and uniqueness of bioactive metabolites synthesis in this strain. In this study, we generated short- and long-reads using the Illumina sequencing and the Oxford Nanopore technology, respectively, and employed this hybrid sequencing technique to complete genome assembly of the strain SMC256T. In general, using only short-read sequencing strategy for genome assembly usually leads to an incomplete assembly of important segments of the genome. Supplementation of long reads can overcome these spontaneous errors and complete crucial details of the genome17. Hence, hybrid technique was applied to fill ambiguous gaps that exist in a draft genome sequence previously obtained from the short-read sequencing, as described especially in many actinomycetes genomes which carry complex repeated DNA segments18. In our case, the hybrid genome assembly improved contiguity and identified more expected genes. Indeed, using hybrid assembly, we were able to obtain circular completed genome (1 contig) while using Illumina short reads alone produced gaps in the assembly, resulted in discontinuous genome contigs (42 contigs) and made some missing annotated genes (CDS, RNA) as seen in supplementary Table S5. By the hybrid approach, the completed genome sequence of the strain SMC256T revealed high G+C content (69.86%) and large genome size (10.41 Mbp) which is a typical characteristic of actinobacterial genomes19. Genome annotation also revealed that half of gene abundances are functionally assigned to the metabolism (1063 genes) probably encoding complex secondary metabolites. Moreover, we observed high numbers of hypothetical proteins (4249 genes), indicating that unknown proteins have not been identified yet (Table 2). Phylogenomic tree analysis of these Kutzneria based on MLST revealed the similar cladogram pattern to those obtained by clustering heatmaps of COGs and secondary metabolite BGCs analyses (antiSMASH). These results, in agreement with the PCA, obviously conveyed the consistency in close relationships among the Kutzneria species. Kutzneria chonburiensis, Kutzneria sp.744, K. kofuensis, Kutzneria sp. CA 103260 and K. buriramensis were grouped in a cluster whereas Kutzneria viridogrisea and Kutzneria albida were in another minor group in the same node of the genus Kutzneria (Figs. 3, 4, 7). Analysis of secondary metabolite gene clusters using bioinformatics tools showed that the most abundant BGCs found among genomes of the Kutzneria were “hybrid clusters” consisting of different types of BGCs which imply that they have a high potential for producing secondary metabolites with high structural diversity (Fig. 5). However, there were a number of clusters identified by antiSMASH with no similarity to the known BGCs in the databases (e.g. MIBiG), which were suggested to be orphan gene clusters. By identifying these genes, it is highly possible to discover novel secondary metabolites20,21. Kutzneria chonburiensis strain SMC256T could be bioinformatically predicted to possess the unique gene clusters which did not exist in genomes of other Kutzneria (Fig. 7). The unique gene clusters include guanidinotides, hybrid clusters of (NRPS/T2PKS/PKS-like/Lanthipeptide class-I), (oligosaccharide/NRPS-like/RRE-containing/NRPS), (thioamide/NRP/NRPS), (transAT-PKS/NRPS/butyrolactone), (PKS-like/phenazine), (T1PKS/ thioamide-NRP), (NRPS/T2PKS) and (T1PKS/NRPS/lanthipeptide class-IV). Of the 9 unique gene clusters, 6 clusters were responsible for biosynthesis of known important biologically active secondary metabolites, including virginiamycin S1, lysolipin I, esmeraldin, rakicidin, aclacinomycin and streptoseomycin (Fig. 9). Virginiamycin S1, originally produced by Streptomyces virginiae in 1950s, is a cyclic depsipeptides antibiotic belonging to the Streptogramin family group B and shows a strong synergistic bactericidal activity (100-fold increase) against a wide range of Gram-positive bacteria, including multidrug-resistant bacteria when combination with virginamycin M1 (Streptogramin family group A). The extraordinary features of the virginiamycin biosynthesis