Abstract
In the effort of isolating novel microbial species, the strain PL0132T was isolated from a fallen leaf under fresh water at a stream, which glided when grown on a tap water medium (without nutrients). The strain was determined to be Gram-negative, strictly aerobic, and rod-shaped, which grew optimally at 25 °C, pH 6–7, and the strain tolerates 1% (w/v) NaCl concentration. The complete genome of strain PL0132T comprises one contig with a sequencing depth of 76×, consisting of 8,853,064 base pairs and the genomic DNA G + C content was 46.7% (genome). 16S rRNA gene sequence analysis revealed that strain PL0132T represents a member of the phylum Bacteroidetes and is affiliated with the genus Spirosoma. Based on genomic, phenotypic, and chemotaxonomic characteristics, the strain PL0132T represents a novel species of the genus Spirosoma, for which the name Spirosoma foliorum sp. nov. is proposed (= KCTC 72228 T = InaCC B1447T).
Similar content being viewed by others
Introduction
Microbial diversity on the earth is affluent, and 99% of microbial species are still unknown1. The limitation of the traditional culture method is one of the drawbacks of isolating novel microbial species and few microorganisms have been cultivated by current techniques2. Therefore, new methods for microbial isolation and cultivation should be invested in and studied more.
The genus Spirosoma belongs to the family Cytophagaceae which is the largest family belonging to the phylum Bacteroidetes3. The genus Spirosoma was originally described by Larkin and Borrall in 19844 and was emended by Finster et al.5 and Ahn et al.6 At the time of writing, the genus Spirosoma comprised 44 valid names (https://lpsn.dsmz.de/genus/spirosoma), including 12 recently described species: Spirosoma taeanense TS118T7, Spirosoma endbachense -24T8, Spirosoma telluris HMF3257T, Spirosoma arboris HMF4905T9, Spirosoma agri S7-3-3T10, Spirosoma terrae 15J9-4T11, Spirosoma jeollabukense S2-3-6T12, Spirosoma pollinicola HA7T13, Spirosoma humi S7-4-1T14, Spirosoma horti S7-3-19T15, Spirosoma harenae 15J8-9T16 and Spirosoma pomorum S7-2-11T17.
The species of the genus Spirosoma have been isolated from soil, dust, air, water, and extreme conditions like Arctic glaciers18,19,20. Furthermore, the characteristics of the genus include Gram-stain-negative, strictly aerobic, non-spore-forming, yellow or orange pigmented bacteria which are characterized as menaquinone MK-7 as the respiratory quinone, phosphatidylethanolamine as the major polar lipid, and summed feature 3 (C16: 1ω6C and/or C16: 1ω7C) C16:1ω5c, iso-C15:0, and C16:0 as the major fatty acids6. In this paper, a gliding bacterium was isolated from a decaying leaf of Acer palmatum in a stream of fresh water. The distinctive characteristics led us to propose a novel species in the genus Spirosoma.
Results and discussion
For phylogenetic characterisation, the comparative 16S rRNA gene sequence results revealed that the strain was phylogenetically affiliated with the genus Spirosoma. The 16S rRNA gene of PL0132T showed similarity values of 97.9%, 97.1%, 96.4% and 96.4% to Spirosoma arboris HMF4905T, Spirosoma litoris 15J16-2T3AT, Spirosoma migulaei 15J9-8T and Spirosoma telluris HMF3257T, respectively (Figs. 1, S1 and S2).
Maximum-likelihood phylogenetic tree, based on 16S rRNA gene sequences, showing the phylogenetic position of Spirosoma foliorum PL0132T among related strains of the genus Spirosoma. Closed circles indicate that the corresponding nodes were also recovered in the tree generated with the neighbor-joining and maximum parsimony algorithm. Bootstrap values in the order ML/NJ/MP are indicated as percentages of 1000 replications datasets, when greater than 70%. The tree was rooted using Bacteroides fragilis ATCC 25285T (X83935) as an outgroup. Bar, 0.05 substitutions per nucleotide position.
