Abstract
Positron emission tomography/computed tomography (PET/CT) with 18F-fluorodeoxyglucose (FDG) is widely used for management of nasopharyngeal carcinoma (NPC). Combining the radiomic features of pre- and post-treatment FDG PET images may improve tumor characterization and prognostic predication. We investigated prognostic value of radiomic features from pre- and post-radiotherapy FDG PET images in patients with NPC. Quantitative radiomic features of primary tumors were extracted from the FDG PET images of 145 NPC patients and the delta values were also calculated. The study population was divided randomly into two groups, the training and test sets (7:3). A random survival forest (RSF) model was adopted to perform analyses of progression-free survival (PFS) and overall survival (OS). There were 37 (25.5%) cases of recurrence and 16 (11.0%) cases of death during a median follow-up period of 54.5 months. Both RSF models with clinical variables and radiomic PET features for PFS and OS showed comparable predictive performance to RSF models with clinical variables and conventional PET parameters. Tumoral radiomic features of pre- and post-treatment FDG PET and the corresponding delta values may predict PFS and OS in patients with NPC.
Similar content being viewed by others
Introduction
Nasopharyngeal carcinoma (NPC) is an uncommon epithelial carcinoma and has characteristics distinct from those of other epithelial head and neck cancers. Globally, NPC shows an unbalanced geographic distribution and the non-keratinizing subtype of NPC, which is associated with Epstein-Barr virus (EBV) infection, is predominant in EBV-endemic areas1. Radiotherapy is mainstay of NPC treatment due to its radio-sensitive nature and anatomic location of the tumors2,3. Improvement of clinical outcomes among patients with NPC has been attributed to advances in radiotherapy techniques4. Meanwhile, the effects of radiotherapy treatment may persist for months and are accompanied by inflammation5, which makes it difficult to evaluate early treatment response.
Positron emission tomography/computed tomography (PET/CT) with 18F-fluorodeoxyglucose (FDG) is a widely used imaging modality for management of NPC. In addition, FDG PET/CT is used to predict the prognosis of patients with NPC. High maximum standardized uptake value (SUV), metabolic tumor volume (MTV) and total lesion glycolysis (TLG), which are major conventional parameters derived from SUV, in pre-treatment FDG PET/CT images of patients with NPC have been associated with adverse prognosis in a meta-analysis6. Also, significant residual FDG uptake in post-radiotherapy FDG PET/CT images has been associated with poor clinical outcomes7. However, the predictive value of various FDG PET parameters, which could provide abundant information about tumor biology, remain to be clarified with regard to treatment response and prognosis.
Intratumoral heterogeneity (ITH) implies subclonal cell variability in time and space, and involves genetic, epigenetic, and metabolic features8. A high degree of ITH is known to be associated with inferior clinical outcomes and may contribute to the development of resistance and treatment failure in cancer patients9. Various FDG PET texture parameters could be used to evaluate metabolic ITH non-invasively10. Furthermore, difference in the features obtained from pre- and post-treatment images could reveal subtle changes in the tumor11. Combining the radiomic features of pre- and post-treatment FDG PET images may improve tumor characterization and prognostic prediction in patients with NPC. However, there has been no previous study using FDG PET to evaluate the radiomic features of NPC tumors both pre- and post-treatment.
In this study, we assessed the prognostic value of radiomic features identified in pre- and post-radiotherapy FDG PET images among patients with NPC.
Methods
Study subjects
All patients included in this study were diagnosed with NPC and underwent FDG PET/CT scans before and after definitive radiotherapy (with or without chemotherapy), between 2008 and 2019. The study population also met the following criteria: (1) subjects were more than 20 years of age; (2) the interval between definitive radiotherapy and follow-up FDG PET/CT was less than 6 months; and (3) the PET images were acquired with the same spatial resolution within the study population. We excluded patients who were treated for recurrence before the follow-up FDG PET/CT. Patients with the PET/CT image data loss were also excluded. Consequently, 145 patients were included in this study. Figure 1 is flow diagram of study subjects and FDG PET processing.
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Samsung Medical Center (protocol code 2020-06-127; date of approval: 15 July 2020). The requirement of written informed consent from enrolled subjects was waived by the Institutional Review Board of Samsung Medical Center due to the retrospective study design.
Medical record review
In this retrospective study, medical records were reviewed for demographic/clinical characteristics and information on recurrence/death among NPC patients. Follow-up of records review included the 60 months after definitive radiotherapy. Progression-free-survival (PFS) was defined as the duration of definitive radiotherapy completion and recurrence in months. Date of recurrence was determined as the date of documented clinical decision based on imaging modalities. Overall survival (OS) was defined as the duration to definitive radiotherapy completion and death from all causes in months. Cancer staging was based on the Sixth Edition of the American Joint Committee on Cancer’s Cancer Staging Manual to 2009, the Seventh Edition from 2010 to 2017, and Eighth Edition after 2018.