is a simultaneous production of both M1 and S1 at a suitable ratio providing their maximum synergistic activity in the suppression of the protein biosynthesis in susceptible microorganisms22,23. Even though the detailed pathway of the Virginiamycin biosynthesis has not been clarified, at least four and twenty plausible genes were identified for Virginiamycin S and Virginiamycin M biosynthesis, respectively. Because virginiamycin is a close structural relative to pristinamycin (a member of the same Streptogramin family) they were proposed to share a common biosynthetic gene cluster24. The identified BGCs (NRPS/transAT-PKS/butyrolactone) of region 166 of strain SMC256T by antiSMASH shared 77% sequence similarity of gene product to the best hit known functional BGCs of virginiamycin S1(NRPS/PKS) in MIBiG database (Fig. 9a). Lysolipin has strong antibacterial activity against a wide variety of multidrug-resistant pathogens and also has tumorstatic activity. This aromatic polyketide was first isolated in 1975 from Streptomyces violaceoniger Tü 96 and from Streptomyces tendae Tü 4042 in 199525,26. Later, the complete lysolipin biosynthetic gene cluster was entirely sequenced and identified27. The cluster encodes a Type II polyketide synthases (T2PKSs), cyclases, methyltransferases, halogenase, amidotransferase, ferredoxin, transporter and regulatory proteins. Moreover, fifteen genes coding for enzymes involved in redox modifications of the polyketide precursor existed in the lysolipin biosynthetic gene cluster. Owing to this great number of oxidoreductases, lysolipin is among the most highly modified aromatic polyketides known so far27. The regions 31 and 35 of strain SMC256T, identified to contain NRPS/T2PKS by antiSMASH, showed 32% and 73% sequence similarities to the BGCs of lysolipin I (T2PKS) of Streptomyces tendae in MIBiG, respectively (Fig. 9b). Esmeraldins (A and B), dark green metabolites isolated from Streptomyces antibioticus Tü 2706 in 1988, are antimicrobial and antitumor metabolites28. A chemical core structure of esmeraldins consists of diphenazines, which show a wide range of bioactivities. The 24 putative genes in biosynthetic gene cluster of esmeraldin production were identified and involved in the complicated synthetic pathways of phenazine-1-carboxylic acid (PCA) and phenazine-1,6-dicarboxylic acid (PDA). Phenazines are electron shuttles that reduce molecular oxygen and generate toxic reactive oxygen species making them broadly inhibit growth of bacteria, fungi and parasites29. The BGCs (PKS-like/phenazine) of region 167 of the strain SMC256T identified by antiSMASH showed 44% sequence similarity of gene to known functional BGCs of esmeraldin (PKS/aminocoumarin) in MIBiG (Fig. 9c). Rakicidins (A and B), a cyclic depsipeptide compound originally discovered from Micromonospora sp. and Streptomyces sp., display antitumor activity selectively to hypoxic cancer cells and stem-like leukemia cells30,31. The rakicidin B differs from rakicidin A by one additional methylene group in the lipid side chain and its cytotoxic activity (IC50 = 200 ng/ml) was less than that of rakicidin A (IC50 = 40 ng/ml). The biosynthetic pathways of rakicidins A and B, involved a hybrid PKS-NRPS biosynthetic gene clusters and being rather different from other types of rakicidins (e.g. rakicidin D), were elucidated in Micromonospora sp. M42 and M. purpureochromogenes NRRL B-267232. The BGCs (thioamide-NRP/NRPS) of region 70 of strain SMC256T identified by antiSMASH shared 22% sequence similarity of gene to known functional gene cluster of rakicidins A/B (NRP: cyclic depsipeptide/T1PKS) in MIBiG (Fig. 9d). Another anticancer/antibiotics drugs belonging to anthracycline are aclacinomycins, which were isolated from Streptomyces galilaeus33. The aromatic polyketide anthracyclines are able to penetrate and accumulate in high concentration in cancer cells in which they act on suppression of the DNA synthesis. It is used in treatments of acute myelogenous and lymphoblastic leukemia, malignant lymphoma, and gastric, lung, breast and ovarian