The complete genome sequence of strain PL0132T comprised one contig with a sequencing depth of 76×; the contig was 8,853,064 nt long with a circular structure. The GC contents of the chromosome were 46.7%. Gene annotation by NCBI Prokaryotic Genome Annotation Pipeline identified 7,408 genes which include 7,126 genes coding protein, 9 rRNAs (5S, 3; 16S, 3; 23S, 3), and 44 tRNAs. The complete genome was deposited in the GenBank/EMBL/DDBJ under the accession number CP059732. The identification of secondary metabolite biosynthesis gene cluster was determined by using antiSMASH21. Secondary metabolite clusters annotated by antiSMASH included terpene synthase genes, polyketide synthase type I and III (T1PKS and T3PKS), unspecified ribosomally synthesized and post-translationally modified peptide product cluster (RiPP-like), RRE-element containing cluster and non-ribosomal peptide synthetase (NRPS) (Fig. S5).
The average nucleotide identity (ANI) scores between the genomic sequence of PL0132T and Spirosoma arboris HMF4905T, Spirosoma migulaei 15J9-8 T, and Spirosoma telluris HMF3257T were 80.81, 79.13 and 79.72%9, respectively, which were determined by the ANI Calculator of EzBiocloud service (www.ezbiocloud.net/tools/ani). Digital DNA-DNA hybridization (dDDH) values were also calculated using the Genome Blast Distance Phylogeny version 2.1 web browser from DSMZ (http://ggdc.dsmz.de). The dDDH values between the whole genome sequences of strain PL0132T and its closest relatives were 24.40, 22.7, and 23.30% for Spirosoma arboris HMF4905T, Spirosoma migulaei 15J9-8T, and Spirosoma telluris HMF3257T, respectively. These values of ANI and dDDH were below the standard cut-off criteria for ANI (95–96%)22 and dDDH (70%)23, indicating that strain PL0132T represents a novel species of the genus Spirosoma.
To determine the taxonomic position of strain PL0132T, a genome-based phylogenetic tree was reconstructed using an up-to-date bacterial core gene (UBCG) set consisting of 92 genes, as described by Na et al.24 Briefly, the core genes identified (hmmsearch; v3.1b2) from predicted CDSs (Prodigal; v2.6.3) were concatenated, and aligned (MAFFT; v7.310), and subjected to construct a UBCG tree. The phylogenomic tree showed that PL0132T formed a clade with S. arboris HMF4905T within the genus Spirosoma (Fig. 2).
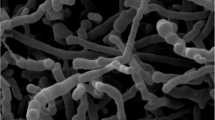
Genome-based phylogenetic tree of strain Spirosoma foliorum PL0132T and other related type strains using UBCGs (concatenated alignment of 92 core genes). Bootstrap values are indicated at nodes. Scale bar, 0.05 substitutions per position; Predicted gene clusters of secondary metabolites biosynthesis annotated in antiSMASH against strain PL0132T complete genome. Analyses provided the identification of six clusters involved in biosynthesis of terpene synthase genes, polyketide synthase type I and III (T1PKS and T3PKS), unspecified ribosomally synthesized and post-translationally modified peptide product cluster (RiPP-like), RRE-element containing cluster and non-ribosomal peptide synthetase (NRPS); Two-dimensional TLC patterns of the total polar lipids of strain Spirosoma foliorum PL0132T with The following spray reagents were used for detection: (A) molybdatophosphoric acid (for total lipids); (B) molybdenum blue (for phospholipids); (C) ninhydrin (for amino lipids); (D) α-naphthol (for glycolipids). Phosphatidylethanolamine (PE), Amino lipid (AL), Amino Phospho Lipid (APL), and unidentified lipids (L1-L4).1st: first dimension; 2nd: second dimension; Scanning electron micrographs (SEM) of strain Spirosoma foliorum PL0132T grown on R2A for 2 days at 25 °C. Bars 3 μm.
The strain PL0132T was Gram-stain-negative, strictly aerobic, non-spore-forming, catalase and oxidase-positive, rod-shaped without flagella, approximately 0.8–1.0 μm wide, 2–6 μm long (Fig. S3). The strain glided when grown on a tap water medium (without nutrients). Colonies grown on R2A for 48 h were circular, convex, and pale yellow. Growth occurred at 4–30 °C (optimum 25 °C), at pH 6.5–8.5 (optimum 7), and the strain did not require NaCl for its growth, but it tolerated at a concentration up to 1% (w/v) NaCl. The isolated strain grew on R2A and NA, and weakly on TSA but did not grow on LB agar. Physiological and biochemical characteristics of the isolated strain were described in the species description. Phenotypic and chemotaxonomic properties that differentiated strain PL0132T from its closest relatives in the genus are listed in Table 1.