FDG PET/CT imaging
Patients fasted for at least 6 h before the injection of FDG, and their blood glucose concentration was confirmed to be less than 200 mg/dL at the time of FDG injection. At 60 min after injection of 5 MBq/kg of FDG, imaging was performed using an STe PET/CT scanner (GE Healthcare) without intravenous or oral contrast. Whole-body CT images were obtained with a continuous spiral 16-slice helical CT technique (140 keV; 30–170 mA; section width, 3.75 mm). An emission PET scan was obtained from the level of the thigh to the skull base. Scanning was performed at 2.5 min per frame in 3-D mode with attenuation-corrected images (3.9 × 3.9 × 3.3 mm) reconstructed using a 3-D ordered-subset expectation maximization algorithm (20 subsets, 2 iterations).
Imaging analyses
Primary tumor segmentation of all pre- and post-treatment FDG PET images was computed using the gradient-based segmentation method (‘PET edge’) of MIM version 6.4 (MIM software, Inc., Cleveland, OH, USA). Only primary tumors were analyzed in this study. A total of 72 quantitative radiomic features were extracted from the PET images using Chang-Gung Image Texture Analysis (CGITA) toolbox12. The relative variation of each radiomic feature was also evaluated in pre- and post-treatment scans:
Statistical analyses
The study population was divided randomly into two groups, the training and test sets (7:3) using the createDataPartition function of caret package in R. The demographic and clinical characteristics were compared between two groups using Fisher’s exact test, the chi-square test, and Wilcoxon rank sum test. Two-sided p values less than 0.05 were considered significant. A random survival forest (RSF) model was adopted to perform survival analyses of PFS and OS in the training set. An RSF is a nonparametric method to analyze right-censored survival data with multiple covariates and there is no need to fulfill any assumptions such as the proportional hazards assumption of classical Cox regression13. An RSF provides a variable importance (VIMP) that demonstrates variables of key role in predicting the survival outcome. First, we developed an RSF model with clinical variables—age, sex, smoking history, p16 status, EBV status, and stage— and conventional PET parameter—maximum SUV, mean SUV, MTV, and TLG—from pre- and post-treatment scans. Next, we made another RSF model with clinical variables and selected 10 relatively important radiomic PET features. Each model was then evaluated in the test set. Brier score (BS; mean squared error) was used to assess the prediction performance of the models in the training and test sets. Time-dependent BS and continuous ranked probability scores (CRPS; integrated BS divided by time) were recorded.
All statistical analyses were performed using R version 4.1.2 (The R Foundation for Statistical Computing, Vienna, Austria), with caret version 6.0.92, dplyr version 1.0.9, psych version 2.2.5, tidyverse version 1.3.1, gmodels version 2.18.1.1., survival version 3.3.1, randomForestSRC version 3.1.0 packages.
Results
Study subjects’ characteristics
The characteristics and clinical outcomes of 145 study subjects are summarized in Table 1. The median age was 52.3 (range 20.2–80.7) years and 108 of 145 (74.5%) subjects were males. There were 37 (25.5%) cases of recurrence and 16 (11.0%) deaths, respectively, during a median follow-up period of 54.5 months. Demographic and clinical characteristics of the training and test groups were not significantly different.
Analyses of PFS
The RSF analyses with clinical variables and conventional PET parameters for PFS using the training set resulted in an out-of-bag (OOB) error rate of 49.2%. On the other hand, Δcode similarity (texture feature coding co-occurrence), Δinverse difference moment (texture feature coding co-occurrence), mean SUVpost-treatment, code similarity (texture feature coding co-occurrence)pre-treatment, high-intensity zone emphasis (intensity-size-zone)post-treatment, high-intensity short-zone emphasis (intensity-size-zone)post-treatment, Δhomogeneity (texture feature coding co-occurrence), entropy (normalized co-occurrence)pre-treatment, inverse difference moment (texture feature coding co-occurrence)post-treatment, and high-intensity run emphasis (voxel-alignment)post-treatment were relatively important in the RSF analyses with all radiomic PET parameters. The VIMP of the radiomic PET parameters is summarized in Table 2. After that, an RSF model with clinical variables and ten selected radiomic PET parameters was developed using the training set, and the OOB error rate was 30.4%.