cancers34. The biosynthesis of aclacinomycins, likely to anthracyclines biosynthetic pathways with some modification, was directed by a hybrid type II PKS/nonribosomal peptide synthetase (NRPS) system35. This complex biosynthesis comprised BGCs of Type II PKS, post-PKS tailoring enzymes and a multi-enzyme cascade including non-ribosomal peptide (NRP) assembly steps. The region 2 of strain SMC256T, identified to contain NRPS/T2PKS/PKS-like/lanthipeptide class-I by antiSMASH, showed 20% sequence similarities to known functional BGCs of aclacinomycin (PKS) of Streptomyces galilaeus in MIBiG (Fig. 9e). The last secondary metabolite predicted in Kutzneria chonburiensis was streptoseomycin. Streptoseomycin, discovered in Streptomyces seoulensis in 2018, is a rare macrodilactone belonging to a small tricyclic macrolactone family with only four members discovered to date. This compound exhibited a potent inhibitory action against 8 microaerophilic bacteria and particularly most highly effective against Helicobacter pylori36. The biosynthesis of streptoseomycin was identified and managed by type I PKS gene cluster (76 kB) haboring giant genes that encode PKS megaenzymes36. The BGC (lanthipeptide class I) of region 109 of strain SMC256T identified by antiSMASH shared 4% sequence similarity of gene to known functional BGCs of streptoseomycin (polyketide) in MIBiG database (Fig. 9f). The abovementioned technologies of hybrid sequencing-assembly, and data analytics of BGCs in genomes would be a crutial alternative approach to accelerate the discovery of new bioactive secondary metabolites hidden in the Kutzneria. To the best of our knowledge, only nine bioactive metabolites have been investigated in genus Kutzneria, of which four kutznerides (cyclic depsipeptides showing antifungal activity) were isolated from Kutzneria sp. 74437, aculeximycin (a macrolide being active against both bacteria and fungi) and huimycin (a pyrrolopyrimidine compound with a broad spectrum of biological activities) were from Kutzneria albida16, epemicins A and B (macrolides antibiotics against methicillin resistant Staphylococcus aureus (MRSA)) were extracted from Kutzneria sp. CA-103266, and the last one, phenol, 2,4-bis (1,1-dimethylethyl) which exhibits antifungal activity were isolated from Kutzneria sp. strain TS II38. Moreover, Kutzneria is still one of an underexplored genus, which shows great individual variation in terms of the total number of BGCs, therefore, the need of using genome mining as an aid to guide structural elucidation of novel compounds would improve the frequencies of drug discovery in this rare actinomycete effortlessly.
In conclusion, genomic variation of Kutzneria chonburiensis strain SMC256T in comparison with other strains/species in the genus has been demonstrated in accordance with the comparative genomics studies. Numerous BGCs of this strain have been shown to be both strain-specific and unidentified, which support the fact that microbes carry out specialized metabolic tasks exclusively for survival in particular ecological environment and potentially construct different metabolic routes for new bioactive secondary metabolite production. These findings contribute the future effective/productive effort to use integration of high throughput screening and bioinformatic based approaches to uncover, target and isolate new bioactive metabolites from the high rich sources of bioactive molecules including Kutzneria species and other rare actinobacteria.
Methods
Bacterial strain and cultivation condition
Kutzneria chonburiensis strain SMC256T was isolated and taxonomically identified as described previously39. Briefly, it was isolated from a soil sample collected in a mountain forest in Chonburi province, Thailand. The sample was taken from the organic layer of the sandy soil with pH of 5.9, and kept at − 20 °C before being air-dried at 37 °C for 7 days. The strain SMC256T was isolated on humic acid-salts vitamin agar supplemented with cycloheximide and nystatin. A pure culture was preserved by freezing at − 80 °C in glycerol (10% v/v) until use.