In the chemotaxonomic analysis, the fatty acids of strain PL0132T were summed feature 3 as (C16: 1 ω6C and/or C16: 1 ω7C) (34.8%), C16:1 ω5c (18.6%), iso-C15:0 (20.0%), C16:0 (8.1%) and iso-C17:0 3-OH (5.6%) (Table 2). The overall fatty acids of strain were similar to those of other Spirosoma species with the major components such as summed feature 3 and C16:1 ω5c. However, there were differences in the percentages of some components, particularly iso-C15:0 and C16:0.
The quinone of strain PL0132T was menaquinone-7, which is similar to in other members of the genus Spirosoma5, 6. The polar lipid profile of PL0132T showed the major lipids phosphatidylethanolamine (PE), two aminophospholipids (APL1-2), and four unidentified lipids (L1–L4) (Fig. S4). The profile was similar to other strains in the genus with major polar lipids, and the profile of the novel isolate strain had more unidentified lipids and minor amounts of aminophospholipid.
Strain PL0132T had biochemical and physiological characteristics that differentiated strain PL0132T from the other species of the genus Spirosoma. One difference was that Spirosoma foliorum PL0132T and S. migulaei KCTC 52028T could grow at 10 °C and on tryptic soy agar while the other closely species of genus Spirosoma do not. Second, S.foliorum PL0132T and S. migulaei KCTC 52028T could produce acid from gentiobiose and inulin while the type strains of S. arboris, S. litoris, and S. telluris could not (Table 1), however, S.foliorum PL0132T could not produce acid from D-arabinose while the other close relative species could produce acid from D-arabinose. Third, it is important to note that glycolipid was found in other closely related species9 but not in S. foliorum PL0132T.
Materials and methods
All experiments were carried out in accordance with relevant institutional, national, and international guidelines and legislation.
Sampling sites and isolation
The strain PL0132T was isolated from a fallen leaf of Acer palmatum under fresh water at a stream in Naejang mountain, in Jeongeup city, South Korea (35°28′48.3′′N 126°53′21.6′′E) in July 2018. The leaf was cut into a small piece of 5 × 5 mm and placed on a tap water agar medium without nutrients and incubated at 20 °C. After gliding motility was observed by a stereoscopic microscope, one strain was purified by transferring on Reasoner’s 2A (R2A) (BD Difco) agar. Finally, a bacterium with yellow color, designated PL0132T, was collected. Then, the strain was preserved in 15% (v/v) glycerol suspension at − 80 °C.
16S rRNA gene phylogeny
The genomic DNA of the strain PL0132T was extracted from cells grown on R2A at 25 °C for 48 h. The 16S rRNA gene was amplified with two primers 27F and 1492R25. The sequence analysis was carried out by Sanger’s sequencing with an ABI3730XL automated sequencer (Applied Biosystems, USA). Then, the 16S rRNA gene sequences were uploaded to the EzBiocloud server26 to collect sequence information. All sequences of corresponding species obtained from the EzBiocloud were aligned and edited by BioEdit27 and CLUSTAL X software28. The phylogenetic trees were reconstructed by using neighbor-joining (NJ)29, maximum-likelihood (ML)30, and maximum parsimony (MP)31 methods in the MEGA 7 program32 with 1000 bootstrap replications.
Genome sequencing and annotation
Whole-genome sequence of strain PL0132T was determined by Nanopore technology. High molecular weight DNA was prepared and libraries were constructed following the native barcoding genomic DNA protocol (with EXP-NBD104, and SGK-LSK109, version NBE_9065_v109_revV_14Aug2019), after which sequencing was carried out. The library was loaded onto a MinION flow cell model FLO-MIN 106 (version R10.3). Then, sequencing was performed on MinKNOW platform (Oxford Nanopore Technologies). The resulting raw reads were quality assessed using pycoQC (2.5.2)33, and preprocessing was performed with porechop (0.2.4) to obtain 2.4 GB of reads. These reads were then processed using a custom pipeline based on Canu 2.034, which was improved by referring to the CCBGpipe (Consensus Circular Bacterial Genome pipeline)35. Sequencing reads were produced through base calling on ONT Guppy software (version 3.2.10, Oxford Nanopore Technologies, Ltd., Oxford, UK), for 4 h. Assembling was done for reads which had good quality of quality scores ≥ 7 and sequence length ≥ 3000 bp. Before filtering, the mean read length was about 3,420 bp, and the read count was 675,410. After filtering, the mean read length was about 16,821 bp and the read count was 39,426. Subsequently, The assembled contigs were corrected and polishing was performed with Medaka36 (version 1.3.2, bacteria_odb10 1.3.2, https://github.com/nanoporetech/medaka) to improve genome quality and the completeness of the Nanopore assembly was evaluated and the quality of the assembled genome was confirmed with BUSCO using BUSCO37 (version 5.1.2, (https://busco.ezlab.org/, score: 94.4) and CheckM (version 1.1.3, score: 99.7)38, and annotations were added using PROKKA (1.1.2)39.