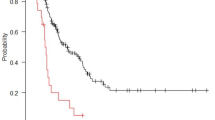
The CRPS of the predictive model for PFS with clinical variables and conventional PET parameters was 0.165 when using the training set and 0.199 when using the test set. In addition, the CRPS of the predictive model for PFS with clinical variables and selected radiomic PET parameters was 0.128 when using the training set and 0.200 when using the test set. The time-dependent BS for each prediction model is provided in Fig. 2.
Time-dependent Brier score (BS; mean-squared error) curve of random survival forest models to predict of progression-free survival (PFS). (a) The predictive performance of the model with clinical variables and selected radiomic PET parameters was superior to that of the predictive model with clinical variables and conventional PET parameters when using the training set. (b) The predictive performance of model with clinical variables and selected radiomic PET parameters was comparable to the model with clinical variables and conventional PET parameters when using the test set.
Analyses for OS
The RSF analyses with clinical variables and conventional PET parameters for OS when using the training set resulted in an OOB error rate of 35.2%. On the other hand, coarseness (neighborhood intensity-difference)pre-treatment, tumor volume (SUV statistics)pre-treatment, inverse difference moment (texture feature coding co-occurrence)post-treatment, run-length variability (voxel-alignment)pre-treatment, code similarity (texture feature coding co-occurrence)post-treatment, zone percentage (intensity-size-zone)post-treatment, minimum SUV (SUV statistics)pre-treatment, inverse difference moment (co-occurrence)pre-treatment, Δlow-intensity zone emphasis (intensity-size-zone), and max spectrum (texture spectrum)pre-treatment were relatively important in RSF analysis with all radiomic PET parameters for OS. The VIMP of the radiomic PET parameters is summarized in Table 3. An RSF model with clinical variables and the selected radiomic PET parameters for OS was developed using the training set and the OOB error rate was 31.6%. The CRPS of the prediction model for OS with clinical variables and conventional PET parameters was 0.043 when using the training set and 0.071 when using the test set. In addition, the CRPS of the predictive model for OS with clinical variables and selected radiomic PET parameters was 0.040 when using the training set and 0.071 when using the test set. The time-dependent BS for each predictive model is demonstrated in Fig. 3.
Time-dependent Brier score (BS; mean-squared error) curve of random survival forest models to predict overall survival (OS). (a) The predictive performance of the model with clinical variables and selected radiomic PET parameters was better than that of the predictive model with clinical variables and conventional PET parameters when using the training set. (b) The predictive performance of the model with clinical variables and selected radiomic PET parameters was similar to that of the model with clinical variables and conventional PET parameters when using the test set.
Discussion
To the best of our knowledge, this is the first study to evaluate the prognostic value of tumoral radiomic features from pre- and post-radiotherapy FDG PET images in patients with NPC. Texture parameters from pre- and post-treatment FDG PET scans had an impact on prediction of the prognosis of NPC patients. RSF models with clinical variables and radiomic PET features showed comparable prediction performance for PFS and OS to RSF models with clinical variables and conventional PET parameters.
The prognostic value of textural features derived from pre-treatment FDG PET/CT imaging of head and neck squamous cell carcinoma (HNSCC) has been evaluated in several previous studies. Yoon et al. conducted univariate and multivariate regression analysis of 119 cases of HNSCC and reported that pre-treatment FDG PET radiomics contain prognostic information14; however, the tumors were mainly located in the oropharynx (71.4%) in that study, while only 5 NPC patients were included. Similarly, Feliciani G. et al. analyzed 90 HNSCC patients and reported the superiority of imaging biomarkers from a pre-treatment FDG PET/CT model over a clinical-variables-alone model for prediction of PFS15; nevertheless, only 13 (15%) NPC patients were included in the study. Given the distinctive character of NPC, we included only NPC patients in this study rather than all HNSCC patients. In addition, Peng et al. developed models to predict treatment failure (locoregional recurrence or distant metastases) in 85 locally advanced NPC patients using machine learning methods16. They reported that a model with only PET parameters demonstrated greater area under curve (AUC) values for predictive performance than models with only clinical variables or with PET and clinical variables. The Peng et al. considered whether adverse events occurred or not, and the follow-up duration was not standardized in the retrospective study. We analyzed PFS and OS to avoid possible selection bias due to loss to follow-up.