Genomic DNA extraction
Genomic DNA (gDNA) of Kutzneria chonburiensis SMC256T was extracted as stated by a modified method of Saito and Miura40 from cells grown in glucose-yeast extract broth at 28 °C, 250 rpm for 5 days. Freeze-dried cells were lysed using grinding with mortar and pestle, instead of lysozyme. The quantity and molecular weight of extracted genomic DNA were measured using the Denovix fluorometer (DeNovix Inc., DE, USA) and Agilent 2100 bioanalyzer (Agilent, CA, USA).
Whole genome sequencing
The genomic DNA library of Kutzneria chonburiensis strain SMC256T was prepared using the QIAGEN FX kit (Qiagen, USA). Briefly, gDNA was fragmented using enzymatic reaction and cleaned with magnetic beads. An adaptor index was ligated to the fragmented DNA. Quality and quantity of the indexed libraries were measured using Agilent 2100 Bioanalyzer and Denovix fluorometer and pooled in equimolar quantity. Cluster generation and paired-end 2 × 250 nucleotide read sequencing were performed on Illumina MiSeq sequencer. The sequencing process was carried out at the Omics Sciences and Bioinformatics Center, Chulalongkorn University, Bangkok, Thailand. The same DNA samples were prepared for long-read sequencing with the Oxford Nanopore Technologies (ONT) Ligation library preparation kit in accordance with the manufacturer’s protocol with the addition of continuous gentle mixing during the ligation incubation step. The libraries were sequenced with the ONT MinION sequencer using rev C R9.4 flow cells. The sequencer was controlled with the MinKNOW v.2.2.12, and sequencing runs were scheduled for 48–60 h, and allowed to run until fewer than ten pores remained functional. Raw base called data was generated using Guppy v.2.3.5, and the raw data was uploaded to the Galaxy web-based bioinformatics analysis tools platform (usegalaxy.eu). The pipeline of Hybrid de novo genome assembly—Nanopore draft Illumina polishing was carried out. First, adapters for Nanopore reads were removed using Porechop (v. 0.2.3)41, and Canu (v1.5) was used to assemble the filtered subreads42. Next, Pilon (v1.2) was used to polish the assembly sequence and improve genomic analyses with Illumina short reads43. In fact, when executing Pilon, three rounds of polishing were run iteratively on the assemblies, with Illumina short reads mapped to the polished assemblies obtained from the previous round using BWA-MEM44. In addition, genome assembly of short (Illumina) reads data alone was attempted in this study. Adaptors and poor-quality reads were removed using fastp (version 0.23.2) with detect_adapter_for_pe and -q 20 options45. Subsequentially, filtered reads were used as an input for SPAdes (version 3.15.4), genome assembly program with default parameters46.
Genome annotation and comparisons
The complete genome sequences of Kutzneria chonburiensis strain SMC256T and other close relatives were downloaded from the National Center for Biotechnology Information (NCBI). The Rapid Annotations using Subsystems Technology (RAST) was used for annotation and functional assignment (protein-encoding sequences (CDS), rRNA and tRNA genes and subsystem) to the genes in the genomes47. The annotated protein sequences were also assigned to clusters of orthologous groups (COGs) of proteins using WebMGA48, and the resulted COGs were compared and visualized in a heat map created by ClusVis49. The genome assembly statistics were evaluated using QUAST50. In addition, completeness estimation of the genome annotations and assemblies was assessed through identification of Benchmarking Universal Single-Copy Orthologs (BUSCO v.5.1.2) using the actinobacteria class dataset odb10 which contains 356 BUSCO markers51. Pairwise genome average nucleotide identity (ANI) was calculated using a web server, JSpeciesWS52. Phylogeny for genome comparison was constructed according to multi-locus sequence analysis (MLSA) technique using autoMLST53.
Identification of the putative secondary metabolite biosynthetic gene clusters
Putative secondary metabolite gene clusters (BGCs) in the genomes of Kutzneria were predicted using antiSMASH (v 6.0) with default settings15. The abundance of BGCs of Kutzneria chonburiensis was compared along with those of other species in this genus. The results were displayed in a heatmap generated as previously mentioned. Additionally, putative bacteriocins were identified in all analyzed genomes using the online web-server of BAGEL 410.