Morphological and phenotypic analyses
Gram-staining was determined with a BD Gram-staining kit. Cells of the strain PL0132T incubated 48 h on R2A (Difco) at 25 °C were used to examine the morphology using a FEI Quanta 250 FEG scanning electron microscope (FEI). Gliding motility was tested by microscopic hanging drop16. The phenotypic features were determined as follows: the optimal temperature for growth was investigated at 4, 10, 20, 25, 30, 37, and 45 °C on R2A agar after incubation for 7 days. Salt tolerance was examined using R2A broth containing 0, 0.5, 1, 2, 3, 4, 5 and 10% (w/v) NaCl. Growth at pH 5 to 10 (at intervals of 0.5 unit) was determined in R2A broth adjusted with various buffers, the concentration of 100 mM, acetate for pH 5–6, phosphate for pH 6.5–8, Tris for pH 8.5–9, and carbonate for pH 9.5–109. Growth in the broth media was observed using OD600. Catalase and oxidase activity were carried out as described by Han et al.40 Growth on various media was assessed on R2A agar (Difco), Luria–Bertani agar (LB; Difco), nutrient agar (NA; Difco), and trypticase soy agar (TSA; Difco) after incubation for 7 days, at 25 °C. Anaerobic growth was determined by cultivation inside an anaerobic chamber (5% CO2, 5% H2, and 90% N2) on R2A supplemented with 10 mM KNO3 at 25 °C for 7 days41. The hydrolysis of macromolecular compounds was determined on 1:10 strength R2A supplemented with skimmed milk (3%, w/v), starch (1%, w/v), dextrin (1%, w/v), and carboxymethyl-cellulose (1%, w/v) at 25°C9. Other enzyme activities and carbon source utilization ability were determined by using API 20NE, API 50CH, and API ZYM kits (bioMérieux).
Chemotaxonomic analyses
To determine the fatty acid composition, cells of strain PL0132T and the reference strains were collected from R2A agar at the same physiological age, by using the method of Sasser42. Fatty acid analyses were analyzed by the Sherlock Microbial Identification System (TSBA; library version 6.0)43. The isolated strain was cultured in R2A broth (Difco, BD) for 48 h to collect cell mass for isoprenoid quinone and polar lipid analysis. The isoprenoid quinone was extracted according to the method of Komagata and Suzuki44 from 100 mg freeze-dried cells using the solution of chloroform/methanol (2:1, v/v). The crude compound was purified by using a preparative TLC (20 mm × 20 mm, silica gel 60 F254 plates, Merck) with petroleum benzene/diethyl ether (9:1, v/v); then the compound was analyzed by using reverse-phase HPLC with a mixture of methanol and isopropyl ether (3:1, v/v), and detected by a UV detector. Polar lipids were extracted by a chloroform/methanol/water system, and then they were developed and separated using two-dimensional TLC45. The polar lipids were identified as described by Han et al.46.
Conclusion
Based on the data for phylogenetic analysis, phenotypic and chemotaxonomic characteristics, PL0132T represents a novel species of the genus Spirosoma. However, several phenotypic differences between the isolated strain and its phylogenetically related strains were summarized in Table 1. Therefore, PL0132T should be classified as a novel species of the genus Spirosoma, for which the name Spirosoma foliorum sp. nov. is proposed.
Description of Spirosoma foliorum sp. nov
Spirosoma foliorum sp. nov. (fo.li.o'rum. L. pl. gen. n. foliorum, of leaves, referring to the isolation of the type of strain from decaying leaves). The novel strain designated PL0132T was isolated from decaying leaves under fresh water at a stream in Naejang mountain, in Jeongeup city, Republic of Korea.