Among radiomic PET features, Δcode similarity (texture feature coding co-occurrence) was an important variable for PFS prediction and coarseness (neighborhood intensity-difference)pre-treatment was an important variable for OS prediction in this study. Code similarity is based on texture feature number (TFN) co-occurrence matrix and demonstrates the density of same TFNs in its 8 connectivity neighborhood17. Lower pretreatment tumor code similarity was reported to be associated with poor survival in patients with esophageal squamous cell carcinoma18,19. On the other hand, coarseness (neighborhood-intensity difference) is based on the differences between each voxel and the neighboring voxels in the adjacent image planes and associated with the human perception of image granularity20. Greater coarseness_neighbouring gray tone difference matrix (NGTDM) was reported to be associated with higher risk of treatment failure in locally advanced NPC patients16. Coarseness was also negatively correlated with OS in a cohort study with various HNSCC patients21.
Delta radiomic features on serial images are longitudinal changes in radiomic features and were introduced to assess treatment response noninvasively when using various imaging modalities to assess various cancers11,22,23,24. A previous study investigated the potential of delta radiomics when using quantitative ultrasound to assess early treatment response during radiotherapy in HNSCC based on the index lymph node25. Response prediction accuracy was 86% using delta radiomics features acquired one week into radiotherapy, though the study had a limitation, in that primary tumors could not be accessed by ultrasound. In this study, we assessed primary tumors that were located at depth using FDG PET/CT, and found delta-radiomic features were important variables, especially for predicting PFS. Delta radiomics with FDG PET/CT may improve prediction of the prognosis of patients with NPC.
Our study has several limitations. First, the study was conducted in a relatively small number of subjects at a single center. Further study with more subjects in multiple centers is needed to develop a more precise predictive model of the survival of NPC patients and generalize the model. We intended to assess the potential prognostic power of tumor radiomic features in this preliminary study. In addition, the follow-up period after definitive treatment in this retrospective study was restricted to 60 months, because most patients who had survived for 5 years without recurrence had no further clinical visits to our institution. Missing data in smoking history, EBV, and p16 status arise from retrospective study design may obscure clinical implication of the variables. Furthermore, pre-treatment plasma EBV deoxyribonucleic acid (DNA) is an established prognostic factor for EBV-related NPC26. However, data regarding plasma EBV DNA titers was not available in this study. We did not consider the differences in detailed treatment strategies between subjects, either. We thus conducted analyses with clinical variables and conventional PET parameters in addition to the analyses of interest, which comprised clinical variables and selected radiomic PET parameters, to serve as a comparison.
Conclusions
Tumoral radiomic features of pre- and post-treatment FDG PET images and the corresponding delta values may contain prognostic values for PFS and OS in patients with NPC.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Chen, Y. P. et al. Nasopharyngeal carcinoma. Lancet 394, 64–80 (2019).
Bossi, P. et al. Nasopharyngeal carcinoma: ESMO-EURACAN clinical practice guidelines for diagnosis, treatment and follow-up(†). Ann. Oncol. 32, 452–465 (2021).
Sun, X. S., Li, X. Y., Chen, Q. Y., Tang, L. Q. & Mai, H. Q. Future of radiotherapy in nasopharyngeal carcinoma. Br. J. Radiol. 92, 20190209. https://doi.org/10.1259/bjr.20190209 (2019).
Lee, A. W. et al. Evolution of treatment for nasopharyngeal cancer-success and setback in the intensity-modulated radiotherapy era. Radiother. Oncol. 110, 377–384 (2014).
McKelvey, K. J., Hudson, A. L., Back, M., Eade, T. & Diakos, C. I. Radiation, inflammation and the immune response in cancer. Mamm. Genome. 29, 843–865 (2018).
Lin, J. et al. Prognostic value of 18F-FDG-PET/CT in patients with nasopharyngeal carcinoma: A systematic review and meta-analysis. Oncotarget 8, 33884–33896 (2017).
Jeong, Y., Jung, I. H., Kim, J. S., Chang, S. K. & Lee, S. W. Clinical significance of the post-radiotherapy (18)F-fludeoxyglucose positron emission tomography response in nasopharyngeal carcinoma. Br. J. Radiol. 92, 20180045. https://doi.org/10.1259/bjr.20180045 (2019).
Jacquemin, V. et al. Dynamic cancer cell heterogeneity: Diagnostic and therapeutic implications. Cancers 14, 280. https://doi.org/10.3390/cancers14020280 (2022).
Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018).
Ha, S., Choi, H., Paeng, J. C. & Cheon, G. J. Radiomics in oncological PET/CT: A methodological overview. Nucl. Med. Mol. Imaging. 53, 14–29 (2019).
Bera, K., Velcheti, V. & Madabhushi, A. Novel quantitative imaging for predicting response to therapy: Techniques and clinical applications. Am. Soc. Clin. Oncol. Educ. Book. 38, 1008–1018 (2018).