Data availability
All data are available under BioProject PRJNA835675, and the GenBank accession number for the genome is CP097263. Other data associated with the research are available as Supplementary Information file.
References
Adamek, M., Spohn, M., Stegmann, E. & Ziemert, N. Mining bacterial genomes for secondary metabolite gene clusters. Methods Mol. Biol. 1520, 23–47 (2017).
Belknap, K. C., Park, C. J., Barth, B. M. & Andam, C. P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 10, 2003 (2020).
Lee, N. et al. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 18, 1548–1556 (2020).
Ding, T., Yang, L. J., Zhang, W. D. & Shen, Y. H. The secondary metabolites of rare actinomycetes: Chemistry and bioactivity. RSC Adv. 9, 21964–21988 (2019).
Wei, B. et al. An atlas of bacterial secondary metabolite biosynthesis gene clusters. Environ. Microbiol. 23, 6981–6992 (2021).
Kontou, E. E. et al. Discovery and characterization of epemicins A and B, new 30-membered macrolides from Kutzneria sp. CA-103260. ACS Chem. Biol. 16, 1456–1468 (2021).
Franco, C. M. M. & Labeda, D. P. The order Pseudonocardiales. In The Prokaryotes: Actinobacteria (eds Rosenberge, E. et al.) 743–860 (Springer, 2014).
Schoch, C. L. et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 1–21. https://doi.org/10.1093/database/baaa062 (2020).
Harrison, J. & Studholme, D. J. Recently published Streptomyces genome sequences. Microb. Biotechnol. 7, 373–380 (2014).
van Heel, A. J. et al. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281 (2018).
Starcevic, A. et al. ClustScan: An integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 36, 6882–6892 (2008).
Weber, T. et al. CLUSEAN: A computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 140, 13–17 (2009).
Li, M. H., Ung, P. M., Zajkowski, J., Garneau-Tsodikova, S. & Sherman, D. H. Automated genome mining for natural products. BMC Bioinform. 10, 185 (2009).
Skinnider, M. A., Merwin, N. J., Johnston, C. W. & Magarvey, N. A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 45, W49–W54 (2017).
Blin, K. et al. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, 29–35 (2021).
Rebets, Y. et al. Complete genome sequence of producer of the glycopeptide antibiotic Aculeximycin Kutzneria albida DSM 43870T, a representative of minor genus of Pseudonocardiaceae. BMC Genom. 15, 885–900 (2014).
Maio, N. D. et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 5, e000294 (2019).
Paszkiewicz, K. & Studholme, D. J. De novo assembly of short sequence reads. Brief. Bioinform. 11, 457–472 (2010).
Ventura, M. et al. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548 (2007).
Singh, M. & Sareen, D. Novel LanT associated lantibiotic clusters identified by genome database mining. PLoS ONE 9, e91352 (2014).
Shi, J. et al. Comparative genome mining and heterologous expression of an orphan NRPS gene cluster direct the production of ashimides. Chem. Sci. 10, 3042–3048 (2019).
Pulsawat, N., Kitani, S. & Nihira, T. Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic Streptomyces virginiae. Gene 393, 31–42 (2007).
Namwat, W. et al. Characterization of virginiamycin S biosynthetic genes from Streptomyces virginiae. Gene 286, 283–290 (2002).
Mast, Y. et al. Characterization of the ‘pristinamycin supercluster’ of Streptomyces pristinaespiralis. Microb. Biotechnol. 4, 192–206 (2011).
Drautz, H., Keller-Schierlein, W. & Zähner, H. Metabolic products of microorganisms, 149. Lysolipin I, a new antibiotic from Streptomyces violaceoniger. Arch. Microbiol. 106, 175–190 (1975).
Blum, S. HPLC-DAD Screening von Streptomyceten-Isolierung und Charakter-Isierung neuer Naturstoffe. PhD thesis at the University of Tübingen (Ger) (1995).
Patricio, L. et al. Isolation of the lysolipin gene cluster of Streptomyces tendae Tü 4042. Gene 461, 5–14 (2010).