Cells are Gram-stain-negative, and strictly aerobic rods, 0.8–1.0 μm wide, and 2–6 μm long. The strain glides when grown on a tap water medium (no nutrient). Colonies are convex, translucent, circular, and pale yellow. Growth occurs at occurred 4–30 °C (optimum 25 °C), at pH 6.5–8.5 (optimum 7), and the strain did not require NaCl for its growth. Tolerates 1% but not 2% (w/v) NaCl. Catalase and oxidase are positive. Starch, casein (skimmed milk), CM-cellulose, and DNA are not hydrolyzed. In API 20 NE tests, positive for aesculin hydrolysis, and β-galactosidase (PNPG test), but negative for gelatin hydrolysis, nitrate reduction, indole production, urease, and arginine dihydrolase. In assimilation of API 20NE, N-acetyl-D-glucosamine, D-glucose, D-maltose, and D-mannose are utilized, but adipate, L-arabinose, capric acid, potassium gluconate, malic acid, D-mannitol, trisodium citrate, and phenylacetate, are not utilized. In API ZYM tests, positive for acid phosphatase, alkaline phosphatase, N-acetyl-β-glucosaminidase, ß-glucuronidase, crystine arylamidase, esterase (C4), esterase (C8), α-galactosidase, ß-galactosidase (w), α-glucosidase (w), ß-glucosidase, leucine arylamidase, α-mannosidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase, but negative for α-chymotrypsin, α-fucosidase, trypsin and lipase (C14). In API 50 CH tests, acid is produced from N-acetyl-glucosamine, amygdalin, L-arabinose, arbutin, cellobiose, aesculin, D-fructose, D-galactose, gentiobiose, D-glucose, inulin, 5-ketogluconate, lactose, D-lyxose, D-mannose, D-maltose, melezitose, melibiose, methyl α-D-glucopyranoside, methyl α-D-mannopyranoside, raffinose, sucrose, salicin, trehalose, turanose, D-xylose but not from D-adonitol, D-arabinose, D-arabitol, L-arabitol, erythritol, D-fucose, L-fucose, gluconate, glycerol, glycogen, inositol, 2-ketogluconate, D-mannitol, methyl- ß -D-xylopyranoside, L-rhamnose, D-ribose, D-sorbitol, L-sorbose, starch, D-tagatose, xylitol, or L-xylose. The major cellular fatty acids of strain PL0132T are summed feature 3 as C16: 1ω6C and/or C16: 1ω7C (45.6%), C16:1ω5c (28.1%), iso-C15:0 (11.4%), C16:0 (3.6%) and iso-C17:0 3-OH (5.4%). The only respiratory quinone of PL0132T is MK-7. Phosphatidylethanolamine, aminolipid, aminophospholipid, and four unidentified lipids are the major polar lipid of PL0132T. The DNA G + C content is 46.7% (determined from the whole genome sequencing).
The type of strain, PL0132T (= KCTC 72228T = InaCC B1447T), was isolated from decayed leaves in Naejang mountain, Jeongeup city, Republic of Korea. The complete genome sequence of Spirosoma foliorum PL0132T comprised one contig with a sequencing depth of 76×; the contig was 8,853,064 nt long with a circular structure. The GC contents of the chromosome were 46.7%.
Data availability
The 16S rRNA gene sequence and whole genome of strain Spirosoma foliorum PL0132T generated and analysed during the current study are deposited the US National Institution of Health Genetic Sequence Database (GenBank), https://www.ncbi.nlm.nih.gov/genbank/; European Molecular Biology Laboratory (EMBL), https://www.ebi.ac.uk/; and the DNA Data Bank of Japan (DDBJ), http://getentry.ddbj.nig.ac.jp/getentry/na/MT076056/?filetype=html under accessions MT076056 and CP059732, respectively.
Abbreviations
- ANI:
-
Average nucleotide identity
- GGDC:
-
Genome-to-genome distance calculator
- SEM:
-
Scanning electron microscopy
- TLC:
-
Thin-layer chromatography
References
Locey, K. J. & Lennon, J. T. Scaling laws predict global microbial diversity. PNAS 113, 5970–5975. https://doi.org/10.1073/pnas.1521291113 (2016).
Hahn, M. W., Koll, U. & Schmidt, J. The Structure and Function of Aquatic Microbial Communities (Springer International Publishing, 2019).
Rosenberg, E., DeLong, E. F., Lory, S., Stackebrandt, E. & Thompson, F. The Prokaryotes: Actinobacteria (Springer, 2014).