Fang, Y. H. et al. Development and evaluation of an open-source software package “CGITA” for quantifying tumor heterogeneity with molecular images. Biomed. Res. Int. 2014, 248505. https://doi.org/10.1155/2014/248505 (2014).
Pickett, K. L., Suresh, K., Campbell, K. R., Davis, S. & Juarez-Colunga, E. Random survival forests for dynamic predictions of a time-to-event outcome using a longitudinal biomarker. BMC Med. Res. Methodol. 21, 216. https://doi.org/10.1186/s12874-021-01375-x (2021).
Yoon, H. et al. Prognostic value of tumor metabolic imaging phenotype by FDG PET radiomics in HNSCC. Ann. Nucl. Med. 2021(35), 370–377 (2021).
Feliciani, G. et al. Radiomic profiling of head and neck cancer: 18F-FDG PET texture analysis as predictor of patient survival. Contrast Media Mol. Imaging. 2018, e3574310. https://doi.org/10.1155/2018/3574310 (2018).
Peng, L. et al. Prediction of local recurrence and distant metastasis using radiomics analysis of pretreatment nasopharyngeal [18F]FDG PET/CT images. Ann. Nucl. Med. 35, 458–468 (2021).
Horng, M. H., Sun, Y. N. & Lin, X. Z. Texture feature coding method for classification of liver sonography. Comput. Med. Imaging Graph. 26, 33–42 (2022).
Chen, Y. H. et al. Combining the radiomic features and traditional parameters of 18F-FDG PET with clinical profiles to improve prognostic stratification in patients with esophageal squamous cell carcinoma treated with neoadjuvant chemoradiotherapy and surgery. Ann. Nucl. Med. 33, 657–670 (2019).
Takahashi, N. et al. Texture analysis of FDG-PET/CT for patients with esophageal SCC treated by chemoradiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 102, e41–e42 (2018).
Cheng, N. M. et al. Textural features of pretreatment 18F-FDG PET/CT images: Prognostic significance in patients with advanced t-stage oropharyngeal squamous cell carcinoma. J. Nucl. Med. 54, 1703–1709 (2013).
Guezennec, C. et al. Prognostic value of textural indices extracted from pretherapeutic 18-F FDG-PET/CT in head and neck squamous cell carcinoma. Head Neck. 41, 495–502 (2019).
Guo, L. et al. Delta-radiomics based on dynamic contrast-enhanced MRI predicts pathologic complete response in breast cancer patients treated with neoadjuvant chemotherapy. Cancers 14, 3515. https://doi.org/10.3390/cancers14143515 (2022).
Basler, L. et al. Radiomics, tumor volume, and blood biomarkers for early prediction of pseudoprogression in patients with metastatic melanoma treated with immune checkpoint inhibition. Clin. Cancer Res. 26, 4414–4425 (2020).
Chang, Y. et al. An investigation of machine learning methods in delta-radiomics feature analysis. PLoS ONE 14, e0226348. https://doi.org/10.1371/journal.pone.0226348 (2019).
Tran, W. T. et al. Quantitative ultrasound delta-radiomics during radiotherapy for monitoring treatment responses in head and neck malignancies. Fut. Sci OA. 6, FSo624. https://doi.org/10.2144/fsoa-2020-0073 (2020).
Guo, R. et al. Proposed modifications and incorporation of plasma epstein-barr virus DNA improve the TNM staging system for epstein-barr virus-related nasopharyngeal carcinoma. Cancer 125, 79–89 (2019).
Funding
This research was funded by a National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT), grant number NRF-2020M2D9A1094072, which had no role in study design, data collection, analysis, interpretation, or writing of the report.
Author information
Authors and Affiliations
Contributions
Conceptualization, S.J.K., J.Y.C., Y.C.A., M.J.A. and S.H.M.; methodology, S.J.K., J.Y.C. and S.H.M; software, S.J.K.; validation, J.Y.C. and S.H.M.; formal analysis, S.J.K, and J.Y.C.; resources, J.Y.C.; data curation, S.J.K., J.Y.C. and S.H.M.; writing—original draft preparation, S.J.K.; writing—review and editing, J.Y.C. Y.C.A., M.J.A. and S.H.M.; visualization, S.J.K.; supervision, J.Y.C. and S.H.M.; project administration, J.Y.C. and S.H.M.; funding acquisition, J.Y.C. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, S.J., Choi, J.Y., Ahn, Y.C. et al. The prognostic value of radiomic features from pre- and post-treatment 18F-FDG PET imaging in patients with nasopharyngeal carcinoma. Sci Rep 13, 8462 (2023). https://doi.org/10.1038/s41598-023-35582-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-35582-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.