Keller-Schierlein, W., Geiger, A., Zähner, H. & Brandl, M. The esmeraldines A and B, green pigments from Streptomyces antibioticus, strain Tü 2706. Helv. Chim. Acta 71, 2058–2070 (1988).
Rui, Z. et al. Insights into a divergent phenazine biosynthetic pathway governed by a plasmid-born esmeraldin gene cluster. Chem. Biol. 19, 1116–1125 (2012).
McBrien, K. D. et al. Rakicidins, new cytotoxic lipopeptides from Micromonospora sp. fermentation, isolation and characterization. J. Antibiot. 48, 1446–1452 (1995).
Hu, J. F. et al. Rakicidin C, a new cyclic depsipeptide from Streptomyces sp.. Eur. J. Org. Chem. 19, 3353–3356 (2000).
Villadsen, N. L. et al. Synthesis of ent-BE-43547A1 reveals a potent hypoxia-selective anticancer agent and uncovers the biosynthetic origin of the APD-CLD natural products. Nat. Chem. 9, 264–272 (2017).
Oki, T. et al. New antitumor antibiotics, aclacinomycins A and B. J. Antibiot. 28, 830–834 (1975).
Wang, J., Maitani, Y. & Takayama, K. Antitumor effects and pharmacokinetics of aclacinomycin A carried by injectable emulsions composed of vitamin E, cholesterol, and PEG-lipid. J. Pharm. Sci. 91, 1128–1134 (2002).
Hulst, M. et al. Anthracyclines: Biosynthesis, engineering and clinical applications. Nat. Prod. Rep. 39, 814–841 (2022).
Zhang, B. et al. Discovery, biosynthesis, and heterologous production of streptoseomycin, an anti-microaerophilic bacteria macrodilactone. Org. Lett. 20, 2967–2971 (2018).
Broberg, A., Menkis, A. & Vasiliauskas, R. Kutznerides 1–4, depsipeptides from the Actinomycete Kutzneria sp. 744 inhabiting mycorrhizal roots of Picea abies seedlings. J. Nat. Prod. 69, 97–102 (2006).
Devi, T. S. et al. Antifungal activity and molecular docking of phenol, 2,4-bis(1,1-dimethylethyl) produced by plant growth-promoting actinobacterium Kutzneria sp. strain TSII from mangrove sediments. Arch. Microbiol. 202, 2855–2864 (2021).
Chanama, M., Thongkrachang, N., Suriyachadkun, C. & Chanama, S. Kutzneria chonburiensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 65, 4169–4174 (2015).
Saito, H. & Miura, K. I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72, 619–629 (1963).
Wick, R. Porechop. In GitHub repository. GitHub. https://github.com/rrwick/Porechop (2017).
Koren, S. et al. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Walker, B. J. et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at http://arXiv.org/1303.3997v2 (2013).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Aziz, R. K. et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 9, 75 (2008).
Wu, S., Zhu, Z., Fu, L., Niu, B. & Li, W. WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genom. 12, 444 (2011).
Metsalu, T. & Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566–W570 (2015).
Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D. & Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34, 142–150 (2018).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Richter, M., Rosselló-Móra, R., Glöckner, F. O. & Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931 (2015).
Alanjary, M., Steinke, K. & Ziemert, N. AutoMLST: An automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 47, 276–282 (2019).
Acknowledgements
This research project is supported by Mahidol University.
Author information
Authors and Affiliations
Contributions
M.C. conceptualized the research, designed and carried out the experiments, analyzed the data, prepared manuscript and did acquisition of financial support. P.P. performed the data analysis and help drafting manuscript. S.C. performed experiment and data analysis, edited and reviewed manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chanama, M., Prombutara, P. & Chanama, S. Comparative genome features and secondary metabolite biosynthetic potential of Kutzneria chonburiensis and other species of the genus Kutzneria. Sci Rep 13, 8794 (2023). https://doi.org/10.1038/s41598-023-36039-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-36039-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.