Larkin, J. M. & Borrall, R. Family I. Spirosomaceae Larkin and Borrall 1978 595AL. In Bergey’s Manual of Systematic Bacteriology. Vol. 1 ( Baltimore: Williams & Wilkins, 1984).
Finster, K. W., Herbert, R. A. & Lomstein, B. A. Spirosoma spitsbergense sp. Nov. and Spirosoma luteum sp. Nov., isolated from a high Arctic permafrost soil, and emended description of the genus Spirosoma. Int. J. Syst. Evol. Microbiol. 59, 839–844. https://doi.org/10.1099/ijs.0.002725-0 (2009).
Ahn, J. H. et al. Spirosoma oryzae sp. nov., isolated from rice soil and emended description of the genus Spirosoma. Int. J. Syst. Evol. Microbiol. 64, 3230–3234. https://doi.org/10.1099/ijs.0.062901-0 (2014).
Lee, J. H. et al. Spirosoma taeanense sp. nov., a radiation resistant bacterium isolated from a coastal sand dune. Antonie Van Leeuwenhoek 114, 151–159. https://doi.org/10.1007/s10482-020-01508-0 (2021).
Rojas, J. et al. Spirosoma endbachense sp. nov., isolated from a natural salt meadow. Int. J. Syst. Evol. Microbiol. https://doi.org/10.1099/ijsem.0.004601 (2021).
Kang, H., Cha, I., Kim, H. & Joh, K. Spirosoma telluris sp. nov. and Spirosoma arboris sp. nov. isolated from soil and tree bark, respectively. Int. J. Syst. Evol. Microbiol. 7, 5355–5362. https://doi.org/10.1099/ijsem.0.004418 (2020).
Li, W., Lee, S. Y., Kang, I. K., Ten, L. N. & Jung, H. Y. Spirosoma agri sp. nov., isolated from apple orchard soil. Curr. Microbiol. 75, 694–700. https://doi.org/10.1007/s00284-018-1434-z (2018).
Ten, L. N. et al. Spirosoma terrae sp. nov., isolated from soil from Jeju Island, Korea. Curr. Microbiol. 75, 492–498. https://doi.org/10.1007/s00284-017-1408-6 (2018).
Li, W., Ten, L. N., Lee, S. Y., Lee, D. H. & Jung, H. Y. Spirosoma jeollabukense sp. nov., isolated from soil. Arch. Microbiol. 200, 431–438. https://doi.org/10.1007/s00203-017-1453-3 (2018).
Ambika Manirajan, B. et al. Spirosoma pollinicola sp. nov., isolated from pollen of common hazel (Corylus avellana L.). Int. J. Syst. Evol. Microbiol. 68, 3248–3254. https://doi.org/10.1099/ijsem.0.002973 (2018).
Weilan, L. et al. Spirosoma humi sp. nov., isolated from soil in South Korea. Curr. Microbiol. 75, 328–335. https://doi.org/10.1007/s00284-017-1384-x (2018).
Li, W., Ten, L. N., Lee, S. Y., Kang, I. K. & Jung, H. Y. Spirosoma horti sp. nov., isolated from apple orchard soil. Int. J. Syst. Evol. Microbiol. 68, 930–935. https://doi.org/10.1099/ijsem.0.002614 (2018).
Ten, L. N. et al. Spirosoma harenae sp. nov., a bacterium isolated from a Sandy Beach. Curr. Microbiol. 75, 179–185. https://doi.org/10.1007/s00284-017-1363-2 (2018).
Li, W., Lee, S. Y., Kang, I. K., Ten, L. N. & Jung, H. Y. Spirosoma pomorum sp. Nov., isolated from apple orchard soil. J. Microbiol. 56, 90–96. https://doi.org/10.1007/s12275-018-7430-y (2018).
Okiria, J. et al. Spirosoma litoris sp. nov., a bacterium isolated from beach soil. Int. J. Syst. Evol. Microbiol. 67, 4986–4991 (2017).
Lee, J.-J. et al. Spirosoma luteolum sp. nov. isolated from water. J. Microbiol. 55, 247–252. https://doi.org/10.1007/s12275-017-6455-y (2017).
Okiria, J. et al. Spirosoma migulaei sp. nov., isolated from soil. J. Microbiol. 55, 927–932. https://doi.org/10.1007/s12275-017-7377-4 (2017).
Blin, K. et al. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucl. Acids Res. 49, W29–W35. https://doi.org/10.1093/nar/gkab335 (2021).
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S. & Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. https://doi.org/10.1007/s10482-017-0844-4 (2017).
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P. & Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 14, 60. https://doi.org/10.1186/1471-2105-14-60 (2013).
Na, S. I. et al. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56, 280–285. https://doi.org/10.1007/s12275-018-8014-6 (2018).
Pheng, S., Han, H. L., Park, D.-S., Chung, C. H. & Kim, S.-G. Lactococcus kimchii sp. nov., a new lactic acid bacterium isolated from kimchi. Int. J. Syst. Evol. Microbiol. 70, 505–510. https://doi.org/10.1099/ijsem.0.003782 (2020).
Yoon, S. H. et al. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. https://doi.org/10.1099/ijsem.0.001755 (2017).
Hall, T. A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98 (1999).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 25, 4876–4882 (1997).
Saitou, N. & Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454 (1987).
Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Fitch, W. M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (1971).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 (2016).
Leonardi, A. & Leonardi, A. pycoQC, interactive quality control for Oxford Nanopore Sequencing. JOSS 4, 1236 (2019).
Koren, S. et al. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. https://doi.org/10.1101/gr.215087.116 (2017).
Liao, Y. C. et al. Completing circular bacterial genomes with assembly complexity by using a sampling strategy from a single MinION run with barcoding. Front. Microbiol. 10, 2068. https://doi.org/10.3389/fmicb.2019.02068 (2019).
Lee, J. Y. et al. Comparative evaluation of Nanopore polishing tools for microbial genome assembly and polishing strategies for downstream analysis. Sci. Rep. 11, 20740. https://doi.org/10.1038/s41598-021-00178-w (2021).
Cantarel, B. L. et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18, 188–196. https://doi.org/10.1101/gr.6743907 (2008).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. https://doi.org/10.1093/bioinformatics/btu153 (2014).
Le Han, H., Nguyen, T. T. H., Li, Z., Shin, N. R. & Kim, S. G. Cellulosimicrobium protaetiae sp. Nov., isolated from the gut of the larva of Protaetia brevitarsis seulensis. Int. J. Syst. Evol. Microbiol. https://doi.org/10.1099/ijsem.0.005296 (2022).
Ten, L. N. et al. Paenibacillus panacisoli sp. nov., a xylanolytic bacterium isolated from soil in a ginseng field in South Korea. Int. J. Syst. Evol. Microbiol. 56, 2677–2681. https://doi.org/10.1099/ijs.0.64405-0 (2006).
Sasser, M. (MIDI technical note 101. Newark, DE: MIDI inc, 1990).
Liu, Y., Le Han, H., Zou, Y. & Kim, S.-G. Flavobacterium ustbae sp. Nov., isolated from rhizosphere soil of Alhagi sparsifolia. Int. J. Syst. Evol. Microbiol. 69, 3955–3960. https://doi.org/10.1099/ijsem.0.003717 (2019).
Komagata, K. & Suzuki, K. I. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–205 (1987).
Minnikin, D. et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241 (1984).
Han, H. L. et al. Halorubrum salinarum sp. Nov., an extremely halophilic archaeon isolated from a saturated brine pond of a saltern. Int. J. Syst. Evol. Microbiol. https://doi.org/10.1099/ijsem.0.005231 (2022).
Acknowledgements
The authors thank Prof. Dr. Bernhard Schink for his suggestion for correct name species and Latin etymology and Dr. Lingmin Jiang for her contribution to building a phylogenomic tree.
Funding
This work was supported by the KRIBB Research Initiative in Korea. This work was also supported by the grant from Ming Chi University of Technology, Taiwan. This research was also funded by University of Science and Technology, The University of Danang, under grant number T2023-02-35.
Author information
Authors and Affiliations
Contributions
H.L.H.: Conceptualization; H.L.H.: Writing - Original Draft; H.L.H., D.A.N., N.M., Y.-J.L., T.T.H.N., S.-G.K. S.S.C., K.S.K., K.W.C., P.L.S., C.Y.C., T.N.T.T., T.D.P.N.: Writing - Review & Editing; H.L.H.: Visualization; P.L.S., T.D.P.N.: Supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Han, H.L., Nurcahyanto, D.A., Muhammad, N. et al. Isolation of Spirosoma foliorum sp. nov. from the fallen leaf of Acer palmatum by a novel cultivation technique. Sci Rep 13, 14684 (2023). https://doi.org/10.1038/s41598-023-35108-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-35108-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.